Abstract
Accurate assessment of forage quality is essential for ensuring optimal animal nutrition. Key parameters, such as Leaf Area Index (LAI) and grass coverage, are indicators that provide valuable insights into forage health and productivity. Accurate measurement is essential to ensure that livestock obtain the proper nutrition during various phases of plant growth. This study evaluated machine learning (ML) methods for non-invasive assessment of grassland development using RGB imagery, focusing on ryegrass and Timothy (Lolium perenne L. and Phleum pratense L.). ML models were implemented to segment and quantify coverage of live plants, dead material, and bare soil at three pasture growth stages (leaf development, tillering, and beginning of flowering). Unsupervised and supervised ML models, including a hybrid approach combining Gaussian Mixture Model (GMM) and Nearest Centroid Classifier (NCC), were applied for pixel-wise segmentation and classification. The best results were achieved in the tillering stage, with R2 values from 0.72 to 0.97 for Timothy (α = 0.05). For ryegrass, the RGB-based pixel-wise model performed best, particularly during leaf development, with R2 reaching 0.97. However, all models struggled during the beginning of flowering, particularly with dead grass and bare soil coverage.
1. Introduction
Forages are critical for animal production systems, particularly for ruminants, due to their role in economic nutrition and environmental conservation. Despite regional variations in their use, forages are vital for sustainable and cost-effective animal production [1]. Furthermore, grasslands cover 26% of the world’s land area and 70% of the global agricultural area, supporting over 800 million people [2,3]. These lands often include unmanaged or partially managed mixed grasses, legumes, and forbs, alongside millions of hectares of highly managed pasture, hay, and silage crops. However, compilation of forage crops’ global acreage and value do not usually encompass all forage crops, and comprehensive data are sometimes lacking [4]. Global climate change poses a significant challenge for forage crop management and breeding, with northern regions expected to experience a faster warming rate than the global average [5]. In Norway, rising winter and summer temperatures may extend the grassland growing season. Precipitation changes, including extreme weather events and unstable winters, further complicate adaptation strategies. These climatic shifts, whose most significant positive and negative changes are predicted to occur in northernmost Europe, will introduce abiotic stresses that could negatively impact forage yields [6]. Hence, given the complexity and contrasting effects of climate change, more accurate information is needed to efficiently manage forage crops in light of the current and forthcoming challenges. Norway, in particular, exhibits one of the highest global demands for quality livestock production of meat, milk, and dairy products [7]. Thus, substantial resources are allocated to forage production, indicating severe environmental damage [8]. Hence, in Scandinavian countries, significant attention has been directed towards the requirements and needs for food production based on ruminants [9]. Forage production demands an increasingly substantial amount of resources, and this situation is considered a bottleneck in the system [8]. Consequently, the relationship between the requirements for grassland production and the degradation of natural resources has been examined [10]. This examination has involved implementing precision agriculture (PA) processes [11] to contrast qualitative information obtained through traditional methods, such as field sampling or supervised visual estimation, which are often destructive and costly.
Efficient management of forage crops and grasslands is crucial for sustainable agriculture. PA techniques facilitate the implementation of different approaches to rapid and accurate estimates of biomass, forage quality, and grassland productivity indicators. These are essential in decision-making about cutting dates, fertilization, and grassland renovation. For example, Dusseux et al. [12] evaluated the potential of Sentinel-2 satellite data to estimate dry grassland biomass using grassland height as a measurement. Within the electromagnetic spectrum, the Red-edge, Near Infra-Red, and Short Wave Infra-Red spectral bands appeared to contain substantial information that could be utilized for the estimation of grassland biomass. Moreover, Rueda-Ayala and Höglind [13], using unmanned aerial vehicles (UAVs), conducted a research study to determine the ideal grass conditions for successfully establishing Trifolium pratense L. through sod-seeding. The study highlighted the challenges of introducing red clover into dense swards and emphasized the importance of site-specific considerations for grassland renovation. Similarly, Rueda-Ayala et al. [14] assessed grass ley fields using UAVs and on-ground methods (RGB-D information). Plant height, biomass, and volume using digital grass models were estimated. The sensing systems accurately determined parameters by comparing estimated values with ground truth, considering basic economic considerations.
Fricke et al. [15] explored the use of ultrasonic sensors, both statically and mounted on vehicles, to estimate sward heights and correlate these measurements with forage mass. This approach provides a non-destructive method for yield mapping in precision agriculture. Additionally, laboratory studies assessed forage crops to predict quality parameters, further enhancing the application of these technologies. Building on this, Berauer et al. [16] introduced visible-near-infrared spectroscopy (vis-NIRS), which demonstrated high accuracy in predicting forage quality parameters, such as ash, fat, and protein, in bulk samples from species-rich montane pastures. This method also proved valuable for detecting the impacts of climate change and land management on forage quality. On the other hand, RGB imagery, while effective for many agricultural applications, faces notable limitations, particularly in scenarios involving leaf overlap. Overlapping leaves can obscure key sections of the plant, reducing the accuracy of feature detection and analysis. This issue is particularly pronounced in grass mixtures, where the uniformity of vegetation can further complicate segmentation and classification tasks. In contrast, distance sensors [16], which operate based on principles like time-of-flight, offer complementary advantages by enabling the measurement of plant height and the indirect estimation of biomass through height–biomass relationships. These sensors are typically categorized into two main types: ultrasonic devices and LiDAR (Light Detection and Ranging) systems. Ultrasonic sensors rely on sound waves to measure distances, while LiDAR employs laser pulses, providing higher resolution and precision [17]. Both ultrasonic and LiDAR technologies have seen widespread adoption in modern agricultural practices due to their versatility and efficiency. Their ability to rapidly collect data across extensive field areas makes them particularly valuable for applications like crop monitoring, yield estimation, and resource optimization. LiDAR can also be implemented by supplementing spectral data with structural and intensity information, enabling higher accuracy in species classification and mapping. This integration enhances the use of LiDAR in ecological studies and its potential to streamline operational species monitoring [18,19].
However, the integration of non-invasive technologies—including vis-NIRS, ultrasonic and depth sensors, UAVs, and satellite sensing—has created a robust foundation for more precise, efficient, and sustainable management of grasslands and forage crops. Furthermore, advancements in artificial intelligence (AI) have driven significant progress in feature extraction techniques, enabling better assessment of key traits, like leaf area index (LAI), plant height, and biomass. These innovations collectively represent a leap forward in optimizing forage crop management. In AI, machine learning (ML) has become the standard for image analysis [19]. Recent ML advances have opened new avenues for enhancing these processes, leveraging remote sensing technologies, advanced algorithms, and diverse datasets to derive valuable insights. In the context of forage crop management, ML algorithms have been increasingly used and achieved remarkable success in various forage crop assessment and management. For example, Oliveira et al. [20] demonstrated the potential of UAV remote sensing for managing and monitoring silage grass swards. Their study utilized drone photogrammetry and spectral imaging to estimate biomass, nitrogen content, and other quality parameters in grasslands. Training machine learning models, such as Random Forest (RF) and multiple linear regression (MLR), with reference measurements achieved promising accuracy in biomass estimation, nitrogen uptake, and digestibility. Similarly, Lussem et al. [21] utilized UAV-based imaging sensors and photogrammetric structure-from-motion processing to estimate dry matter yield (DMY) and nitrogen uptake in temperate grasslands. They compared linear regression, Random Forest (RF), support vector machine (SVM), and partial least squares (PLS) regression models. Combining structural and spectral features improved prediction accuracy across all models, with RF and SVM outperforming PLS. This study underscored the efficacy of integrating various data features for robust grassland monitoring. In contrast, Xu et al. [22] focused on the use of terrestrial laser scanning (TLS) for estimating aboveground biomass (AGB) in grasslands. TLS provides detailed canopy structural information, which can be used to build regression models. Their study compared four regression methods: simple regression (SR), stepwise multiple regression (SMR), Random Forest (RF), and artificial neural network (ANN). The SMR model achieved the highest prediction accuracy, indicating that incorporating multiple structural variables from TLS data can significantly enhance biomass estimation. Chen et al. [23] explored the integration of Sentinel-2 imagery with advanced ML techniques for estimating pasture biomass on dairy farms. Their sequential neural network model incorporated time-series satellite data, field observations, and climate variables. The model achieved a reasonable prediction accuracy, with an R2 of ≈0.6, suggesting that high spatio-temporal resolution satellite data, when combined with ML models, can provide valuable insights for pasture management. On the other hand, other authors have employed alternative techniques, such as multispectral imaging. Zwick et al. [24] focused on remote sensing-based approaches for forage monitoring in rural Colombia. Using multispectral bands from Planetscope acquisitions and various vegetation indices (VIs), they developed models to predict crude protein (CP) and dry matter (DM). The study highlighted the importance of site-specific model optimization, with different regression algorithms performing best at different sites. The integration of multiple regression techniques and feature selection methods demonstrated the adaptability required for diverse climatic conditions. Interestingly, Defalque et al. [25] introduced a novel approach by incorporating cattle parameters and environmental and spectral data to estimate biomass and dry matter in grazing systems. Their study utilized Pearson’s correlation analysis and Recursive Feature Elimination (RFE) for variable selection. Non-linear models achieved the best prediction results, particularly Extreme Gradient Boosting (XGB) and Support Vector Regressor (SVR). This research highlighted the significant impact of herd characteristics on pasture quantity estimation, providing a comprehensive approach to pasture monitoring.
