GA-Optimized Sampling for Soil Type Mapping in Plain Areas: Integrating Legacy Maps and Multisource Covariates
Abstract
1. Introduction
2. Materials and Methodology
2.1. Research Area Overview
2.2. Selection and Processing of Environmental Covariates
2.3. Models for Sampling Design
2.3.1. Genetic Algorithm Sampling
2.3.2. The K-Nearest Neighbors Sampling
2.3.3. Fuzzy C-Means Clustering Sampling
2.3.4. Field Calibration
2.4. Random Forest Model
2.5. Evaluation of Soil Map Accuracy
3. Results and Discussion
3.1. Selection of Environmental Covariates
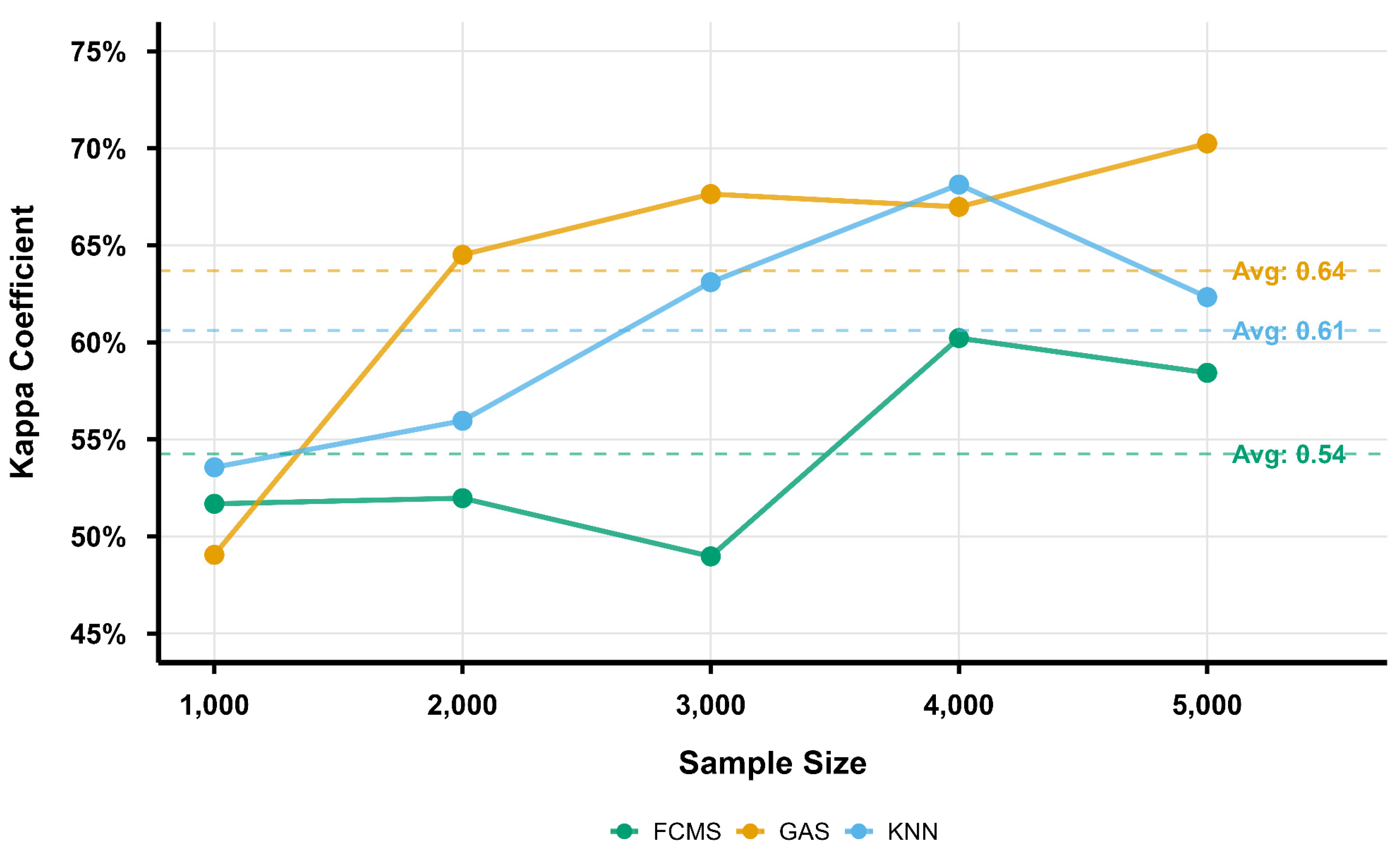
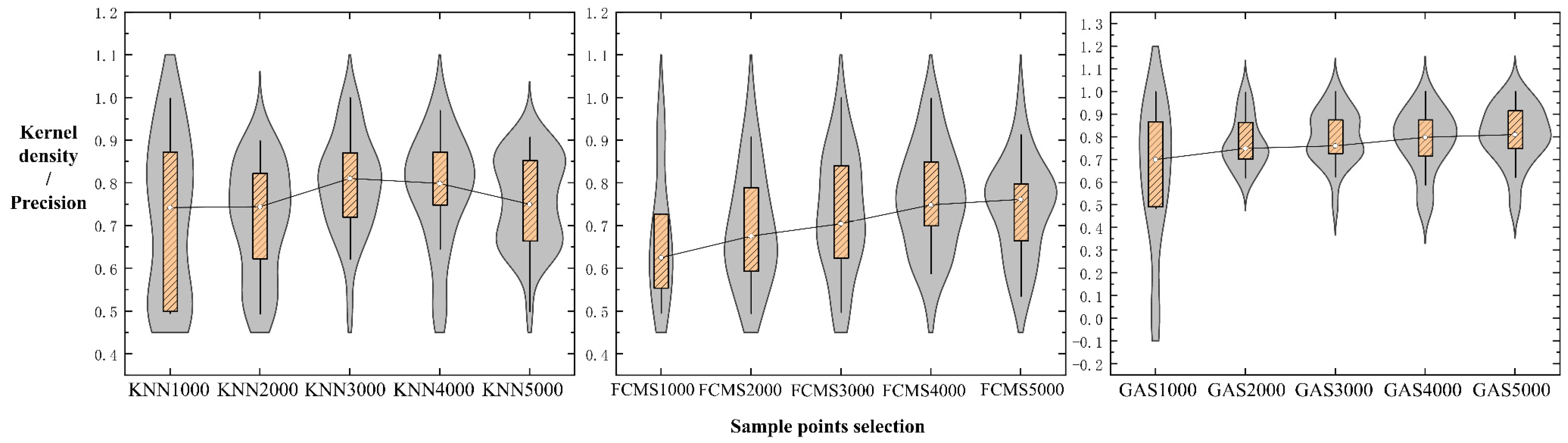
3.2. Comparison of Soil Mapping Accuracy
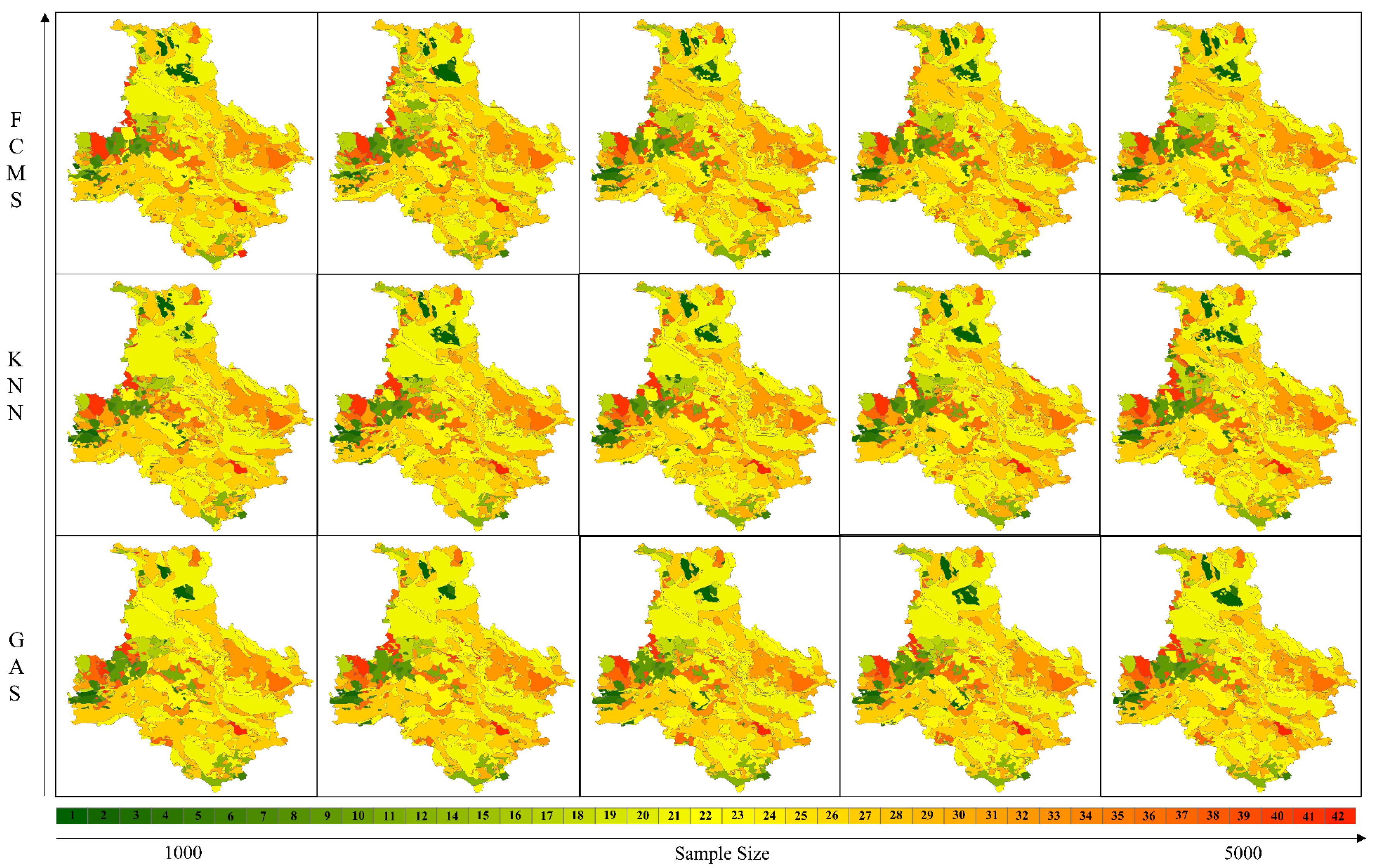
3.3. Accuracy Range Analysis for Soil Types
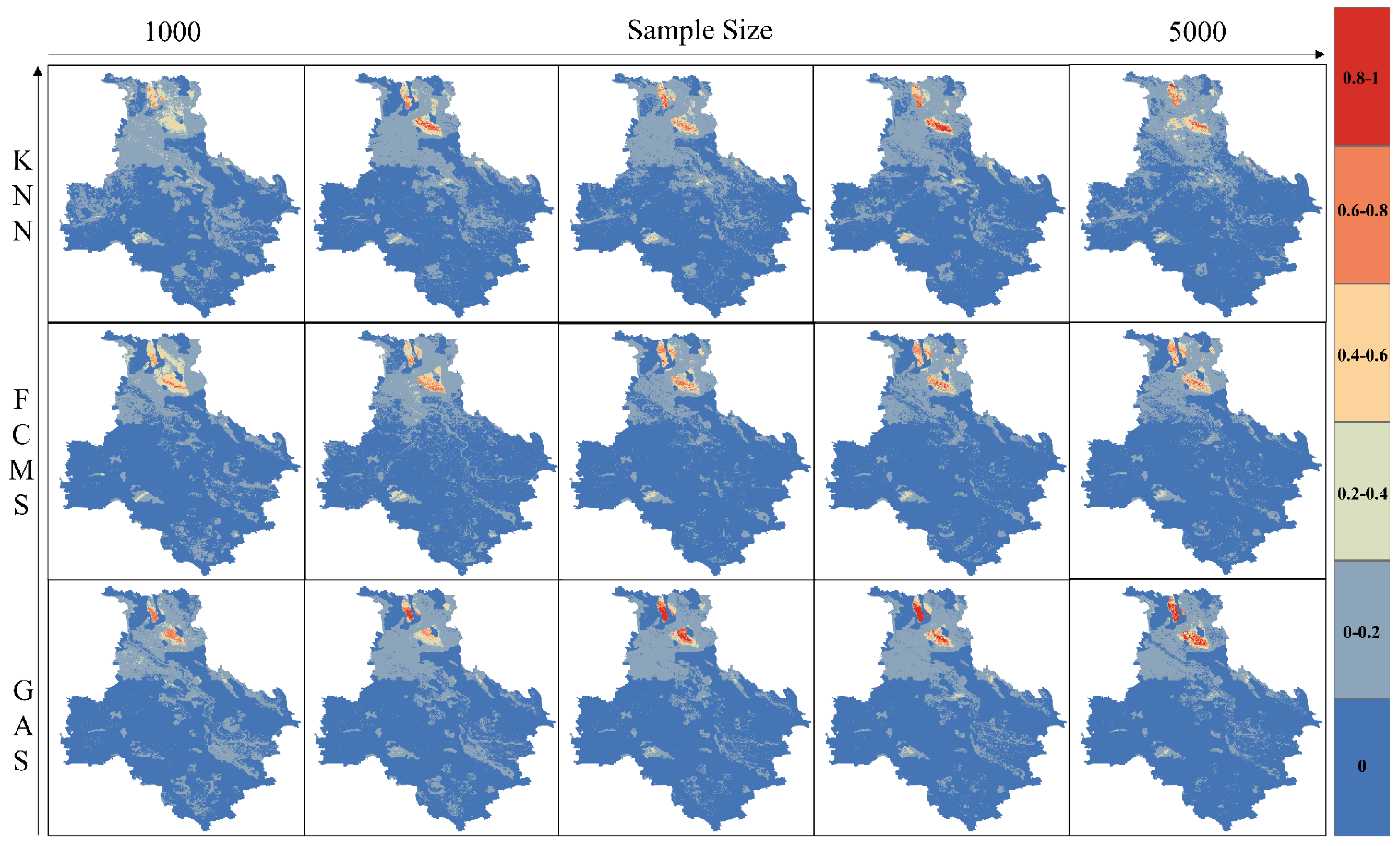
3.4. Comparisons of Mapping Results
3.5. Status of Field Calibration
3.6. Discussions for Sampling Algorithm Differences
3.7. Improving the Utilization of Soil Resources in Agriculture
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sulaeman, Y.; Minasny, B.; McBratney, A.B.; Sarwani, M.; Sutandi, A. Harmonizing Legacy Soil Data for Digital Soil Mapping in Indonesia. Geoderma 2013, 192, 77–85. [Google Scholar] [CrossRef]
- Zeraatpisheh, M.; Bakhshandeh, E.; Hosseini, M.; Alavi, S.M. Assessing the Effects of Deforestation and Intensive Agriculture on the Soil Quality through Digital Soil Mapping. Geoderma 2020, 363, 114139. [Google Scholar] [CrossRef]
- Chen, S.; Arrouays, D.; Leatitia Mulder, V.; Poggio, L.; Minasny, B.; Roudier, P.; Libohova, Z.; Lagacherie, P.; Shi, Z.; Hannam, J.; et al. Digital Mapping of GlobalSoilMap Soil Properties at a Broad Scale: A Review. Geoderma 2022, 409, 115567. [Google Scholar] [CrossRef]
- Petrovskaia, A.; Ryzhakov, G.; Oseledets, I. Optimal Soil Sampling Design Based on the Maxvol Algorithm. Geoderma 2021, 402, 115362. [Google Scholar] [CrossRef]
- Yang, L.; Brus, D.J.; Zhu, A.-X.; Li, X.; Shi, J. Accounting for Access Costs in Validation of Soil Maps: A Comparison of Design-Based Sampling Strategies. Geoderma 2018, 315, 160–169. [Google Scholar] [CrossRef]
