Genome-Wide Identification of Heavy Metal ATPase Family in Aegilops tauschii and Functional Verification of AetHMA4 and AetHMA8
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Physicochemical Characterization of the HMA Gene in A. tauschii
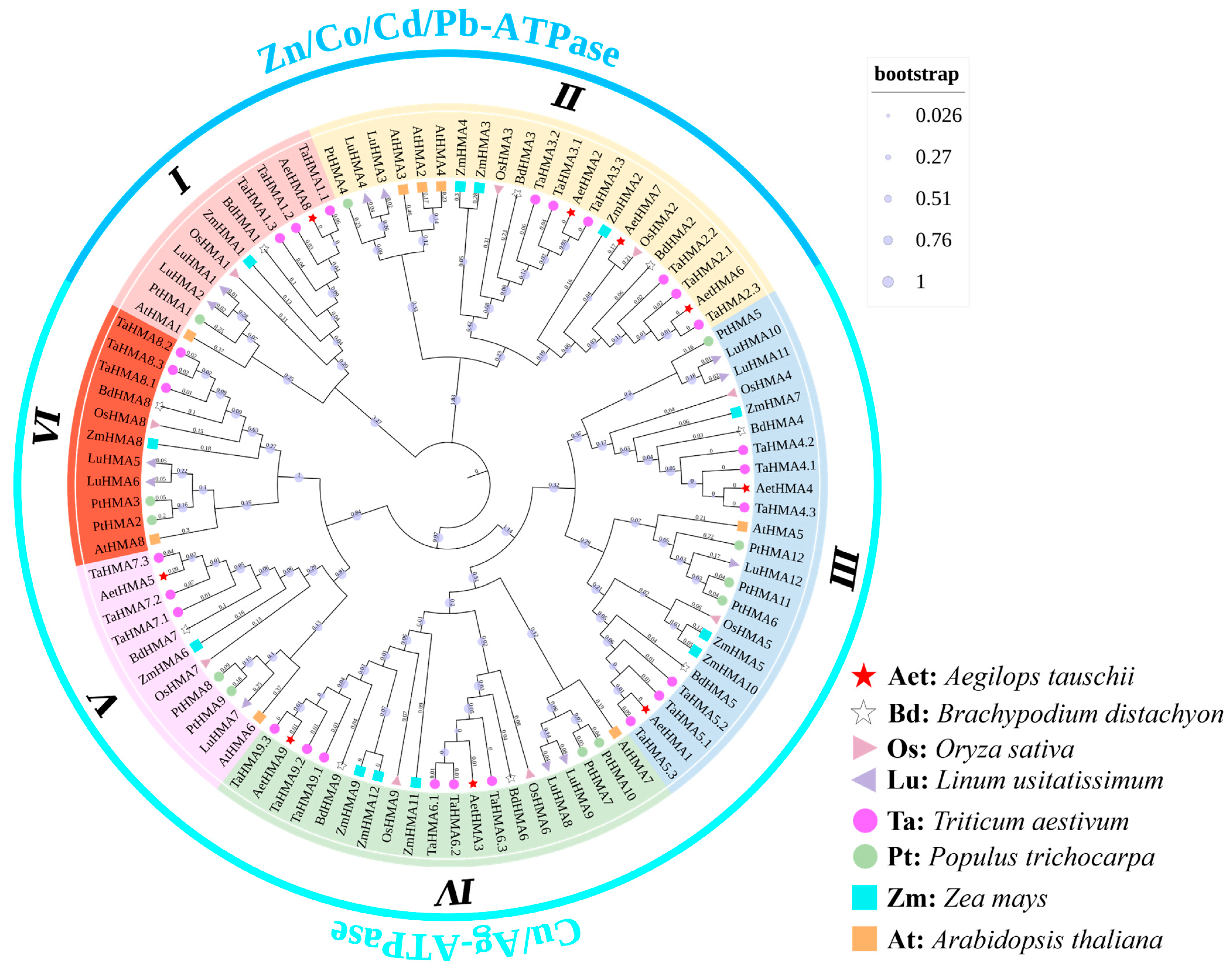
2.2. Phylogenetic Tree Construction of HMAs
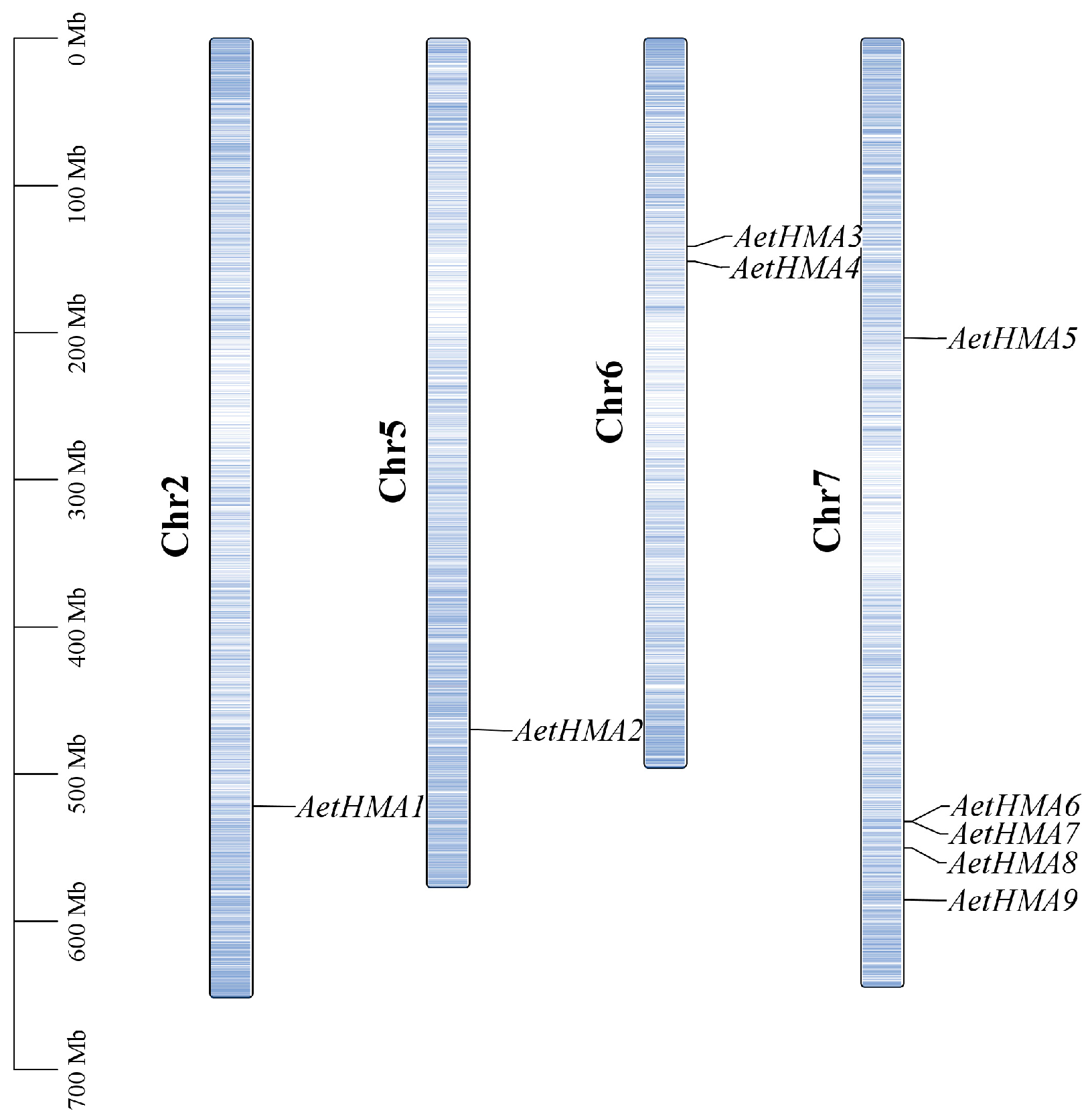
2.3. Chromosomal Mapping of the HMA Gene in A. tauschii
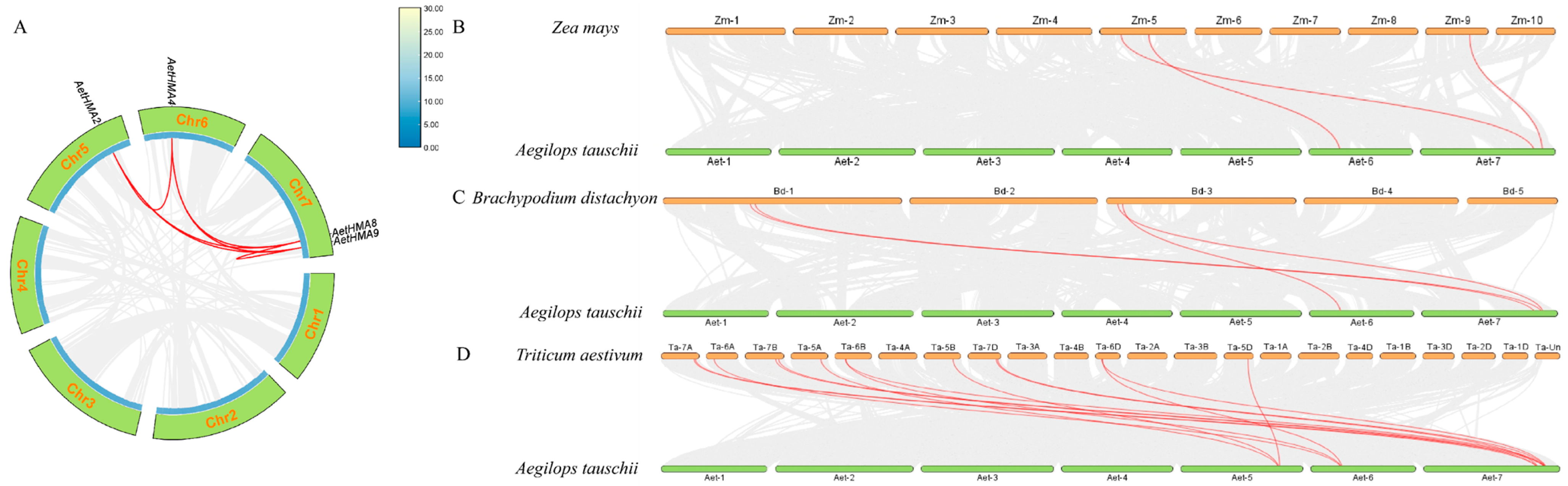
2.4. The Collinearity of the A. tauschii HMA Genes with Other Species