The aforementioned literature highlights the potential of ML to significantly enhance the assessment and evaluation of forage crops and grasslands within the agricultural sector. However, there is a recognized need for further development, particularly concerning ground truth measurements. This advancement specifically aims to reduce reliance on traditional field-based methods, such as destructive sampling and visual assessment, which are known to be labor-intensive, time-consuming, and prone to error [22]. This study presents a low-cost and rapid non-destructive estimation of grassland productivity parameters using ML-based analysis of RGB imagery through artificially generated sward images, specifically focusing on the forage crops ryegrass and Timothy (Lolium perenne L. and Phleum pratense L., respectively). The proposed methodology enabled the accurate segmentation and quantification of live plant material, dead plant material, and bare soil cover coverage. Furthermore, this approach facilitated the determination of LAI, a critical biophysical indicator of vegetation health and photosynthetic capacity. By leveraging ML algorithms, this study addresses the challenges of traditional field sampling, making the process more efficient and accurate. Hence, the utilization of RGB imagery allowed for rapid and cost-effective data acquisition, enabling frequent monitoring of grassland dynamics.
2. Materials and Methods
2.1. Methodology Overview
In this study, a hybrid approach combining Gaussian Mixture Model (GMM) and Nearest Centroid Classifier (NCC) algorithms was employed for image segmentation and classification (Figure 1). The process begins with acquiring images capturing vegetative structures, followed by image preprocessing to enhance the quality of the input data. Preprocessing ensures that the images are cleaned and prepared for analysis, removing any distortions or noise that might affect the accuracy of the results. Next, the features from the images are extracted using various segmentation parameters. Three different approaches are employed for feature extraction: in method A, the pixel values are derived from RGB and Y’CbCr color spaces alongside CMYK components. Method B introduces the Modified Excess Green Index (MExG) to enhance the identification of green vegetation. In contrast, method C further simplifies the feature extraction by focusing on RGB and Y’CbCr, paired with MExG. Following feature extraction, Principal Component Analysis (PCA) is applied for dimensionality reduction, streamlining the dataset by reducing the number of variables, making it easier to process without compromising essential information. Once the features are extracted, the workflow moves into the segmentation phase, where GMM and NCC are applied to cluster and classify the pixels into categories, such as bare soil, living grass, and withered grass. The GMM algorithm helps identify pixel groupings based on the extracted features, while NCC fine-tunes the classification by comparing the pixel values against pre-defined centroids. Although GMM is typically used for unsupervised tasks, its incorporation into this supervised pipeline demonstrates its versatility in hybrid frameworks. Similar methods have shown that GMM can function well as a preliminary segmentation tool, increasing overall accuracy by using pixel groupings as intermediate inputs for supervised classifiers [26,27]. For example, GMM has demonstrated superiority in detecting homogenous pixel regions in Object-Based Image Analysis (OBIA) before the application of supervised techniques, like Random Forest classifiers or SVM [27]. The segmentation results are used to generate artificial images that simulate the sward structures, which are further utilized for training the machine learning models. In the final stage, the models are evaluated through regression analyses to assess their ability to predict key vegetation parameters. Specifically, the models aim to estimate the LAI, which provides insights into vegetation density and health, as well as the coverage of bare soil, living grass, and withered grass. The results from these analyses are used to validate the models’ performance for accurately segmenting and quantifying vegetative structures in ryegrass and timothy crops, ensuring that the machine learning algorithms provide reliable estimates for practical agricultural applications. In the following chapters, each process step, including model training, evaluation, and segmentation accuracy, will be explained in detail.
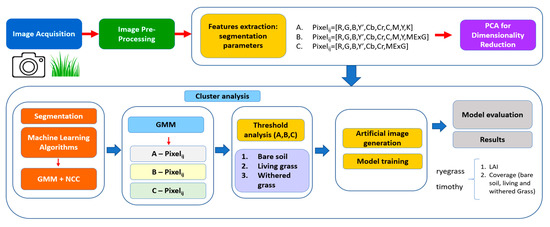
Figure 1.
Workflow for an automated image analysis pipeline for segmenting and evaluating grassland images. The pipeline leverages image processing, machine learning, and statistical analysis to accurately classify and quantify different grass structures.
2.2. Study Site and Experimental Design
The study was carried out at the NIBIO Særheim research station (Klepp Stasjon, Norway, 58°46′22″ N; 5°40′38″ E) at three fields dedicated to permanent ley grass production [13]. Four field trials were implemented based on the most commonly used grass species in the region: ryegrass (Lolium perenne L.) and Timothy (Phleum pratense L.). After the first cut, one field (25.25 m × 12.50 m) with ryegrass and one of the same dimensions with Timothy were evaluated during late summer 2017. Moreover, after the first cut, one ryegrass field (16.8 m × 10.5 m) and another of the same dimensions for Timothy were evaluated during early spring 2018. Each experiment had 4 replication blocks and 10 levels of initial grass plant cover were tested, ranging between 0 and 100%, achieved with a low-dose application of glyphosate [11]. The total amount of precipitation for the late-summer 2017 lapse was 402 mm, and for the early-spring 2018 lapse was 138 mm. Further site characteristics, such as soil type, climatic conditions during the experiment, management practices, and experimental design, are described in [13,14]. Data collection took place in mid-July through mid-September 2017 (late summer), during pasture regrowth after the first cut, and in mid-April through May 2018 (spring), during the beginning of the growing season until the end of vegetative development stage, before the first cut. About 400 images were acquired per experiment, totaling 1605; the images were mainly taken aiming at cloudy and low-sun days to avoid shadows, which could affect the segmentation processes (see Table 1). To establish ground truth measurements of each plot, a 1 m × 1 m field quadrat, representing the region of interest (ROI), was randomly selected across the planned initial coverage plot. Field measurements were conducted immediately after image acquisition to obtain ground-truth data, i.e., visual plant, dead material and bare soil coverages and destructive samples for LAI determination. LAI, which characterizes plant canopies, was determined using a systematic approach with image analysis software, following the approach by Nielsen et al. [28] relating LAI and crop canopy. It is defined as the total leaf area relative to the ground area. The process began by selecting a 1 m × 1 m quadrant and placing it on a representative area of the grassland. The grass within the quadrant was then cut at ground level, ensuring all leaf material within the sampled area, i.e., the ROI, was collected. After harvesting, the leaves were separated from other plant parts, such as stems and flowers, to focus solely on leaf material for the LAI calculation. Grass samples were collected exclusively from within the designated quadrat to maintain consistency in the analysis, even if some plant parts extended beyond its boundaries. This approach ensured standardized sample sizes and prevented overestimation. The collected samples were then dried in a 65 °C oven until they reached a constant weight, which removes moisture and allows for precise measurement of the dry matter content. The leaves were then spread out on a white sheet of millimeter graph paper, carefully arranged to avoid overlap and ensure even distribution, and marked with a bold black line of 10 cm as reference. This prepared sheet with leaves was placed on a flatbed scanner and scanned at high resolution to capture detailed images of the leaves. These images were saved in a digital PNG format. The scanned images were analyzed using FIJI image analysis software (version 2.9.0) to automatically calculate the area covered by the leaves by processing the number of pixels corresponding to the leaf material in the image. This calculation provided a precise measurement of the total leaf area in cm2. Finally, the LAI was determined by comparing the total leaf area, as measured by the software, to the 1 m2 sampled area, and the values were extrapolated to the area in m2. Ryegrass and Timothy sward coverages were also assessed by visual estimation. Images were acquired at the three developmental stages of Timothy and ryegrass: leaf development (initial leaf formation), tillering (full leaf formation and tillering), and beginning of flowering (end of vegetative growth).

Table 1.
Field-based image acquisition specifications for data collection setup and conditions for grass species ryegrass and timothy imaging.
2.3. Sensors and Computing Environment
Image acquisition was performed using two consumer-grade cameras. The cameras were mounted on agricultural machinery, enabling a standardized and quantifiable approach to data collection. This setup ensured consistent data acquisition at a height of 1.75 m, which is particularly advantageous for research applications and large-scale data analysis requiring precision and reproducibility. Images were captured with a SONY DSC-HX60V. Additionally, Digital Hemispherical Photography (DHP) images were obtained using a low-cost camera (APEMAN A60) positioned at a similar height. The SONY camera captured high-resolution still images, featuring a 20.4 MP CMOS sensor and a 24–720 mm equivalent focal range. The APEMAN A60, equipped with a wide-angle (170°) fish-eye lens and producing 12 MP still images, provided complementary hemispherical perspectives. All images had an approximate final resolution of 2736 × 2736 pixels (further details see Table 1). The experiments were conducted in a standard, cost-effective computing environment using a Dell Tower with an Intel Core i7-7700k processor and 16 GB of RAM. The system operated on Linux 18.10, a free and open-source operating system, ensuring accessibility and flexibility. All data processing, analysis, and model implementation were carried out using Python 2.7, a reliable and widely used programming language, and Visual Studio Code (VS Code), a robust and versatile programming platform. This configuration was intentionally selected to demonstrate the practicality and accessibility of the proposed method, showcasing its capability to operate on moderately powered, commercially available hardware without reliance on high-performance computing clusters or specialized equipment such as GPUs.