- Brus, D.J. Balanced Sampling: A Versatile Sampling Approach for Statistical Soil Surveys. Geoderma 2015, 253–254, 111–121. [Google Scholar] [CrossRef]
- Krumbein, W.C. Factors of Soil Formation: A System of Quantitative Pedology. Hans Jenny. J. Geol. 1942, 50, 919–920. [Google Scholar] [CrossRef]
- Minasny, B.; McBratney, A.B. Digital Soil Mapping: A Brief History and Some Lessons. Geoderma 2016, 264, 301–311. [Google Scholar] [CrossRef]
- Yang, L.; Li, X.; Shi, J.; Shen, F.; Qi, F.; Gao, B.; Chen, Z.; Zhu, A.-X.; Zhou, C. Evaluation of Conditioned Latin Hypercube Sampling for Soil Mapping Based on a Machine Learning Method. Geoderma 2020, 369, 114337. [Google Scholar] [CrossRef]
- Stumpf, F.; Schmidt, K.; Behrens, T.; Schönbrodt-Stitt, S.; Buzzo, G.; Dumperth, C.; Wadoux, A.; Xiang, W.; Scholten, T. Incorporating Limited Field Operability and Legacy Soil Samples in a Hypercube Sampling Design for Digital Soil Mapping. J. Plant Nutr. Soil Sci. 2016, 179, 499–509. [Google Scholar] [CrossRef]
- Zhu, A.X.; Yang, L.; Li, B.; Qin, C.; English, E.; Burt, J.E.; Zhou, C. Purposive Sampling for Digital Soil Mapping for Areas with Limited Data. In Digital Soil Mapping with Limited Data; Hartemink, A.E., McBratney, A., Mendonça-Santos, M.d.L., Eds.; Springer: Dordrecht, The Netherland, 2008; pp. 233–245. [Google Scholar]
- Barthold, F.K.; Wiesmeier, M.; Breuer, L.; Frede, H.-G.; Wu, J.; Blank, F.B. Land Use and Climate Control the Spatial Distribution of Soil Types in the Grasslands of Inner Mongolia. J. Arid Environ. 2013, 88, 194–205. [Google Scholar] [CrossRef]
- Wadoux, A.M.J.-C.; Brus, D.J.; Heuvelink, G.B.M. Sampling Design Optimization for Soil Mapping with Random Forest. Geoderma 2019, 355, 113913. [Google Scholar] [CrossRef]
- Kempen, B.; Brus, D.J.; Heuvelink, G.B.M.; Stoorvogel, J.J. Updating the 1:50,000 Dutch Soil Map Using Legacy Soil Data: A Multinomial Logistic Regression Approach. Geoderma 2009, 151, 311–326. [Google Scholar] [CrossRef]