2.5. Identification of Cis-Regulatory Elements in the HMA Genes of A. tauschii
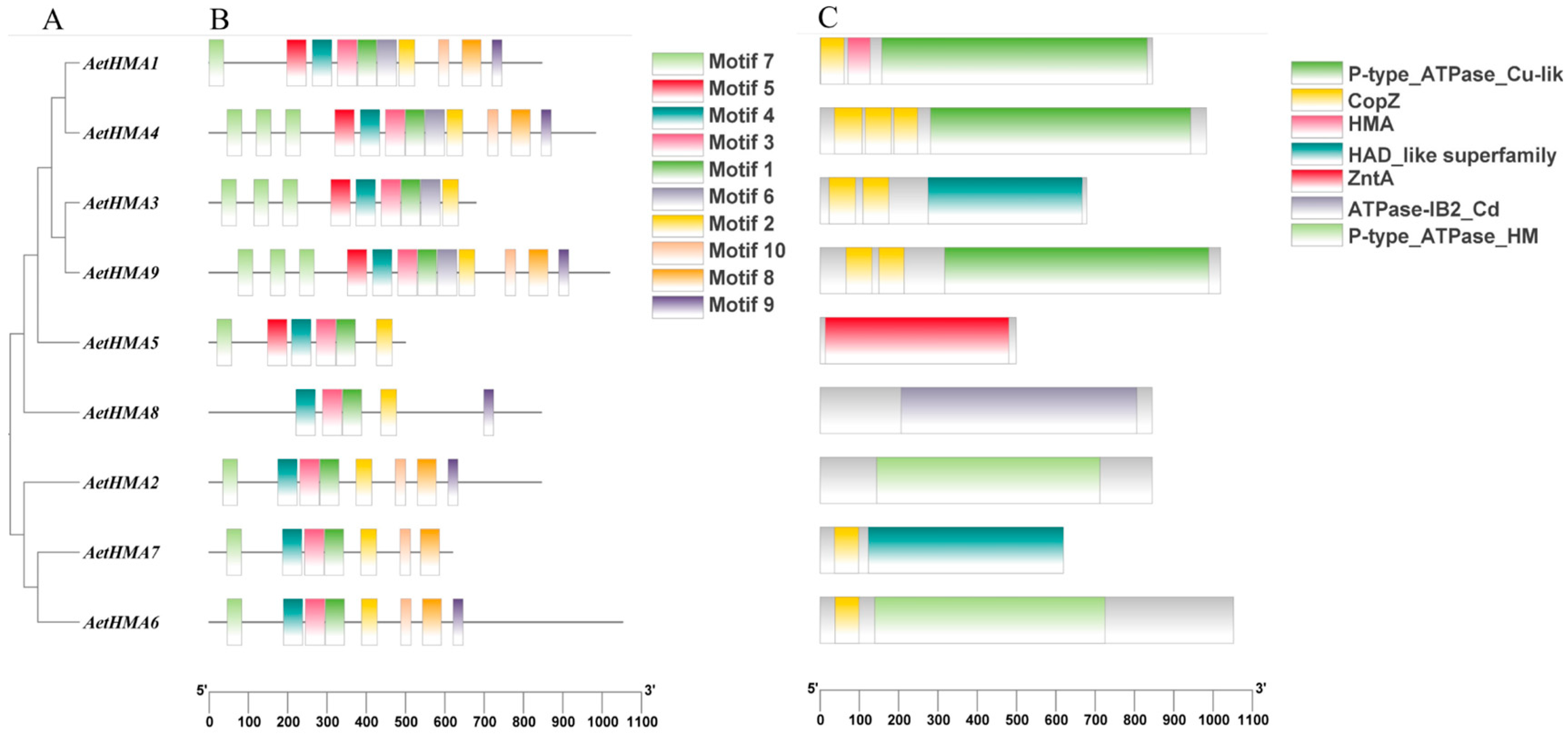
2.6. AetHMAs Gene Domain Analysis
2.7. Plant Materials and Growth Conditions
2.8. qRT-PCR and Data Analysis
2.9. The Heterologous Expression of AetHMA4 and AetHMA8 in Yeast
3. Results
3.1. Identification and Physicochemical Properties of the AetHMA Gene Family
3.2. Phylogenetic Analysis of Proteins of the HMA Family Genes in A. tauschii
3.3. Analysis of Chromosomal Locations of HMA Genes in A. tauschii
3.4. Analysis of Collinearity in the HMA Gene Family of A. tauschii