2.4. Image Pre-Procesing
In preparation for the analysis of the images acquired by the consumer-grade cameras SONY DSC-HX60V and APEMAN A60 cameras, the latter equipped with a fish-eye lens, a meticulous preprocessing stage is implemented. This crucial phase serves to rectify distortions inherent to the fish-eye lens and enhance overall image quality. The preprocessing workflow encompasses several key steps. Initially, camera calibration is executed to ascertain the intrinsic parameters of each camera, including focal length and lens distortion coefficients. This calibration is facilitated through the utilization of a calibration grid or checkerboard pattern. The resultant parameters are subsequently employed to rectify images and ameliorate lens distortions. The ROI encapsulates the analysis target and is delineated following calibration. This delineation can be performed manually or automated through image segmentation techniques. To streamline further processing, the ROI is partitioned into four quadrants. Corner detection algorithms are applied within each quadrant to pinpoint distinct and stable feature points. These identified corners function as reference points for subsequent transformations and measurements. The final stage involves orthorectification, a process that geometrically corrects the ROI and mitigates distortions arising from camera tilt and perspective, particularly crucial for images captured with the fish-eye lens. An overview of this preprocessing workflow is visually represented in Figure 2.

Figure 2.
Structured Workflow for Image Preprocessing, covering all essential steps from defining the ROI to ensuring accurate orthorectification.
The image preprocessing workflow begins with camera calibration as the initial step. This step is crucial to ensure accurate measurements and interpretations from the captured images. The RGB cameras in this workflow provide high-resolution images with an average size of 2736 × 2736 pixels, offering substantial detail and information. One of the key distinctions between the two cameras employed is the presence of “fish-eye” distortion in the second camera. This distortion is inherent to wide-angle lenses, such as those commonly used in action cameras. The “fish-eye” distortion causes straight lines to appear curved and objects closer to the edges of the image to be stretched, resulting in a characteristic “bulging” effect. This distortion can significantly impact the accuracy of measurements and analysis if not properly accounted for. Figure 3 effectively illustrates the difference between the images captured by the two cameras.
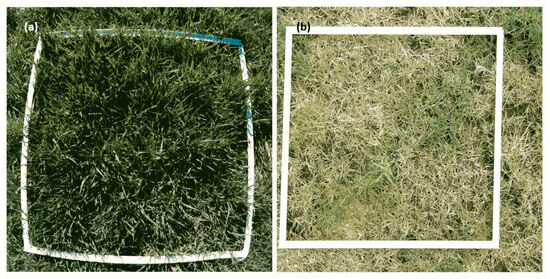
Figure 3.
Comparison of sward field images captured with Apeman A60 (a) and Sony DSC-HX60V (b) cameras.
In Figure 3, the left image, captured by the camera without barrel distortion, also known as the “fish-eye” lens effect, exhibits straight lines and consistent object proportions throughout the image. In contrast, the right image, captured by the camera with barrel distortion, displays curved lines and distorted object proportions, particularly near the edges of the image. The “fish-eye” distortion, evident as a bulging effect that elongates objects near the edges, is a critical factor in image preprocessing. Correcting this distortion involves calibrating and quantifying distortion parameters, which are then applied to transform images for accurate analysis. An adjustment procedure, based on a proposed calibration method by [29], was developed to address this issue effectively. The algorithm for detecting intersection points and correcting distortion in the calibration process was implemented according to [29] to ensure precision and robustness. Multiple images of a 14 × 25 cm checkerboard calibration pattern were captured from various viewpoints to account for diverse perspectives. The algorithm began by detecting intersection points, then images were converted to grayscale to reduce complexity while preserving geometric features, and the Shi-Tomasi corner detection method [30] was applied to identify prominent corners based on eigenvalues of the gradient covariance matrix. In cases where parts of the pattern were obscured by grass or other elements, a convex hull algorithm reconstructed missing intersection points by connecting the outermost detected features. To further refine the localization of corners, the Canny edge detection method [31] was used, enhancing robustness in challenging conditions. Once the intersection points were detected, a homography matrix was computed to align these points with the undistorted reference checkerboard pattern, assuming zero distortion. To address the assumption of zero distortion during the initial calculation of intrinsic and extrinsic parameters, preliminary tests were conducted to evaluate the validity of this approach. These tests were performed using a standard checkerboard-type calibration pattern, captured from multiple views to account for varying perspectives and potential distortion effects. This alignment corrected geometric inconsistencies in the images. Subsequently, intrinsic and extrinsic parameters of the camera were calculated to achieve accurate system calibration. The RANSAC algorithm was used to refine point matching, eliminating outliers and ensuring robust accuracy against noise or partial occlusions. The distortion correction process, while effective, resulted in slight pixel displacements near the image edges. However, this secondary distortion was negligible for the analysis, since the ROI was confined to a 1 m2 area within the visible white frame. The ROI was defined through a series of image segmentation steps. Initially, the images were transformed into the HSV color scale, followed by binarization. Erosion and dilation operations were performed as part of opening and closing morphological operations, using a kernel matrix of 3 × 5 and 5 × 5 pixels, respectively. The Shi-Tomasi method was then applied to delineate the ROI polygon accurately, and when vegetation obscured parts of the frame, the convex hull algorithm ensured completeness. Finally, the ROI underwent ortho-rectification using a vertical plane, where a refined homography matrix was calculated with the support of the RANSAC algorithm [32]. The resulting area was cropped, and any residual pixels from the white frame were removed.
2.5. Sward Image Feature Extraction Models
The methodology employed for plant structure segmentation integrates advanced image processing, feature extraction, and the proposed ML techniques. Pixel-wise segmentation using color information was implemented, leveraging diverse color representations, including the Y’CbCr color space, CMYK color model, and the MExG, to enhance spectral information and feature discrimination. Three models were developed for feature extraction: Model A combined RGB, Y’CbCr, and CMYK values; Model B added the MExG index to RGB, Y’CbCr, and CMYK values; and Model C combined Y’CbCr values with the MExG index. Vector data were scaled using a Min–Max Scaler to fit within a 0–255 range, eliminating the need for explicit normalization while retaining key features. A hybrid machine learning approach was employed, utilizing the GMM for unsupervised probability distribution modeling and the NCC for supervised pixel classification. Dimensionality reduction using PCA isolated two principal components: PC1 captured variance related to vegetation health (e.g., chlorophyll content, canopy cover), while PC2 captured variance related to soil conditions (e.g., moisture, structure). PCA addressed multicollinearity among variables like C, M, Yk, R, G, and Y’, optimizing the feature set and ensuring effective data representation. Correlation and statistical analyses further refined the methodology. Correlation analysis detected redundancies, while ANOVA identified influential variables, the vegetation index being the most impactful, followed by Y’, C, M, R, and G channels. The multi-step image processing pipeline, which included transformations of RGB data into various color spaces and calculated vegetation indices, enabled comprehensive feature extraction and effective classification. This approach facilitated accurate segmentation and representation of living plants, dead vegetation, and bare soil, despite the inherent complexities and variability in the data.
2.6. GMM–NCC Machine Learning Hybrid Approach
This study explored a hybrid approach combining GMM and NCC to effectively classify plant structures. The GMM was utilized, also considering a smoother version of the Kmeans algorithm. The use of GMM was implemented in the three possibilities for data generation, i.e., feature extraction models A, B, and C, for the initial classification of plant structures. In search of better differentiation, an attempt was made to implement a supervised classification method that uses the information provided by the GMM segmentation. Due to this, a variant of the KNN algorithm, the nearest centroid classifier (NCC), was adapted [33]. This algorithm assigns the class based on the centroid closest to it, said centroid, depending on the training samples. The procedure followed for adapting the GMM–NCC algorithm is explained in the following sequence: GMM was applied to input images, grouping pixels into live plant structures, dead material, and soil. To improve the accuracy of these clusters, class boundaries were refined through pixel-level corrections using an image manipulation program, resulting in precise reference masks. These refined masks served as the basis for a synthetic data augmentation process, through which synthetic images were generated to expand the dataset. The augmented dataset preserved the original class assignments while simulating a broader range of conditions. The mean values of each cluster from these synthetic images were then calculated and used as centroids to train the NCC. The application of this procedure sought to improve classification, especially in the boundaries between classes, where possible misclassifications occur.
The initial segmentation was applied by applying the GMM algorithm based on the artificial images, which were then validated on both forage species. The mean values of each cluster were obtained (live plant structures, dead ones, and soil). With the mean value of each cluster, the NCC algorithm was applied, in which said mean value was the centroid used for discrimination. The application of this procedure sought to improve classification, especially in the boundaries between classes, where possible misclassifications occur. The initial segmentation was performed using the GMM algorithm, which was applied to artificial images (i.e., computer-generated images) obtained through a synthetic data augmentation process. This process expanded the variability of the input data by simulating realistic scenarios based on previously segmented structures. From these clusters, the mean values of each were applied by applying the GMM algorithm based on the artificial images, i.e., computer-generated images, and then validated on both forage species. The mean values of each cluster were obtained (live plant structures, dead ones, and soil). With the mean value of each cluster, the NCC algorithm was applied, in which said mean value was the centroid used for discrimination. The application of this procedure sought to improve classification, especially in the boundaries between classes, where possible misclassifications occur.