- Zeng, C.; Qi, F.; Zhu, A.-X.; Liu, F. Construction of Land Surface Dynamic Feedback for Digital Soil Mapping Considering the Spatial Heterogeneity of Rainfall Magnitude. CATENA 2020, 191, 104576. [Google Scholar] [CrossRef]
- Deb, K. Multi-Objective Optimisation Using Evolutionary Algorithms: An Introduction. In Multi-Objective Evolutionary Optimisation for Product Design and Manufacturing; Wang, L., Ng, A.H.C., Deb, K., Eds.; Springer: London, UK, 2011; pp. 3–34. [Google Scholar]
- Goldberg, D.E.; Holland, J.H. Genetic Algorithms and Machine Learning. Mach. Learn. 1988, 3, 95–99. [Google Scholar] [CrossRef]
- The Third National Soil Survey Work Plan. Available online: https://www.gov.cn/xinwen/2022-02/24/content_5675442.htm (accessed on 8 April 2024).
- Heung, B.; Ho, H.C.; Zhang, J.; Knudby, A.; Bulmer, C.E.; Schmidt, M.G. An Overview and Comparison of Machine-Learning Techniques for Classification Purposes in Digital Soil Mapping. Geoderma 2016, 265, 62–77. [Google Scholar] [CrossRef]
- Xu, Y.; Li, K.; Hu, J.; Li, K. A Genetic Algorithm for Task Scheduling on Heterogeneous Computing Systems Using Multiple Priority Queues. Inf. Sci. 2014, 270, 255–287. [Google Scholar] [CrossRef]
- Sun, Y.; Xue, B.; Zhang, M.; Yen, G.G.; Lv, J. Automatically Designing CNN Architectures Using the Genetic Algorithm for Image Classification. IEEE Trans. Cybern. 2020, 50, 3840–3854. [Google Scholar] [CrossRef]
- Soleimani, H.; Kannan, G. A Hybrid Particle Swarm Optimization and Genetic Algorithm for Closed-Loop Supply Chain Network Design in Large-Scale Networks. Appl. Math. Model. 2015, 39, 3990–4012. [Google Scholar] [CrossRef]
- Deng, W.; Zhao, H.; Zou, L.; Li, G.; Yang, X.; Wu, D. A Novel Collaborative Optimization Algorithm in Solving Complex Optimization Problems. Soft Comput. 2017, 21, 4387–4398. [Google Scholar] [CrossRef]
- Mumali, F.; Kałkowska, J. Intelligent Support in Manufacturing Process Selection Based on Artificial Neural Networks, Fuzzy Logic, and Genetic Algorithms: Current State and Future Perspectives. Comput. Ind. Eng. 2024, 193, 110272. [Google Scholar] [CrossRef]