3.5. Analysis of Cis-Acting Elements in the AetHMA Gene Family of A. tauschii
3.6. Analysis of HMA Gene Family Structure in A. tauschii
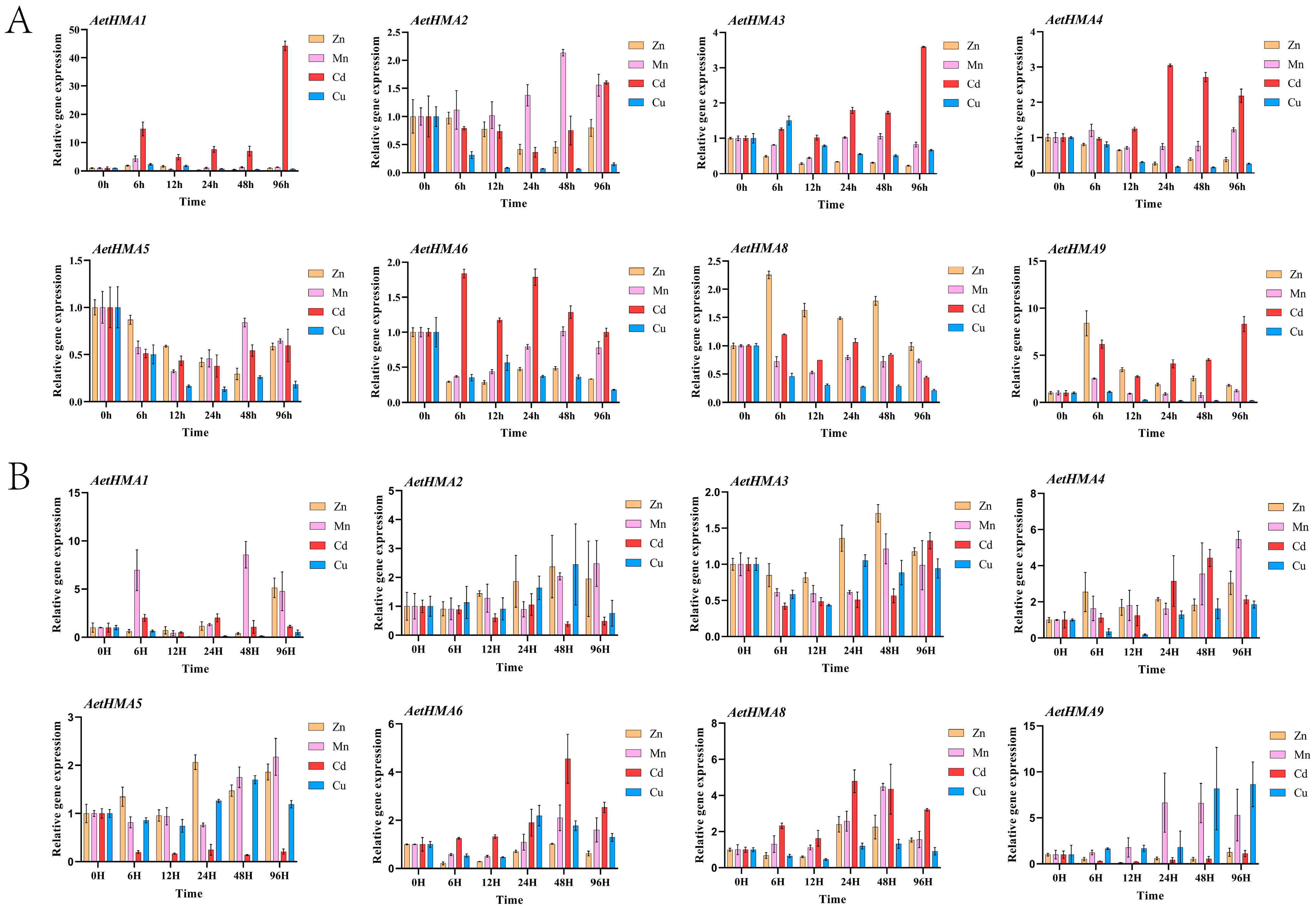
3.7. Expression Analysis of the HMA Gene Family in A. tauschii Under Different Heavy Metal Stresses
3.8. Heterologous Expression of AetHMA8 in Yeast to Enhance Tolerance to Metal Ions
4. Discussion
4.1. Analysis of the HMA Gene Family Members in A. tauschii
4.2. The AetHMA Gene Family Influences the Heavy Metal Tolerance of A. tauschii
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, F.; Dong, W.; Zhao, Z.; Wang, H.; Li, W.; Chen, G.; Wang, F.; Zhao, Y.; Huang, J.; Zhou, T. Heavy Metal Pollution in Urban River Sediment of Different Urban Functional Areas and Its Influence on Microbial Community Structure. Sci. Total Environ. 2021, 778, 146383. [Google Scholar] [CrossRef] [PubMed]
- CCICED. Special Policy Study on Soil Pollution Management. China Council for International Cooperation on Environment and Development. 2015, p. 1. Available online: https://cciced.eco/wp-content/uploads/2020/06/Soil-Pollution-Management-Special-Study-Report-2015.pdf (accessed on 12 March 2025).