2.7. Artificial Sward Images Generation
An automated color image processing tool capable of segmenting intricate grass structures in ley swards, including green leaves, straw, and bare soil, which are considered in the calculation of the LAI using artificial intelligence techniques, was developed. Utilizing data from color scale transformations applied to all RGB channels, including transformations to the color space achromatic luminance and blue and red chromatic channels (Y’CbCr) and the color model cyan, magenta, yellow, and key (CMYK), the GMM in conjunction with NCC achieved excellent classification outcomes. Implementing the GMM algorithm created a dataset of 30 artificial images (Figure 4). For this purpose, an algorithm was created to convert the images of the GMM grouping into binary masks and later generate a label for each pixel value. These labeled images will serve as input for training the supervised NCC algorithm, which will ultimately be used to estimate parameters such as vegetation coverage and support the calculation of the LAI in subsequent analyses.

Figure 4.
This figure displays a sample from the dataset of 30 artificial sward images, which includes two forage crop species, ryegrass (Lolium perenne L.) and Timothy (Phleum pratense L.).
Therefore, image processing and segmentation were conducted utilizing unsupervised and supervised machine learning algorithms, including a combination of both. Unsupervised algorithms were implemented initially due to the absence of a previously classified image dataset. As aforementioned, the GMM based on the expectation–maximization algorithm with random samples was employed [34]. Original images exceeding 2000 pixels in dimension were rescaled to 912 × 912 to concentrate analysis on the ROI. The resulting groups from GMM were arranged into a set of 30 artificial images, created pixel by pixel through the computational combination of 912 × 912 pixels for each artificial image. This artificial dataset served as training data and binary masks for labeling living plants, dead plants, and bare soil classes. Each computer-generated image consists of labeled segments resulting from GMM-based segmentation. While the GMM provided an initial segmentation, minor inaccuracies required refinement, particularly in boundary regions. For this purpose, labeling was performed using GIMP version 2.10.10 (developed by the GIMP Development Team) to apply targeted pixel-level corrections under the visual supervision of the known classes. The initial segmentation, generated by GMM, served as an objective reference, and corrections were limited to specific boundary regions where ambiguities were identified. To minimize operator error, we adhered to a standardized protocol that included systematic visual refinement, cross-validation using a Python algorithm to assign class values (0 to bare soil, 1 to living structures, and 2 to dead material), and multiple reviews of the corrected regions to ensure accuracy and consistency. It is important to note that GIMP was used exclusively for these focused adjustments and not for full-scale manual labeling (Figure 5).
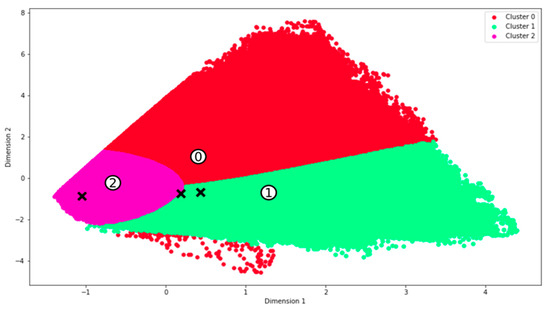
Figure 5.
Clustering of three classes using the unsupervised GMM classifier. The data was first reduced using PCA, and numbers indicate the centroids of each cluster. Black “x” marks represent the transformed sample data points. The clusters are categorized as follows: Cluster 0 represents bare soil, Cluster 1 corresponds to living structures, and Cluster 2 indicates dead material.
The implementation of GMM as an unsupervised classifier allowed the artificial creation of images with labels for the three classes: living plant, dead plant, and bare soil. This class separation was acceptable and especially accurate for the living plant class (Figure 6), keeping an undistorted plant shape when segmenting the classes.
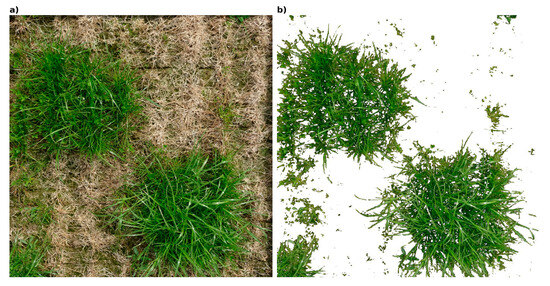
Figure 6.
GMM implementation after supervised labeling on a sample timothy image. (a) Original image; (b) Segmentation of the living plants class.
2.8. NCC Model Training
Once the artificially generated images were obtained, they were used as “labeled segments” to test various supervised classification models, aiming to improve decision boundary definition, particularly in edge cases where the GMM clustering was less effective. Among the models tested, the NCC outperformed the others. A detailed description of this process can be found in Rueda-Ayala et al. [14]. To ascertain the independence of the algorithm’s class allocation efficacy from both the image to which it is applied and the quantity of each plant structure present within the image, experiments were conducted utilizing images of the same experiment captured at three distinct coverage stages of the same crop (leaf development, tillering, beginning of flowering). Following the evaluation of the segmentation’s efficacy at the various coverage stages, the GMM–NNC algorithm was employed. Figure 7 illustrates the segmentation outcome acquired after implementing the GMM–NNC algorithm as an example of the analyzed images.
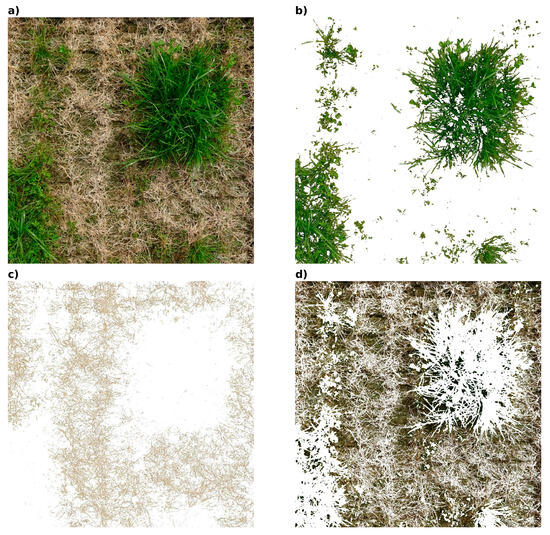
Figure 7.
The figure consists of four images: (a) the original image, (b) the segmentation of living structures, (c) the segmentation of dead structures, and (d) the segmentation of bare soil. These images demonstrate the effectiveness of the GMM–NCC algorithm in distinguishing between different components within the scene.
Models A, B, and C were utilized as supervised machine learning models and subsequently analyzed in accordance with the threshold of the MExG. Model A entailed the conversion of RGB color values into Y’CbCr and CMYK color spaces to facilitate the capture of a more comprehensive range of color information. Model B combined RGB, Y’CbCr, and CMYK color values with the MExG to enhance green areas and enable more precise differentiation of plant material. Model C directly converted RGB values to the Y’CbCr color space and subsequently calculated the MExG index based on these Y’CbCr values, thereby combining luminance and chrominance data with vegetation-focused green intensity. Employing the GMM, the mean values of each cluster (live plant structures, dead plant structures, and soil) were obtained. Subsequently, utilizing the mean value of each cluster, the NCC algorithm was applied, wherein this mean value served as the centroid for discrimination purposes.
Conversely, the analysis of RGB images, encompassing the color spaces Y’CbCr and the CMYK model, necessitates substantial computational power if the analysis is conducted on a pixel-by-pixel basis. This requirement is attributable to the number of features calculated per pixel in each image, i.e., features resulting from the combination of R, G, B, Y’, Cb, Cr, C, M, Y, and K. For instance, if nine features were selected per pixel, a 912 × 912 image would yield approximately 7,500,000 features to be analyzed. Reducing the dimensionality to only two features per pixel in a 912 × 912 image resulted in 1,500,000 features per image that underwent analysis and segmentation, thereby significantly reducing the requisite computational power. When classification is executed at the pixel level, for the combination GMM–NCC, the classification was initially conducted in accordance with the spatial location of pixels, whereby neighboring pixels with information about one class were grouped. Subsequently, the classification was executed based on the probability that a group of pixels belongs to the same class.
2.9. Model Comparison
To assess the effectiveness of the implemented algorithms, a comprehensive comparison was conducted between the model estimated percentage results for live structures, dead structures, soil, and LAI against the visual coverage and LAI destructively calculated. This comparison was performed for all models tested and across the three grass growth stages of both grass species in the following manner. First, linear regression analyses (with an α = 0.05) comparing the model coverage estimations of live plant, dead material, and bare soil versus their corresponding visual coverage assessments, each at leaf development, tillering, and beginning of flowering. Similarly, destructive LAI measurements (per m2) were compared with their model estimations at the same grass development stages. The resulting R2 coefficients indicated the model estimation accuracy. Then, to further compare between models, a linear mixed-effects model, fitted by the restricted maximum likelihood approach (REML) was applied using the statistical software R, version 4.4.1 [35] and the package ‘nlme’ [36]. The models (A, B, and C) and the grass development stages (leaf development, tillering, and beginning of flowering) were assigned as fixed factors. The 10 initially planned grass coverage levels nested in the replication blocks were assigned as random. Marginal means were adjusted with the Tukey HSD (α = 0.05) method and their corresponding 95% confidence limits were calculated with the ‘emmeans’ package [37]. Marginal means compared allowed grouping models by their statistical difference.