- Borcard, D.; Gillet, F.; Legendre, P. Numerical Ecology with R; Use R! Springer International Publishing: Cham, Switzerland, 2018; ISBN 978-3-319-71403-5. [Google Scholar]
- Sánchez-Mercado, A.Y.; Ferrer-Paris, J.R.; Franklin, J. Mapping Species Distributions: Spatial Inference and Prediction. Oryx 2010, 44, 615. [Google Scholar] [CrossRef]
- Guisan, A.; Zimmermann, N.E. Predictive Habitat Distribution Models in Ecology. Ecol. Model. 2000, 135, 147–186. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very High Resolution Interpolated Climate Surfaces for Global Land Areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Rogers, D.J.; Randolph, S.E. Studying the Global Distribution of Infectious Diseases Using GIS and RS. Nat. Rev. Microbiol. 2003, 1, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, M.S.; Anderson, C.D. PASSaGE: Pattern Analysis, Spatial Statistics and Geographic Exegesis. Version 2. Methods Ecol. Evol. 2011, 2, 229–232. [Google Scholar] [CrossRef]
- Luo, X.; Wang, H.; Wu, D.; Chen, C.; Deng, M.; Huang, J.; Hua, X.-S. A Survey on Deep Hashing Methods. ACM Trans. Knowl. Discov. Data 2023, 17, 1–50. [Google Scholar] [CrossRef]
- Li, J.; Lin, S.; Yu, K.; Guo, G. Quantum K-Nearest Neighbor Classification Algorithm Based on Hamming Distance. Quantum Inf. Process 2021, 21, 18. [Google Scholar] [CrossRef]
- Mohammadrezapour, O.; Kisi, O.; Pourahmad, F. Fuzzy C-Means and K-Means Clustering with Genetic Algorithm for Identification of Homogeneous Regions of Groundwater Quality. Neural Comput. Applic 2020, 32, 3763–3775. [Google Scholar] [CrossRef]
- Horta, A.; Malone, B.; Stockmann, U.; Minasny, B.; Bishop, T.F.A.; McBratney, A.B.; Pallasser, R.; Pozza, L. Potential of Integrated Field Spectroscopy and Spatial Analysis for Enhanced Assessment of Soil Contamination: A Prospective Review. Geoderma 2015, 241–242, 180–209. [Google Scholar] [CrossRef]