- FAO. Soil Pollution a Hidden Reality. 2018, p. 1. Available online: https://openknowledge.fao.org/server/api/core/bitstreams/3f7e6959-db0b-44d3-971e-109bcfe78195/content (accessed on 12 March 2025).
- Jogawat, A.; Yadav, B.; Chhaya; Narayan, O.P. Metal Transporters in Organelles and Their Roles in Heavy Metal Transportation and Sequestration Mechanisms in Plants. Physiol. Plant. 2021, 173, 259–275. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Z.; Shi, G. Genome-Wide Identification and Expression Profiling of Heavy Metal ATPase (HMA) Genes in Peanut: Potential Roles in Heavy Metal Transport. Int. J. Mol. Sci. 2024, 25, 613. [Google Scholar] [CrossRef] [PubMed]
- Shukla, U.C.; Singh, J.; Joshi, P.C.; Kakkar, P. Effect of Bioaccumulation of Cadmium on Biomass Productivity, Essential Trace Elements, Chlorophyll Biosynthesis, and Macromolecules of Wheat Seedlings. Biol. Trace Elem. Res. 2003, 92, 257–273. [Google Scholar] [CrossRef] [PubMed]
- Sandalio, L.M.; Dalurzo, H.C.; Gómez, M.; Romero-Puertas, M.C.; del Río, L.A. Cadmium-induced Changes in the Growth and Oxidative Metabolism of Pea Plants. J. Exp. Bot. 2001, 52, 2115–2126. [Google Scholar] [CrossRef]
- Qin, P.; Wang, L.; Liu, K.; Mao, S.; Li, Z.; Gao, S.; Shi, H.; Liu, Y. Genomewide Association Study of Aegilops tauschii Traits under Seedling-Stage Cadmium Stress. Crop J. 2015, 3, 405–415. [Google Scholar] [CrossRef]
- Bhardwaj, E.; Shukla, R.; Das, S. Plant Roots and Mineral Nutrition: An Overview of Molecular Basis of Uptake and Regulation, and Strategies to Improve Nutrient Use Efficiency (NUE). In Plant Stress Biology; Springer Nature: Berlin, Germany, 2021; pp. 131–184. [Google Scholar] [CrossRef]
- Li, G.; Song, H.; Li, B.; Kronzucker, H.J.; Shi, W. Auxin Resistant1 and PIN-FORMED2 Protect Lateral Root Formation in Arabidopsis under Iron Stress. Plant Physiol. 2015, 169, 2608–2623. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K. Heavy Metals Toxicity in Plants: An Overview on the Role of Glutathione and Phytochelatins in Heavy Metal Stress Tolerance of Plants. S. Afr. J. Bot. 2010, 76, 167–179. [Google Scholar] [CrossRef]
- Baran, A. Assessment of Zea Mays Sensitivity to Toxic Content of Zinc in Soil. Pol. J. Environ. Stud. 2013, 22, 77–83. [Google Scholar]
- Xu, J.; Yin, H.; Li, Y.; Liu, X. Nitric Oxide Is Associated with Long-Term Zinc Tolerance in Solanum nigrum. Plant Physiol. 2010, 154, 1319–1334. [Google Scholar] [CrossRef]
- Marastoni, L.; Tauber, P.; Pii, Y.; Valentinuzzi, F.; Astolfi, S.; Simoni, A.; Brunetto, G.; Cesco, S.; Mimmo, T. The Potential of Two Different Avena sativa L. Cultivars to Alleviate Cu Toxicity. Ecotoxicol. Environ. Saf. 2019, 182, 109430. [Google Scholar] [CrossRef] [PubMed]
- Alejandro, S.; Höller, S.; Meier, B.; Peiter, E. Manganese in Plants: From Acquisition to Subcellular Allocation. Front. Plant Sci. 2020, 11, 300. [Google Scholar] [CrossRef]
- Zhang, W.; Munyaneza, V.; Kant, S.; Wang, S.; Wang, X.; Cai, H.; Wang, C.; Shi, L.; Wang, S.; Xu, F.; et al. Transcription Factor AtNAC002 Positively Regulates Cu Toxicity Tolerance in Arabidopsis thaliana. J. Hazard. Mater. 2024, 480, 136186. [Google Scholar] [CrossRef]
- Shabbir, Z.; Sardar, A.; Shabbir, A.; Abbas, G.; Shamshad, S.; Khalid, S.; Natasha; Murtaza, G.; Dumat, C.; Shahid, M. Copper Uptake, Essentiality, Toxicity, Detoxification and Risk Assessment in Soil-Plant Environment. Chemosphere 2020, 259, 127436. [Google Scholar] [CrossRef] [PubMed]