3. Results and Discussion
3.1. Feature Extraction and Initial Model Training
This section presents the outcomes and insights derived from the application of the proposed ML techniques for plant structure segmentation, encompassing living and dead vegetative structures as well as bare soil. The analysis builds upon data vectors representing features extracted from processed images, which were utilized by the ML algorithms to LAI and sward coverage. The performance of these algorithms was benchmarked against ground truth data to evaluate their accuracy and reliability. Given the significant variability in the shape, dimensions, and structural complexity of living plants, traditional segmentation methods based solely on shape or structure recognition were found to be inadequate. The results highlight the effectiveness of the employed methodologies in overcoming these challenges, providing a robust framework for segmenting complex vegetative structures. The presence of overlapping structures further complicated the segmentation process, making it challenging to delineate individual structures accurately. A pixel-wise segmentation approach utilizing color information was employed to overcome these limitations. Therefore, in the image analysis stage aimed at feature extraction, a multi-faceted approach was adopted to extract meaningful features and segment plant coverage accurately. A diverse set of color representations, including the Y’CbCr color space, the CMYK color model, and the MExG, were combined to capture a wide range of spectral information relevant to identification, i.e., living and dead vegetative structures and bare soil. This approach leveraged the strengths of different color spaces and vegetation indices to enhance feature discrimination. These proposed models for image processing and segmentation pipeline are involved in the next stage of a hybrid approach utilizing both unsupervised and supervised machine learning algorithms. The GMM, initialized with random samples, was employed to model the underlying probability distribution of the image data. The GMM algorithm better solves the classification stage, being highly used for the unsupervised classification of multi-class information. Moreover, the GMM algorithm presents better results for differentiating the background from the objects of interest [38]. Due to these advantages, the use of GMM was determined, in the three possibilities of data generation, i.e., feature extraction models A, B, and C, for the initial classification of plant structures. The NCC algorithm was then used to assign pixels to different classes based on their feature vectors and proximity to pre-defined cluster centers or labeled training data. Thus, prior to image segmentation, three different methods were tested for feature extraction to optimize color information and enhance image analysis. The first method involved transforming the RGB color values of each pixel into two additional color spaces: Y’CbCr and CMYK. The YCbCr and YCMYK color scales were selected owing to their inherent advantages in discerning an object from the backdrop of an image. These advantages are attributable to the distinct separation of luminance and chrominance values within these scales [39]. The Y’CbCr color space separates image luminance (Y’) from chrominance components, with Cb representing blue and Cr representing red, making it perceptually uniform. This meant that equal changes in these channels correspond to equal changes in perceived color. The CMYK model, used primarily in printing, is based on subtractive color mixing and represents cyan, magenta, yellow, and black (key). By converting RGB data into these two additional color spaces, this method captures a broader range of color information, critical for detailed analysis, as described by Equation (1), representing model A henceforth. In contrast, the RGB, Y’CbCr, and CMYK color values were further enhanced by calculating the MExG in the second model. The MExG is an effective vegetation index, particularly useful in highlighting green areas by amplifying the green channel while minimizing the influence of red and blue channels. This technique allowed for more precise differentiation of plant material within the image by combining the detailed color representation of the three color models with the focused green intensity provided by the MExG index. Hence, this model (Equation (3), i.e., model B) is a combination that provides a richer set of data for image analysis, with the calculations following equations 1 and 2, as defined by Burgos-Artizzu et al. [40]. The third model (model C) involved a direct conversion of RGB values to the Y’CbCr color space, followed by calculating the MExG index based on these Y’CbCr values. This technique effectively combined the luminance and chrominance data from Y’CbCr with the vegetation-focused green intensity provided by the MExG index. This method enabled the extraction of color and luminance information, offering a comprehensive image content analysis outlined in Equation (4). Notably, all vector data generated through these methods were scaled using a Min–Max Scaler to fit within a standard range of 0–255, eliminating the need for explicit normalization [41]. This approach preserves the inherent characteristics of certain variables, such as vegetation indices, which could be distorted by standard normalization techniques [42]. This is advantageous as it allows the data to be directly used in subsequent analyses without requiring additional preprocessing steps while ensuring that key features are retained.
Model A: Pixelij = [R, G, B, Y′, Cb, Cr, C, M, Y, K]
MExG = 1.262G − 0.884R − 0.311B
Model B: Pixelij = [R, G, B, Y′, Cb, Cr, C, M, Y, MExG]
Model C: Pixelij = [R, G, B, Y′, Cb, Cr, MExG]
Dimensionality reduction was performed using PCA to optimize the feature set and address multicollinearity issues. PCA, a statistical technique for identifying patterns in high-dimensional data, was applied to isolate the most influential vectors from the MExG feature set. This ensured that critical information was retained while reducing redundancy. PCA revealed two principal components that are pivotal in categorizing the three classes of interest: living plants, dead plants, and bare soil. The first principal component (PC1) primarily captured variance related to vegetation health and vitality, emphasizing features such as chlorophyll content, leaf area index, and canopy cover. High values indicated healthy vegetation, while low values corresponded to dead or sparse vegetation. The second principal component (PC2) captured variance associated with soil conditions and moisture levels, distinguishing bare soil from living and dead plants. Features such as soil structure, moisture content, and surface roughness were highlighted, with high values indicating dry, compacted soil and low values representing moist, porous soil. Correlation analysis among variables in the dataset identified high negative correlations among the C, M, Yk, R, G, and Y’ channels in the color scales, indicating multicollinearity. To refine the analysis, a subset of images was manually labeled, and a correlation analysis was conducted between independent variables and the target classes (living plants, dead plants, and soil). ANOVA results demonstrated that the vegetation index was the most influential variable, followed by the Y’, C, M, R, and G channels. PCA reduced the dimensionality of the feature set, minimizing redundancy and improving interpretability. This reduction was essential due to the increased data volume per image, allowing effective classification and enhanced data representation for subsequent analyses.
3.2. Evaluation of Supervised and Unsupervised Learning Outcomes
The hybrid approach used in this study, integrating an unsupervised GMM with a supervised NCC, effectively segmented and classified the grass components of live plant material, dead plant material, and soil. The GMM was initially employed for clustering and segmenting pixels based on the extracted features, i.e., models A, B, and C, which provided an unsupervised classification of RGB imagery. This stage identified the preliminary groupings of different elements in the grassland ecosystem. Artificial sward images were generated from those groups to enhance the supervised learning stage and avoid manual labeling of grass structures. These artificial images, created by pixel-wise segmentation, simulated real-world sward structures and were used to train the NCC. The supervised stage of the NCC algorithm refined the segmentation by assigning pixels to specific classes—live grass, dead grass, or soil—based on their proximity to pre-defined cluster centroids derived from the GMM output. The artificial images were generated by manually correcting, on a pixel-by-pixel basis, the groups identified by the GMM. These corrected images were then employed as ‘labelled segments’ to subsequently test various supervised classification models as complementary approaches to the GMM. A detailed description of this process can be found in Rueda-Ayala et al. [14]. Among the evaluated supervised algorithms, the NCC yielded favorable results, demonstrating speed and precision. Furthermore, we chose the NCC approach to highlight the practical and accessible nature of the proposed methodology. Specifically, the classification tasks were conducted on a moderately powered, commercially available computer, without the need for high-performance computing clusters or specialized GPU equipment. Table 2, Table 3, Table 4 and Table 5 show the models’ estimation accuracy, indicated by the R2 values of the model estimations versus the ground truth measurements at different Timothy and ryegrass growth stages.

Table 2.
Stage-wise Comparison of LAI derived from feature models and LAI measured as ground truth (R2). Cursive letters indicate the grouping of marginal means of estimations. If two or more share the same letters, they are rather comparable and not statistically different (α = 0.05).

Table 3.
Stage-wise Comparison of Living Grass Structure Coverage: Features Models vs. Visual Estimation as ground truth (R2). Cursive letters indicate the grouping of marginal means of estimations. Upper-case letters show model comparisons; lower-case show comparisons among plant growth stages. If two or more of share the same letters, they are rather comparable and not statistically different (α = 0.05).

Table 4.
Stage-wise Comparison of dead Grass Structure Coverage: Features Models vs. Visual Estimation as ground truth (R2). Cursive letters indicate the grouping of marginal means of estimations. Upper-case letters show model comparisons; lower-case show comparisons among plant growth stages. If two or more share the same letters, they are rather comparable and not statistically different (α = 0.05).

Table 5.