- Minasny, B.; McBratney, A.B. A Conditioned Latin Hypercube Method for Sampling in the Presence of Ancillary Information. Comput. Geosci. 2006, 32, 1378–1388. [Google Scholar] [CrossRef]
- Hengl, T.; Heuvelink, G.B.M.; Stein, A. A Generic Framework for Spatial Prediction of Soil Variables Based on Regression-Kriging. Geoderma 2004, 120, 75–93. [Google Scholar] [CrossRef]
- Zhu, A.X.; Hudson, B.; Burt, J.; Lubich, K.; Simonson, D. Soil Mapping Using GIS, Expert Knowledge, and Fuzzy Logic. Soil. Sci. Soc. Am. J. 2001, 65, 1463–1472. [Google Scholar] [CrossRef]
- Mulder, V.L.; de Bruin, S.; Schaepman, M.E.; Mayr, T.R. The Use of Remote Sensing in Soil and Terrain Mapping—A Review. Geoderma 2011, 162, 1–19. [Google Scholar] [CrossRef]
- Guo, P.-T.; Li, M.-F.; Luo, W.; Tang, Q.-F.; Liu, Z.-W.; Lin, Z.-M. Digital Mapping of Soil Organic Matter for Rubber Plantation at Regional Scale: An Application of Random Forest plus Residuals Kriging Approach. Geoderma 2015, 237–238, 49–59. [Google Scholar] [CrossRef]
- Chen, W.; Xie, X.; Wang, J.; Pradhan, B.; Hong, H.; Bui, D.T.; Duan, Z.; Ma, J. A Comparative Study of Logistic Model Tree, Random Forest, and Classification and Regression Tree Models for Spatial Prediction of Landslide Susceptibility. CATENA 2017, 151, 147–160. [Google Scholar] [CrossRef]
- Brungard, C.W.; Boettinger, J.L.; Duniway, M.C.; Wills, S.A.; Edwards, T.C. Machine Learning for Predicting Soil Classes in Three Semi-Arid Landscapes. Geoderma 2015, 239–240, 68–83. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Were, K.; Bui, D.T.; Dick, Ø.B.; Singh, B.R. A Comparative Assessment of Support Vector Regression, Artificial Neural Networks, and Random Forests for Predicting and Mapping Soil Organic Carbon Stocks across an Afromontane Landscape. Ecol. Indic. 2015, 52, 394–403. [Google Scholar] [CrossRef]
- Schmidt, K.; Behrens, T.; Scholten, T. Instance Selection and Classification Tree Analysis for Large Spatial Datasets in Digital Soil Mapping. Geoderma 2008, 146, 138–146. [Google Scholar] [CrossRef]