- Rascio, N.; Navari-Izzo, F. Heavy Metal Hyperaccumulating Plants: How and Why Do They Do It? And What Makes Them so Interesting? Plant Sci. 2011, 180, 169–181. [Google Scholar] [CrossRef]
- Solioz, M.; Vulpe, C. CPx-Type ATPases: A Class of P-Type ATPases That Pump Heavy Metals. Trends Biochem. Sci. 1996, 21, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Hussain, D.; Haydon, M.J.; Wang, Y.; Wong, E.; Sherson, S.M.; Young, J.; Camakaris, J.; Harper, J.F.; Cobbett, C.S. P-Type ATPase Heavy Metal Transporters with Roles in Essential Zinc Homeostasis in Arabidopsis. Plant Cell 2004, 16, 1327–1339. [Google Scholar] [CrossRef]
- Chan, H.; Babayan, V.; Blyumin, E.; Gandhi, C.; Hak, K.; Harake, D.; Kumar, K.; Lee, P.; Li, T.T.; Liu, H.Y.; et al. The P-Type ATPase Superfamily. J. Mol. Microbiol. Biotechnol. 2010, 19, 5–104. [Google Scholar] [CrossRef]
- Argüello, J.M.; Eren, E.; González-Guerrero, M. The Structure and Function of Heavy Metal Transport P1B-ATPases. BioMetals 2007, 20, 233–248. [Google Scholar] [CrossRef]
- Williams, L.E.; Mills, R.F. P1B-ATPases—An Ancient Family of Transition Metal Pumps with Diverse Functions in Plants. Trends Plant Sci. 2005, 10, 491–502. [Google Scholar] [CrossRef]
- Baxter, I.; Tchieu, J.; Sussman, M.R.; Boutry, M.; Palmgren, M.G.; Gribskov, M.; Harper, J.F.; Axelsen, K.B. Genomic Comparison of P-Type ATPase Ion Pumps in Arabidopsis and Rice. Plant Physiol. 2003, 132, 618–628. [Google Scholar] [CrossRef]
- Rensing, C.; Ghosh, M.; Rosen, B.P. Families of Soft-Metal-Ion-Transporting ATPases. J. Bacteriol. 1999, 181, 5891–5897. [Google Scholar] [CrossRef] [PubMed]
- Axelsen, K.B.; Palmgren, M.G. Inventory of the Superfamily of P-Type Ion Pumps in Arabidopsis. Plant Physiol. 2001, 126, 696–706. [Google Scholar] [CrossRef]
- Cobbett, C.S.; Hussain, D.; Haydon, M.J. Structural and Functional Relationships between Type 1B Heavy Metal-Transporting P-Type ATPases in Arabidopsis. New Phytol. 2003, 159, 315–321. [Google Scholar] [CrossRef]
- Zhiguo, E.; Tingting, L.; Chen, C.; Lei, W. Genome-Wide Survey and Expression Analysis of P 1B-ATPases in Rice, Maize and Sorghum. Rice Sci. 2018, 25, 208–217. [Google Scholar] [CrossRef]
- Khan, N.; You, F.M.; Datla, R.; Ravichandran, S.; Jia, B.; Cloutier, S. Genome-Wide Identification of ATP Binding Cassette (ABC) Transporter and Heavy Metal Associated (HMA) Gene Families in Flax (Linum usitatissimum L.). BMC Genom. 2020, 21, 722. [Google Scholar] [CrossRef]
- Boutigny, S.; Sautron, E.; Finazzi, G.; Rivasseau, C.; Frelet-Barrand, A.; Pilon, M.; Rolland, N.; Seigneurin-Berny, D. HMA1 and PAA1, Two Chloroplast-Envelope PIB-ATPases, Play Distinct Roles in Chloroplast Copper Homeostasis. J. Exp. Bot. 2014, 65, 1529–1540. [Google Scholar] [CrossRef] [PubMed]
- Seigneurin-Berny, D.; Gravot, A.; Auroy, P.; Mazard, C.; Kraut, A.; Finazzi, G.; Grunwald, D.; Rappaport, F.; Vavasseur, A.; Joyard, J.; et al. HMA1, a New Cu-ATPase of the Chloro Plast Envelope, Is Essential for Growth under Adverse Light Conditions*. J. Biol. Chem. 2006, 281, 2882–2892. [Google Scholar] [CrossRef]
- Li, S.; Li, X.; Li, S.; Liu, Y.; Zang, T.; Hao, M.; Zhang, L.; Huang, L.; Jiang, B.; Yuan, Z.; et al. Variation in the Tonoplast Cadmium Transporter Heavy Metal ATPase 3 (HMA3) Homolog Gene in Aegilops tauschii. PLoS ONE 2023, 18, e0279707. [Google Scholar] [CrossRef]
- Shikanai, T.; Müller-Moulé, P.; Munekage, Y.; Niyogi, K.K.; Pilon, M. PAA1, a P-Type ATPase of Arabidopsis, Functions in Copper Transport in Chloroplasts. Plant Cell 2003, 15, 1333–1346. [Google Scholar] [CrossRef]