Stage-wise Comparison of bare soil Coverage: Features Models vs. Visual Estimation as ground truth (R2). Cursive letters indicate the grouping of marginal means of estimations. Upper-case letters show model comparisons; lower-case show comparisons among plant growth stages. If two or more share the same letters, they are rather comparable and not statistically different (α = 0.05).
The comparison of Models A, B, and C in predicting various vegetation metrics for timothy and ryegrass revealed key insights into the performance of each model across different growth stages. The analysis was based on four key parameters: LAI, living grass structure coverage, dead grass structure coverage, and bare soil coverage. In each case, the models were compared against ground truth data measured in the field. Regarding growth stages, i.e., grass coverage, the proposed data collection method is designed to provide standardized and objective measurements, addressing the limitations of human perception as identified in the literature. Andújar et al. [43] demonstrated that, while visual estimations of weed cover are generally accurate for broad assessments, they can be inconsistent and subject to observer bias. Their study found a good correlation between visual estimates of weed cover and objective parameters, such as actual weed cover and biomass. Additionally, their analyses of reliability and repeatability revealed no significant differences in visual estimations made by different observers, or the same observer, at different times, regardless of the scale used. Thus, in our study, by using a camera-based, standardized methodology (the cameras were mounted in agricultural machinery, ensuring a constant height), this approach minimizes subjective variability and enhances the accuracy and repeatability of data collection, making it well-suited for both scientific research and practical agricultural applications. On the other hand, in contrast to the SONY camera, the choice of the APEMAN A60 fish-eye camera was intentional to demonstrate the flexibility and adaptability of the proposed system, highlighting that it is not constrained by the type or cost of the camera used. The system is designed to function effectively with any camera, ranging from high-end professional models to affordable, widely available consumer-grade devices. The inclusion of a fish-eye camera in this study serves as a clear example of its adaptability to diverse optical configurations, showcasing its robustness across different imaging setups. While fish-eye cameras require additional post-processing to correct for distortion, this is a minor trade-off compared to the significant advantage of enabling a wide range of users to implement the system using the equipment they already have access to. This approach ensures that the system remains accessible and scalable, meeting the needs of both resource-constrained and well-equipped users alike.
In Table 2, the performance of the models in predicting LAI for Timothy and ryegrass was examined. For Timothy, all three models performed similarly during the initial and middle stages, with R2 values ranging from 0.52 to 0.79, showing that they all captured the LAI effectively during these early growth periods. The REML analysis showed no difference among models but a decrease in accuracy when estimates are done on images at the beginning of flowering (R2 values of 0.52). This indicated that estimating LAI became more challenging as the grass finished the vegetative growth, likely due to the increased complexity, density and overlapping of leaves. In contrast, the performance for ryegrass showed a poor performance in general, except for models A and B at tillering stage, ranking first on their marginal means grouping. Models A, B and C performed poorly with R2 ranging between 0.16 and 0.40. This suggested that Model A struggled to capture LAI for ryegrass accurately in its early and full vegetative development stages. At the beginning of flowering the estimation becomes challenging, since ryegrass does not have a completely vertical development by the end of its vegetative period. At this stage, many leaves and tillers overlap, producing a shadowing effect that is captured as a darker color in the acquired image, which was misclassified as bare soil. All models had an acceptable performance on Timothy at leaf development and tillering, being statistically similar (R2 s > 0.72 and similar grouping, Table 2). The models performed poorly at the beginning of floweringR2 ranging between 0.22 to 0.32). Although Timothy has a vertical growth at the end of the vegetative stage, leaves overlap much, thus resulting in the aforementioned issue. This issue does not influence destructive LAI assessment, because the observer removes the true number of leaves present in the sample. This issue poses a huge limitation to ML algorithms in estimating coverage and LAI during advanced grass growth stages. On the other hand, Table 3 presents the models’ accuracies to estimate living grass coverage against visual estimation. For Timothy, all models performed similarly and consistently well across the growth stages, with R2 values around 0.75 to 0.84. A slight decrease in model performance was observed for the stages of leaf development and tillering (b marginal means grouping), being different than at the beginning of flowering (a grouping). In contrast, the performance for ryegrass coverage estimation was more variable. At leaf development, Model A performed poorly (R2 = 0.33; grouping Bb), while Models B and C delivered significantly better results (R2 = 0.71 and 0.74, respectively; grouping Aa). At tillering stage, models A and B performed exceptionally well (R2 > 0.96), being statistically different than Model C (R2 = 0.18; grouping Bb). All models declined in their performance at the beginning of flowering (R2 ranging from 0.40 to 0.52; grouping Bb).
In Table 4, the accuracy of the models in predicting dead grass structure coverage was summarized. A similar pattern was observed for Timothy’s live plant coverage estimation. All models performed similarly at all grass growth stages, but particularly at tillering (R2 between 0.79 and 0.83). For ryegrass at leaf development, Models B and C outperformed Model A. At tillering, the accuracy of Model C was significantly poor (R2 = 0.03), while Models A and B achieved the best results (R2 = 0.97). However, all models declined in accuracy at the beginning of flowering, but there were no differences among them. Estimates of bare soil coverage (Table 5) were of acceptable accuracy at leaf development and beginning of flowering for timothy (group a) and good at tillering for ryegrass (grouping Aa). In the remaining stages, all models showed a similarly poor performance. This can be explained by the fact that thresholding methodologies are characterized by their relative simplicity but may not exhibit adequate adaptability and robustness in dynamic field environments and multitemporal instances, especially for images acquired under diverse illumination conditions [44].
3.3. Practical Implications and Future Directions
Canopy cover estimation has previously utilized ML-based classification methods. Unsupervised methods, such as k-means clustering, and supervised methods, like Decision Tree (DT), SVM, and Random Forest (RF), have been employed. These methods have often outperformed the thresholding method. However, classification methods necessitate a degree of human intervention, hindering automation. Moreover, sample selection can be time-consuming, and model training and application may be computationally intensive [45]. In contrast to ML approaches that rely on pixel-level features, certain scenarios exist where color alone is insufficient for classification. For instance, soil may appear green when partially covered by vegetation or brown due to plant aging, which involves visual changes in vegetation transitioning from green to yellow, beige, or brown. This overlap in color ranges further complicates the distinction between soil, crop residues, and aging vegetation, presenting significant challenges for color-based classification approaches. To overcome these challenges, incorporating textural and contextual information becomes necessary for improved segmentation of vegetation and background in RGB images. One approach to enhance the results of our study could involve methodologies based on color–texture–shape characteristics. These methods incorporate contextual and spatial information alongside pixel values extracted from images. Initially, researchers employed handcrafted features, such as Bag of Words, SIFT, GLCM, and Canny Edge Detectors, to address the limitations of pixel-level features. However, the high dimensionality of these features required substantial amounts of data for effective training of algorithms that distinguish between vegetation and background [46]. Although these techniques enhanced segmentation quality, their complexity and high-dimensional nature demanded large, meticulously curated datasets and substantial parameter fine-tuning, making them less adaptable to diverse conditions commonly found in agricultural imagery [47]. Deep learning (DL) has revolutionized feature extraction by enabling automated, end-to-end learning directly from raw input data, eliminating the need for manual feature selection based on domain expertise and leveraging multiple layers of abstraction to identify relevant representations [48,49]. Furthermore, the advent of DL has revolutionized feature extraction, allowing for the automated extraction of essential features from datasets. DL approaches, particularly Convolutional Neural Networks (CNNs), excel at learning multi-scale and task-specific representations directly from raw data. While deep learning models are also heavily dependent on large, high-quality datasets, recent advancements in generative models, such as Stable Diffusion and Generative Adversarial Networks (GANs), offer a viable solution to mitigate data limitations. These methods can produce artificial imagery that mimics real-world patterns, effectively expanding training datasets and enhancing model generalization. By augmenting real data with synthetic samples, deep learning models can achieve robust performance even when real data availability is constrained. This advancement highlights a distinct advantage of deep learning approaches over traditional methods, particularly in domains where data collection is re-source-intensive or limited by environmental factors [50,51]. DL approaches outperform traditional handcrafted features and ML techniques in vegetation segmentation tasks, making them a promising solution [52]. State-of-the-art approaches, such as Extreme Learning Machines (ELM) [53] custom Convolutional Neural Network (cCNN) [54], and semi-supervised Multilayer Perceptron models (MLP) [55], have achieved significant accuracy improvements. Nonetheless, these approaches often rely on extensive labeled datasets, specialized computational infrastructures (e.g., GPUs), and complex feature engineering pipelines. These dependencies present practical limitations in real-world field conditions, where such resources may not be readily available. On the other hand, the calculation of intrinsic and extrinsic camera parameters is a critical aspect of distortion correction, as errors in estimating these parameters significantly influence the effectiveness of the correction applied to images. ML algorithms allow for a better differentiation between live vegetation, dead structures, and soil, compared to using vegetation indices for segmentation. In the generation of data for the segmentation of plant structures, the YCbCr color scale plays a fundamental role in achieving notable separability between live vegetation structures and the image background due to the YCbCr scale’s ability to better represent chrominance. The combined use of vegetation indices with different color scales provides the necessary data for structure segmentation through the application of ML algorithms. While this approach required higher computational resources compared to shape-based methods, it effectively minimized the risk of erroneously assigning a larger area of pixels to a particular class. Thus, by classifying each pixel independently, this method allows for fine-grained analysis. This strategy ensured greater precision and accuracy in segmenting complex living structures, allowing researchers to gain valuable insights into their organization and function. However, due to the implementation of PCA, data dimensionality reduction was essential for decreasing processing time, potentially reducing the time from 90 s to 17 s when processing an image. A ratio of 0.9 for the cumulative explained variance was used during PCA implementation, ensuring that the majority of the original information was preserved while minimizing computational overhead [56]. This method ensures that no important details are lost during dimensionality reduction, allowing efficiency and data quality to be balanced. The GMM algorithm offers better class separability and greater data uniformity within each class. In contrast, supervised algorithms allow for a segmentation of plant structures that closely approximates field truth. Still, they also distort the shapes present in the resulting segmentation as they learn on a pixel scale. The high correlation between visual estimation and the measured LAI suggests a dependent relationship between these variables, with slight differences arising because, in visual estimation, weeds are not discarded, as they are during LAI measurement through field sampling. This demonstrates that weeds in the analyzed images directly impact the estimation of the Leaf Area Index, representing an advantage of using Machine Learning algorithms for plant structure segmentation over the traditional field sampling method. The main differences in correlation values between coverage obtained by algorithms and visual estimation occur during the early stages of coverage, as human perception of structure percentage is subjective. In contrast, values obtained through any of the analyzed ML algorithms provide greater certainty in determining the percentage of plant structures. Similarly, visual estimation and the field measurement method of LAI do not exclude early-stage plant structures, which is why using ML algorithms for coverage estimation provides a result closer to ground truth.