- Keskin, H.; Grunwald, S.; Harris, W.G. Digital Mapping of Soil Carbon Fractions with Machine Learning. Geoderma 2019, 339, 40–58. [Google Scholar] [CrossRef]
- Stum, A.K.; Boettinger, J.L.; White, M.A.; Ramsey, R.D. Random Forests Applied as a Soil Spatial Predictive Model in Arid Utah. In Digital Soil Mapping: Bridging Research, Environmental Application, and Operation; Boettinger, J.L., Howell, D.W., Moore, A.C., Hartemink, A.E., Kienast-Brown, S., Eds.; Springer: Dordrecht, The Netherland, 2010; pp. 179–190. [Google Scholar]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Tharwat, A. Classification Assessment Methods. Appl. Comput. Inform. 2021, 17, 168–192. [Google Scholar] [CrossRef]
- Brus, D.J. Sampling for Digital Soil Mapping: A Tutorial Supported by R Scripts. Geoderma 2019, 338, 464–480. [Google Scholar] [CrossRef]
- Hengl, T.; Nussbaum, M.; Wright, M.N.; Heuvelink, G.B.M.; Gräler, B. Random Forest as a Generic Framework for Predictive Modeling of Spatial and Spatio-Temporal Variables. PeerJ 2018, 6, e5518. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Heap, A.D.; Potter, A.; Daniell, J.J. Application of Machine Learning Methods to Spatial Interpolation of Environmental Variables. Environ. Model. Softw. 2011, 26, 1647–1659. [Google Scholar] [CrossRef]
- Lagacherie, P.; Arrouays, D.; Bourennane, H.; Gomez, C.; Martin, M.; Saby, N.P.A. How Far Can the Uncertainty on a Digital Soil Map Be Known?: A Numerical Experiment Using Pseudo Values of Clay Content Obtained from Vis-SWIR Hyperspectral Imagery. Geoderma 2019, 337, 1320–1328. [Google Scholar] [CrossRef]
- Arrouays, D.; Leenaars, J.G.B.; Richer-de-Forges, A.C.; Adhikari, K.; Ballabio, C.; Greve, M.; Grundy, M.; Guerrero, E.; Hempel, J.; Hengl, T.; et al. Soil Legacy Data Rescue via GlobalSoilMap and Other International and National Initiatives. GeoResJ 2017, 14, 1–19. [Google Scholar] [CrossRef]
- Caubet, M.; Román Dobarco, M.; Arrouays, D.; Minasny, B.; Saby, N.P.A. Merging Country, Continental and Global Predictions of Soil Texture: Lessons from Ensemble Modelling in France. Geoderma 2019, 337, 99–110. [Google Scholar] [CrossRef]