- Verret, F.; Gravot, A.; Auroy, P.; Leonhardt, N.; David, P.; Nussaume, L.; Vavasseur, A.; Richaud, P. Overexpression of AtHMA4 Enhances Root-to-Shoot Translocation of Zinc and Cadmium and Plant Metal Tolerance. FEBS Lett. 2004, 576, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Morel, M.; Crouzet, J.; Gravot, A.; Auroy, P.; Leonhardt, N.; Vavasseur, A.; Richaud, P. AtHMA3, a P1B-ATPase Allowing Cd/Zn/Co/Pb Vacuolar Storage in Arabidopsis. Plant Physiol. 2009, 149, 894–904. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Kuroda, K.; Kimura, K.; Southron-Francis, J.L.; Furuzawa, A.; Kimura, K.; Iuchi, S.; Kobayashi, M.; Taylor, G.J.; Koyama, H. Amino Acid Polymorphisms in Strictly Conserved Domains of a P-Type ATPase HMA5 Are Involved in the Mechanism of Copper Tolerance Variation in Arabidopsis. Plant Physiol. 2008, 148, 969–980. [Google Scholar] [CrossRef]
- Andrés-Colás, N.; Sancenón, V.; Rodríguez-Navarro, S.; Mayo, S.; Thiele, D.J.; Ecker, J.R.; Puig, S.; Peñarrubia, L. The Arabidopsis Heavy Metal P-type ATPase HMA5 Interacts with Metallochaperones and Functions in Copper Detoxification of Roots. Plant J. 2006, 45, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Woeste, K.E.; Kieber, J.J. A Strong Loss-of-Function Mutation in RAN1 Results in Constitutive Activation of the Ethylene Response Pathway as Well as a Rosette-Lethal Phenotype. Plant Cell 2022, 12, 443–455. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Müller-Moulé, P.; Niyogi, K.K.; Pilon, M.; Shikanai, T. Two P-Type ATPases Are Required for Copper Delivery in Arabidopsis thaliana Chloroplasts. Plant Cell 2005, 17, 1233–1251. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Abbas, A.; Hameed, R.; Saeed, M.; Shahani, A.A.A.; Huang, P.; Du, D.; Zulfiqar, U.; Alamri, S.; Alfagham, A.T. Investigating the Dynamic Responses of Aegilops tauschii Coss. to Salinity, Drought, and Nitrogen Stress: A Comprehensive Study of Competitive Growth and Biochemical and Molecular Pathways. Front. Plant Sci. 2023, 14, 1238704. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Han, X.; Zhang, Y.; Yu, M.; Zhang, J.; Xu, D.; Lu, Z.; Qiao, G.; Qiu, W.; Zhuo, R. Transporters and Ascorbate–Glutathione Metabolism for Differential Cadmium Accumulation and Tolerance in Two Contrasting Willow Genotypes. Tree Physiol. 2020, 40, 1126–1142. [Google Scholar] [CrossRef]
- Cao, Y.; Zhao, X.; Liu, Y.; Wang, Y.; Wu, W.; Jiang, Y.; Liao, C.; Xu, X.; Gao, S.; Shen, Y.; et al. Genome-Wide Identification of ZmHMAs and Association of Natural Variation in ZmHMA2 and ZmHMA3 with Leaf Cadmium Accumulation in Maize. PeerJ 2019, 7, e7877. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Wang, L.; Deng, X.; Wang, P.; Ma, Q.; Nian, H.; Wang, Y.; Yang, C. Genome-Wide Characterization of Soybean P1B-ATPases Gene Family Provides Functional Implications in Cadmium Responses. BMC Genom. 2016, 17, 376. [Google Scholar] [CrossRef]
- Li, N.; Xiao, H.; Sun, J.; Wang, S.; Wang, J.; Chang, P.; Zhou, X.; Lei, B.; Lu, K.; Luo, F.; et al. Genome-Wide Analysis and Expression Profiling of the HMA Gene Family in Brassica Napus under Cd Stress. Plant Soil 2018, 426, 365–381. [Google Scholar] [CrossRef]
- Wang, X.; Li, H.; Xu, J. Bioinformatics Analysis of the Heavy Metal Transporting ATPase Gene Family in Poplar Genome. Plant Physiol. J. 2014, 50, 891–900. [Google Scholar] [CrossRef]
- Lee, S.; Kim, Y.-Y.; Lee, Y.; An, G. Rice P1B-Type Heavy-Metal ATPase, OsHMA9, Is a Metal Efflux Protein. Plant Physiol. 2007, 145, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-Y.; Choi, H.; Segami, S.; Cho, H.-T.; Martinoia, E.; Maeshima, M.; Lee, Y. AtHMA1 Contributes to the Detoxification of Excess Zn(II) in Arabidopsis. Plant J. 2009, 58, 737–753. [Google Scholar] [CrossRef]