Manually segmenting vegetation structures in grasslands is highly time-consuming, making labeling of each image a challenging and resource-intensive task. The current approach—employing a clustering algorithm (GMM) followed by a fine-tuning step using an NCC—was specifically chosen to enable image segmentation on a moderately powered, commercially available computer without requiring dedicated GPU resources. This methodological choice provides a practical solution that can be broadly implemented without the need for high-end computational infrastructures. While recent deep learning segmentation models have demonstrated remarkable performance in various domains of image analysis, this study focused on establishing a baseline method that is less complex and computationally demanding. Furthermore, the novelty of the proposed algorithm lies in its ability to operate without an initially labeled dataset. The GMM + NCC generates synthetic images that enable accurate plant structure segmentation on conventional computing hardware by applying minimal pixel-level corrections. Compared to traditional supervised methods (e.g., DT, SVM, RF), which yield higher accuracy but demand extensive and time-consuming labeling, the GMM + NCC approach is resource-efficient. Evenly, cutting-edge methods, such as ELM, cCNN, or MLP variations, achieve outstanding accuracy, they also require more specialized computational resources and large volumes of labeled data. In contrast, the proposed approach offers a balanced strategy between performance and practical feasibility.
4. Conclusions
In Norway, Sweden, and Denmark, both ryegrass (Lolium perenne L.) and timothy grass (Phleum pratense L.) are particularly significant as forage crops, owing to their adaptability to the region’s cool temperate climates and where dairy and beef production are prevalent. Similarly, ryegrass holds a notable position in Canada’s forage industry, thriving in Asian countries predominantly due to the cultivation of timothy grass. In the United States, these grasses are essential, with ryegrass and timothy grass being widely cultivated across various regions as primary sources of livestock feed. Additionally, Timothy grass is significant in New Zealand, and ryegrass is notable in Brazil. Thus, gaining insights into key parameters, such as LAI and grass coverage, are essential for effectively managing these forage crops. These parameters provide critical information about the health, productivity, and sustainability of forage systems, enabling farmers and researchers to make informed decisions that optimize crop performance, animal nutrition, and land use. This study successfully developed an automated image processing tool capable of segmenting intricate grass structures in ley swards using a combination of artificial intelligence techniques to assess LAI and grass coverage. By leveraging both unsupervised and supervised machine learning algorithms, specifically GMM and NCC, the tool demonstrated strong performance in distinguishing between living plants, dead material, and bare soil across various growth stages of Timothy and ryegrass. The integration of multiple color spaces (RGB, Y’CbCr, CMYK) and vegetation indices, such as the MExG, further enhanced the model’s ability to segment plant structures accurately. The results highlighted the utility of GMM–NCC for ground coverage and LAI estimation, providing reliable segmentation outcomes at leaf development and tillering stages of crop development. However, the study also identified challenges with model performance during the beginning of the flowering stage due to increased vegetation density and structure complexity, particularly for ryegrass. Model B emerged as the most consistent among the evaluated models, although improvements are necessary for greater accuracy in later stages. Future work should focus on refining the models by incorporating additional spectral bands, advanced vegetation indices like NDVI, and deep learning techniques for more robust feature extraction and segmentation. Enhancing the model’s ability to process multitemporal data and handle varying environmental conditions would further improve segmentation reliability. Overall, this study underscores the potential of machine learning algorithms for automated vegetation analysis, offering a significant advantage over traditional manual methods in speed, precision, and scalability. On the other hand, the study employs a GMM–NCC approach for grassland image segmentation, offering a practical and computationally efficient solution on standard hardware, avoiding the complexity and resource demands of deep learning models. Furthermore, new research lines can focus on integrating and comparing advanced DL approaches into this pipeline. The segmented images obtained through the proposed solution can efficiently generate training datasets to support the development, training, and evaluation of state-of-the-art deep learning models.
Author Contributions
Conceptualization, H.M. and D.A.; methodology, H.M., C.R.-A. and D.A.; software, C.R.-A. and C.R.; validation, H.M. and D.A.; formal analysis, H.M.; investigation, H.M. and C.R.-A.; resources, D.A. and A.R.; data curation, C.R.-A. and V.R.-A.; writing—original draft preparation, H.M. and C.R.-A.; writing—review and editing, H.M., C.R.-A. and D.A.; visualization, H.M.; supervision, H.M. and D.A.; project administration, D.A. and A.R.; funding acquisition, D.A. and A.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the grant PID2023-150108OB-C33 funded by the MCIU/AEI/10.13039/501100011033 and FEDER, EU and by the grant TED2021-130031B-I00 funded by MICIU/AEI/10.13039/501100011033, as appropriate, by the European Union NextGenerationEU/PRTR.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Acknowledgments
Mats Höglind, and the Department of Grassland and Livestock, NIBIO, Norway.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Chaudhry, A. Forage based animal production systems and sustainability, an invited keynote. Rev. Bras. Zootec. -Braz. J. Anim. Sci. 2008, 37, 78–84. [Google Scholar] [CrossRef]
- Ramesh, T.; Bolan, N.S.; Kirkham, M.B.; Wijesekara, H.; Kanchikerimath, M.; Srinivasa Rao, C.; Sandeep, S.; Rinklebe, J.; Ok, Y.S.; Choudhury, B.U.; et al. Chapter One–Soil organic carbon dynamics: Impact of land use changes and management practices: A review. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 156, pp. 1–107. [Google Scholar]
- Zhou, Q.; Daryanto, S.; Xin, Z.; Liu, Z.; Liu, M.; Cui, X.; Wang, L. Soil phosphorus budget in global grasslands and implications for management. J. Arid Environ. 2017, 144, 224–235. [Google Scholar] [CrossRef]
- Putnam, D.H.; Orloff, S.B. Forage Crops. In Encyclopedia of Agriculture and Food Systems; Van Alfen, N.K., Ed.; Academic Press: Oxford, UK, 2014; pp. 381–405. [Google Scholar]
- Rantanen, M.; Karpechko, A.Y.; Lipponen, A.; Nordling, K.; Hyvärinen, O.; Ruosteenoja, K.; Vihma, T.; Laaksonen, A. The Arctic has warmed nearly four times faster than the globe since 1979. Commun. Earth Environ. 2022, 3, 168. [Google Scholar] [CrossRef]
- Hellton, K.H.; Amdahl, H.; Thorarinsdottir, T.; Alsheikh, M.; Aamlid, T.; Jørgensen, M.; Dalmannsdottir, S.; Rognli, O.A. Yield predictions of timothy (Phleum pratense L.) in Norway under future climate scenarios. Agric. Food Sci. 2023, 32, 80–93. [Google Scholar] [CrossRef]
- Skeie, S.B. Quality aspects of goat milk for cheese production in Norway: A review. Small Rumin. Res. 2014, 122, 10–17. [Google Scholar] [CrossRef]
- Eide, M.H. Life cycle assessment (LCA) of industrial milk production. Int. J. Life Cycle Assess. 2002, 7, 115–126. [Google Scholar] [CrossRef]
- Lesschen, J.P.; van den Berg, M.; Westhoek, H.J.; Witzke, H.P.; Oenema, O. Greenhouse gas emission profiles of European livestock sectors. Anim. Feed Sci. Technol. 2011, 166–167, 16–28. [Google Scholar] [CrossRef]