- Piikki, K.; Söderström, M.; Stadig, H. Local Adaptation of a National Digital Soil Map for Use in Precision Agriculture. Adv. Anim. Biosci. 2017, 8, 430–432. [Google Scholar] [CrossRef]
- Padarian, J.; Minasny, B.; McBratney, A.B. Machine Learning and Soil Sciences: A Review Aided by Machine Learning Tools. SOIL 2020, 6, 35–52. [Google Scholar] [CrossRef]
- Li, X.; Gu, H.; Tang, R.; Zou, B.; Liu, X.; Ou, H.; Chen, X.; Song, Y.; Luo, W.; Wen, B. A Fusion XGBoost Approach for Large-Scale Monitoring of Soil Heavy Metal in Farmland Using Hyperspectral Imagery. Agronomy 2025, 15, 676. [Google Scholar] [CrossRef]
- Li, Y.; Yao, G.; Li, S.; Dong, X. Predicting and Mapping of Soil Organic Matter with Machine Learning in the Black Soil Region of the Southern Northeast Plain of China. Agronomy 2025, 15, 533. [Google Scholar] [CrossRef]



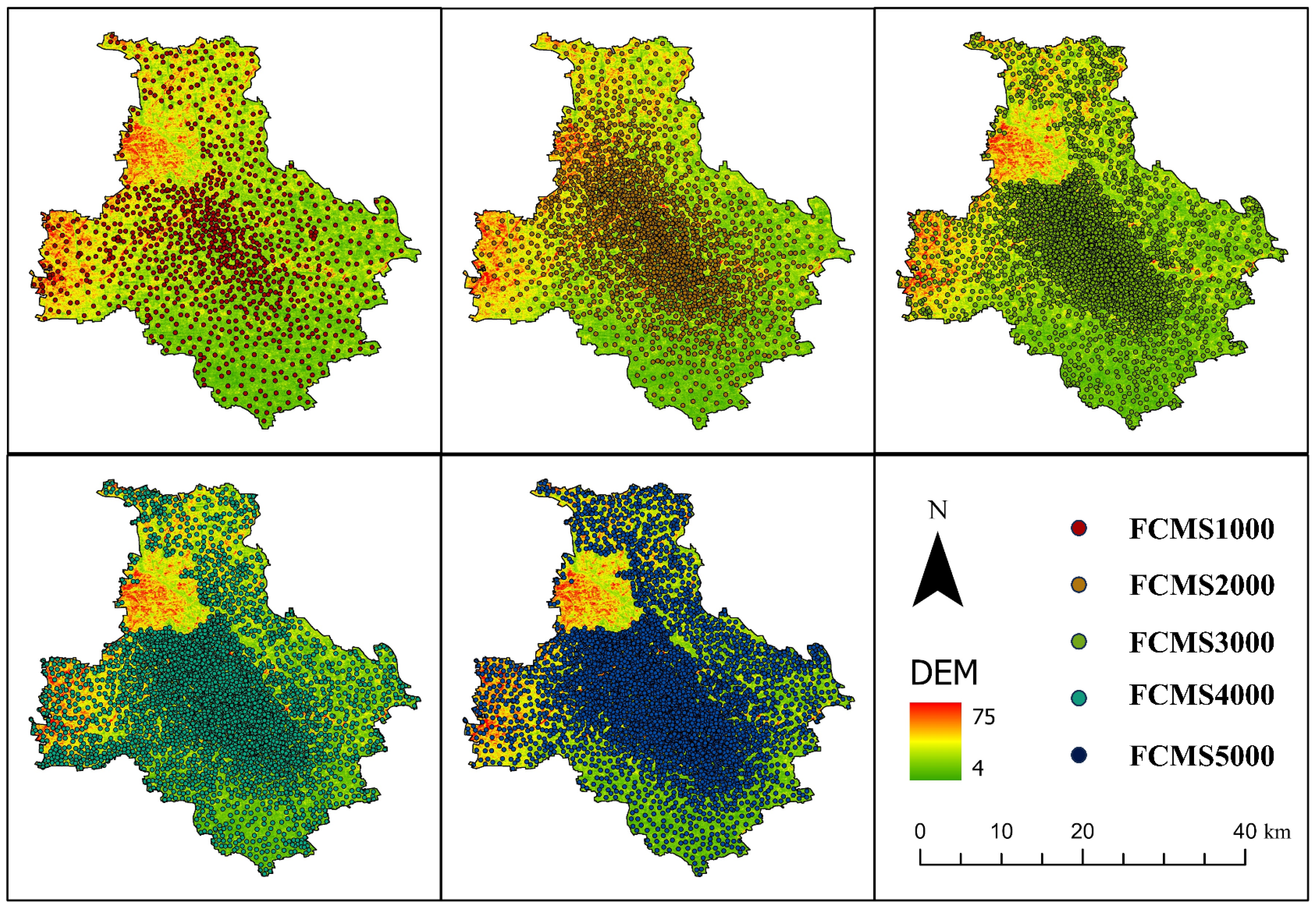
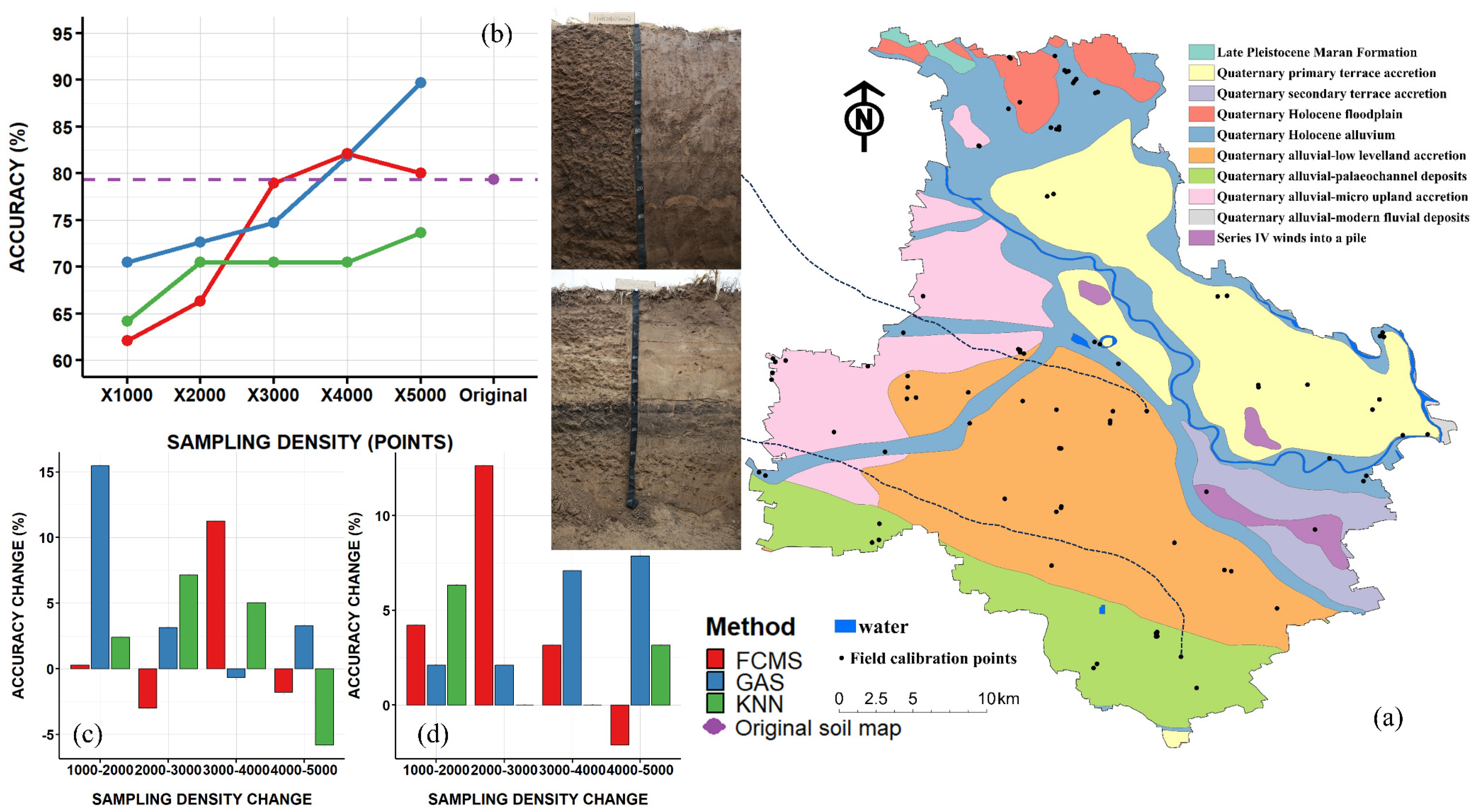

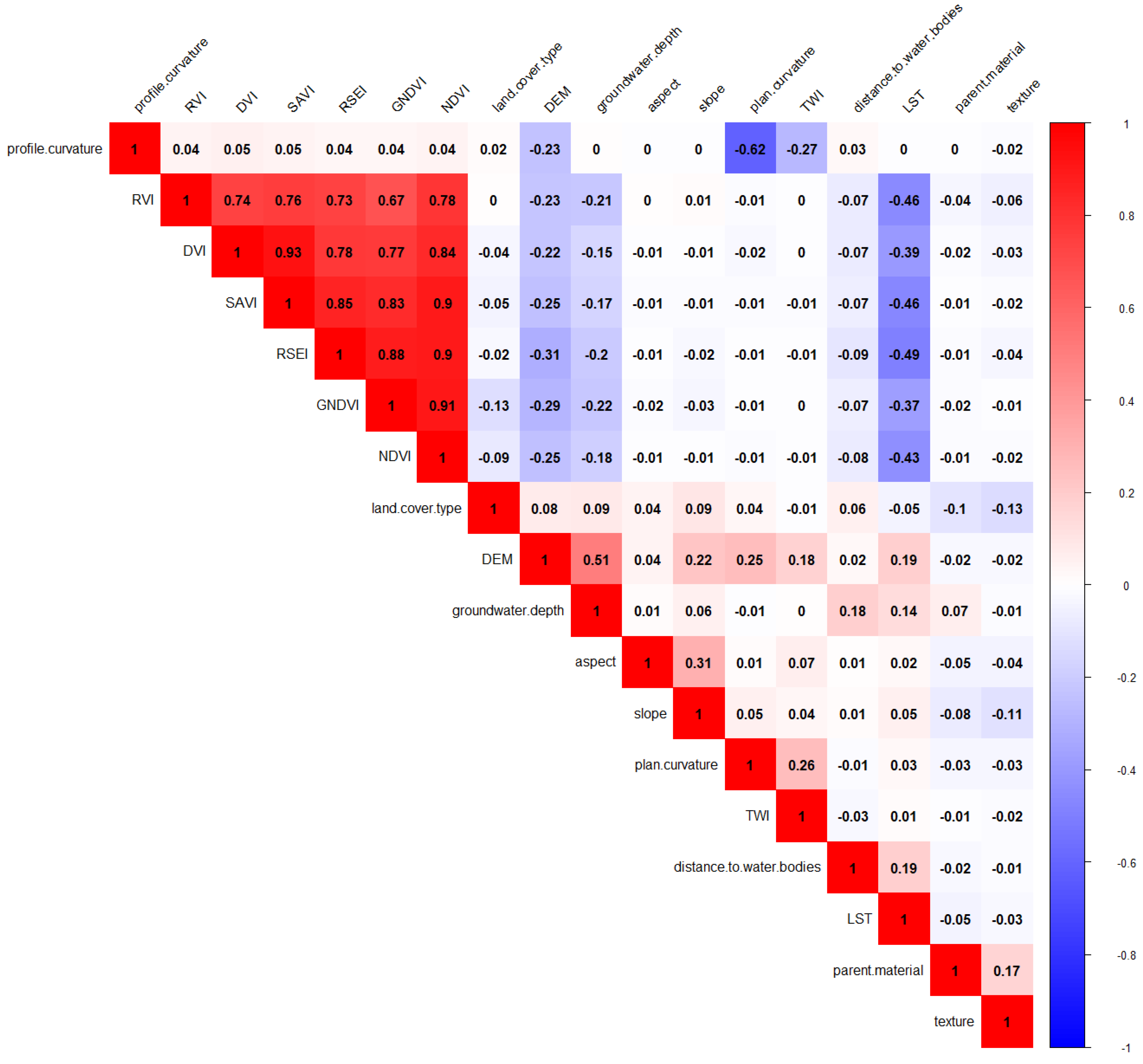
| No. | Name | Abbreviation | Resolution | Source |
|---|---|---|---|---|
| 1 | Parent Material | - | 1:25,000 | Digitized geological map of China |
| 2 | Texture | - | - | Indoor calibration soil map |
| 3 | Land Cover Type | - | 30 m | GlobeLand30 |
| 4 | Elevation | DEM | 12.5 m | National Science & Technology Infrastructure of China (http://www.geodata.cn) |
| 5 | Slope | - | Derived from DEM | Processed using ArcGIS |
| 6 | Aspect | - | Derived from DEM | Processed using ArcGIS |
| 7 | Planar Curvature | - | Derived from DEM | Processed using ArcGIS |
| 8 | Profile Curvature | - | Derived from DEM | Processed using ArcGIS |
| 9 | Topographic Wetness Index | TWI | Derived from DEM | Processed using ArcGIS |
| 10 | Distance to water bodies | - | - | Processed using ArcGIS and water system data |
| 11 | Groundwater Depth | - | - | Water Affairs Bureau of Tongzhou District, interpolated using inverse distance weighting |
| 12 | Risk-Screening Environmental Indicators | RSEI | 30 m | Landsat imagery processed using Google Earth Engine |