- Ghori, N.-H.; Ghori, T.; Hayat, M.Q.; Imadi, S.R.; Gul, A.; Altay, V.; Ozturk, M. Heavy Metal Stress and Responses in Plants. Int. J. Environ. Sci. Technol. 2018, 16, 1807–1828. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Q.; Xu, W.; Zhao, H.; Guo, F.; Wang, P.; Wang, Y.; Ni, D.; Wang, M.; Wei, C. Identification of MTP Gene Family in Tea Plant (Camellia sinensis L.) and Characterization of CsMTP8.2 in Manganese Toxicity. Ecotoxicol. Environ. Saf. 2020, 202, 110904. [Google Scholar] [CrossRef]
- Cailliatte, R.; Schikora, A.; Briat, J.-F.; Mari, S.; Curie, C. High-Affinity Manganese Uptake by the Metal Transporter NRAMP1 Is Essential for Arabidopsis Growth in Low Manganese Conditions. Plant Cell 2010, 22, 904–917. [Google Scholar] [CrossRef]
- Sasaki, A.; Yamaji, N.; Ma, J.F. Overexpression of OsHMA3 Enhances Cd Tolerance and Expression of Zn Transporter Genes in Rice. J. Exp. Bot. 2014, 65, 6013–6021. [Google Scholar] [CrossRef]
- Huang, X.-Y.; Deng, F.; Yamaji, N.; Pinson, S.R.M.; Fujii-Kashino, M.; Danku, J.; Douglas, A.; Guerinot, M.L.; Salt, D.E.; Ma, J.F. A Heavy Metal P-Type ATPase OsHMA4 Prevents Copper Accumulation in Rice Grain. Nat. Commun. 2016, 7, 12138. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | No. of aa | MW a | pI b | Instability Index | Aliphatic Index | GRAVY c | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| AetHMA1 | AET2Gv20910400 | 845 | 91,936.68 | 5.86 | 36.2 | 103.83 | 0.207 | Cell membrane |
| AetHMA2 | AET5Gv20895000 | 845 | 88,162.02 | 6.01 | 47.22 | 100.36 | 0.276 | Cell membrane |
| AetHMA3 | AET6Gv20398800 | 678 | 72,819.31 | 5.83 | 35.36 | 103.36 | 0.312 | Cell membrane |
| AetHMA4 | AET6Gv20415800 | 982 | 105,753.48 | 5.4 | 36.54 | 101.86 | 0.173 | Cell membrane |
| AetHMA5 | AET7Gv20587600 | 498 | 52,501.78 | 6.23 | 38.59 | 101.63 | 0.294 | Cell membrane |
| AetHMA6 | AET7Gv21017000 | 1051 | 114,152.8 | 6.74 | 44.39 | 85.56 | −0.193 | Cell membrane, nucleus |
| AetHMA7 | AET7Gv21017700 | 619 | 66,059.26 | 6.62 | 38.9 | 104.3 | 0.255 | Cell membrane |
| AetHMA8 | AET7Gv21057700 | 844 | 89,543.31 | 7.51 | 36.75 | 99.68 | 0.15 | Chloroplast |
| AetHMA9 | AET7Gv21175500 | 1,018 | 108,744.97 | 5.62 | 38.46 | 103.64 | 0.294 | Cell membrane |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, X.; Zhang, X.; Li, Y.; Ding, Y.; Li, H.; Hao, Z.; Wang, N.; Han, X. Genome-Wide Identification of Heavy Metal ATPase Family in Aegilops tauschii and Functional Verification of AetHMA4 and AetHMA8. Agronomy 2025, 15, 714. https://doi.org/10.3390/agronomy15030714
Liang X, Zhang X, Li Y, Ding Y, Li H, Hao Z, Wang N, Han X. Genome-Wide Identification of Heavy Metal ATPase Family in Aegilops tauschii and Functional Verification of AetHMA4 and AetHMA8. Agronomy. 2025; 15(3):714. https://doi.org/10.3390/agronomy15030714
Chicago/Turabian StyleLiang, Xiaolin, Xiaofang Zhang, Yibo Li, Yifan Ding, Hongying Li, Ziyuan Hao, Ning Wang, and Xiaojiao Han. 2025. "Genome-Wide Identification of Heavy Metal ATPase Family in Aegilops tauschii and Functional Verification of AetHMA4 and AetHMA8" Agronomy 15, no. 3: 714. https://doi.org/10.3390/agronomy15030714
APA StyleLiang, X., Zhang, X., Li, Y., Ding, Y., Li, H., Hao, Z., Wang, N., & Han, X. (2025). Genome-Wide Identification of Heavy Metal ATPase Family in Aegilops tauschii and Functional Verification of AetHMA4 and AetHMA8. Agronomy, 15(3), 714. https://doi.org/10.3390/agronomy15030714