| 13 | Land Surface Temperature | LST | 30 m | Landsat imagery processed using Google Earth Engine |
| 14 | Soil-Adjusted Vegetation Index | SAVI | 30 m | Landsat imagery processed using Google Earth Engine |
| 15 | Normalized Difference Vegetation Index | NDVI | 30 m | Landsat imagery processed using Google Earth Engine |
| 16 | Difference Vegetation Index | DVI | 30 m | Landsat imagery processed using Google Earth Engine |
| 17 | Ratio Vegetation Index | RVI | 30 m | Landsat imagery processed using Google Earth Engine |
| 18 | Green Normalized Difference Vegetation Index | GNDVI | 30 m | Landsat imagery processed using Google Earth Engine |
| Step | Description |
|---|---|
| 1 | Overlay the environmental variable dataset in space. |
| 2 | Perform FCM clustering analysis on the data and divide the data into multiple classes. |
| 3 | Iterate to optimize the cluster centers, train 100 times. |
| 4 | Randomly select sampling points within each optimized class. |
| 5 | Repeat step 4 until the number of sampling points reaches the required amount. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Li, Y.; Wu, K.; Hao, S. GA-Optimized Sampling for Soil Type Mapping in Plain Areas: Integrating Legacy Maps and Multisource Covariates. Agronomy 2025, 15, 963. https://doi.org/10.3390/agronomy15040963
Wu X, Li Y, Wu K, Hao S. GA-Optimized Sampling for Soil Type Mapping in Plain Areas: Integrating Legacy Maps and Multisource Covariates. Agronomy. 2025; 15(4):963. https://doi.org/10.3390/agronomy15040963
Chicago/Turabian StyleWu, Xiangyuan, Yan Li, Kening Wu, and Shiheng Hao. 2025. "GA-Optimized Sampling for Soil Type Mapping in Plain Areas: Integrating Legacy Maps and Multisource Covariates" Agronomy 15, no. 4: 963. https://doi.org/10.3390/agronomy15040963
APA StyleWu, X., Li, Y., Wu, K., & Hao, S. (2025). GA-Optimized Sampling for Soil Type Mapping in Plain Areas: Integrating Legacy Maps and Multisource Covariates. Agronomy, 15(4), 963. https://doi.org/10.3390/agronomy15040963