- Brunet, J.; Felton, A.; Lindbladh, M. From wooded pasture to timber production–Changes in a European beech (Fagus sylvatica) forest landscape between 1840 and 2010. Scand. J. For. Res. 2012, 27, 245–254. [Google Scholar] [CrossRef]
- Schellberg, J.; Hill, M.; Gerhards, R.; Rothmund, M.; Braun, M. Precision agriculture on grassland: Applications, perspectives and constraints. Eur. J. Agron. 2008, 29, 59–71. [Google Scholar] [CrossRef]
- Dusseux, P.; Guyet, T.; Pattier, P.; Barbier, V.; Nicolas, H. Monitoring of grassland productivity using Sentinel-2 remote sensing data. Int. J. Appl. Earth Obs. Geoinf. 2022, 111, 102843. [Google Scholar] [CrossRef]
- Rueda-Ayala, V.P.; Höglind, M. Determining Thresholds for Grassland Renovation by Sod-Seeding. Agronomy 2019, 9, 842. [Google Scholar] [CrossRef]
- Rueda-Ayala, V.P.; Peña, J.M.; Höglind, M.; Bengochea-Guevara, J.M.; Andújar, D. Comparing UAV-Based Technologies and RGB-D Reconstruction Methods for Plant Height and Biomass Monitoring on Grass Ley. Sensors 2019, 19, 535. [Google Scholar] [CrossRef] [PubMed]
- Fricke, T.; Richter, F.; Wachendorf, M. Assessment of forage mass from grassland swards by height measurement using an ultrasonic sensor. Comput. Electron. Agric. 2011, 79, 142–152. [Google Scholar] [CrossRef]
- Berauer, B.J.; Wilfahrt, P.A.; Reu, B.; Schuchardt, M.A.; Garcia-Franco, N.; Zistl-Schlingmann, M.; Dannenmann, M.; Kiese, R.; Kühnel, A.; Jentsch, A. Predicting forage quality of species-rich pasture grasslands using vis-NIRS to reveal effects of management intensity and climate change. Agric. Ecosyst. Environ. 2020, 296, 106929. [Google Scholar] [CrossRef]
- Andújar, D.; Escolà, A.; Rosell-Polo, J.R.; Fernández-Quintanilla, C.; Dorado, J. Potential of a terrestrial LiDAR-based system to characterise weed vegetation in maize crops. Comput. Electron. Agric. 2013, 92, 11–15. [Google Scholar] [CrossRef]
- Marcinkowska-Ochtyra, A.; Jarocińska, A.; Bzdęga, K.; Tokarska-Guzik, B. Classification of Expansive Grassland Species in Different Growth Stages Based on Hyperspectral and LiDAR Data. Remote Sens. 2018, 10, 2019. [Google Scholar] [CrossRef]
- Moreno, H.; Andújar, D. Proximal sensing for geometric characterization of vines: A review of the latest advances. Comput. Electron. Agric. 2023, 210, 107901. [Google Scholar] [CrossRef]
- Oliveira, R.A.; Näsi, R.; Niemeläinen, O.; Nyholm, L.; Alhonoja, K.; Kaivosoja, J.; Jauhiainen, L.; Viljanen, N.; Nezami, S.; Markelin, L.; et al. Machine learning estimators for the quantity and quality of grass swards used for silage production using drone-based imaging spectrometry and photogrammetry. Remote Sens. Environ. 2020, 246, 111830. [Google Scholar] [CrossRef]
- Lussem, U.; Bolten, A.; Kleppert, I.; Jasper, J.; Gnyp, M.L.; Schellberg, J.; Bareth, G. Herbage Mass, N Concentration, and N Uptake of Temperate Grasslands Can Adequately Be Estimated from UAV-Based Image Data Using Machine Learning. Remote Sens. 2022, 14, 3066. [Google Scholar] [CrossRef]
- Xu, K.; Su, Y.; Liu, J.; Hu, T.; Jin, S.; Ma, Q.; Zhai, Q.; Wang, R.; Zhang, J.; Li, Y.; et al. Estimation of degraded grassland aboveground biomass using machine learning methods from terrestrial laser scanning data. Ecol. Indic. 2020, 108, 105747. [Google Scholar] [CrossRef]
- Chen, Y.; Guerschman, J.; Shendryk, Y.; Henry, D.; Harrison, M.T. Estimating Pasture Biomass Using Sentinel-2 Imagery and Machine Learning. Remote Sens. 2021, 13, 603. [Google Scholar] [CrossRef]
- Zwick, M.; Cardoso, J.A.; Gutiérrez-Zapata, D.M.; Cerón-Muñoz, M.; Gutiérrez, J.F.; Raab, C.; Jonsson, N.; Escobar, M.; Roberts, K.; Barrett, B. Pixels to pasture: Using machine learning and multispectral remote sensing to predict biomass and nutrient quality in tropical grasslands. Remote Sens. Appl. Soc. Environ. 2024, 36, 101282. [Google Scholar] [CrossRef]
- Defalque, G.; Santos, R.; Bungenstab, D.; Echeverria, D.; Dias, A.; Defalque, C. Machine learning models for dry matter and biomass estimates on cattle grazing systems. Comput. Electron. Agric. 2024, 216, 108520. [Google Scholar] [CrossRef]
- Zhao, J.; Xie, G.; Li, D.; Song, M. Improved GMM-based method for target detection. IET Netw. 2020, 9, 7–11. [Google Scholar] [CrossRef]
- Ferro, M.V.; Sørensen, C.G.; Catania, P. Comparison of different computer vision methods for vineyard canopy detection using UAV multispectral images. Comput. Electron. Agric. 2024, 225, 109277. [Google Scholar] [CrossRef]
- Nielsen, D.C.; Miceli-Garcia, J.J.; Lyon, D.J. Canopy Cover and Leaf Area Index Relationships for Wheat, Triticale, and Corn. Agron. J. 2012, 104, 1569–1573. [Google Scholar] [CrossRef]
- Zhang, Z. A flexible new technique for camera calibration. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 1330–1334. [Google Scholar] [CrossRef]
- Shi, J.; Tomasi, C. Good Features to Track. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 21–23 June 1994. [Google Scholar] [CrossRef]
- Canny, J. A Computational Approach to Edge Detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, 6, 679–698. [Google Scholar] [CrossRef]
- Vincent, E.; Laganiere, R. Detecting planar homographies in an image pair. In Proceedings of the ISPA 2001, Proceedings of the 2nd International Symposium on Image and Signal Processing and Analysis, Pula, Croatia, 19–21 June 2001; pp. 182–187. [Google Scholar]
- Tibshirani, R.; Hastie, T.; Narasimhan, B.; Chu, G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. USA 2002, 99, 6567–6572. [Google Scholar] [CrossRef]
- Reynolds, D. Gaussian Mixture Models. In Encyclopedia of Biometrics; Li, S.Z., Jain, A.K., Eds.; Springer: Boston, MA, USA, 2015; pp. 827–832. [Google Scholar]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024. [Google Scholar]
- Pinheiro, J.; Bates, D.; Team, R.C. nlme: Linear and Nonlinear Mixed Effects Models, R Package Version 3.1-162: 2023. Available online: https://svn.r-project.org/R-packages/trunk/nlme/ (accessed on 17 April 2024).
- Lenth, R.V. emmeans: Estimated Marginal Means, aka Least-Squares Means, R Package Version 1.8.5: 2023. Available online: https://rvlenth.github.io/emmeans/ (accessed on 17 April 2024).
- Zivkovic, Z. Improved adaptive Gaussian mixture model for background subtraction. In Proceedings of the 17th International Conference on Pattern Recognition (ICPR 2004), Cambridge, UK, 26 August 2004; Volume 22, pp. 28–31. [Google Scholar]
- Riegler-Nurscher, P.; Prankl, J.; Bauer, T.; Strauss, P.; Prankl, H. A machine learning approach for pixel wise classification of residue and vegetation cover under field conditions. Biosyst. Eng. 2018, 169, 188–198. [Google Scholar] [CrossRef]
- Burgos-Artizzu, X.P.; Ribeiro, A.; Guijarro, M.; Pajares, G. Real-time image processing for crop/weed discrimination in maize fields. Comput. Electron. Agric. 2011, 75, 337–346. [Google Scholar] [CrossRef]
- Shajahan, S.; Igathinathane, C.; Saliendra, N.; Hendrickson, J.R.; Archer, D. Color calibration of digital images for agriculture and other applications. ISPRS J. Photogramm. Remote Sens. 2018, 146, 221–234. [Google Scholar] [CrossRef]
- Kefauver, S.C.; Romero, A.G.; Buchaillot, M.L.; Vergara-Díaz, O.; Fernandez-Gallego, J.A.; El-Haddad, G.; Akl, A.; Araus, J.L. Open-Source Software for Crop Physiological Assessments Using High Resolution RGB Images. In Proceedings of the (IGARSS 2020) 2020 IEEE International Geoscience and Remote Sensing Symposium, Virtual, 26 September–2 October 2020; pp. 4359–4362. [Google Scholar]
- Andújar, D.; Ribeiro, A.; Carmona, R.; Fernández-Quintanilla, C.; Dorado, J. An assessment of the accuracy and consistency of human perception of weed cover. Weed Res. 2010, 50, 638–647. [Google Scholar] [CrossRef]
- Sagan, V.; Maimaitijiang, M.; Sidike, P.; Bhadra, S.; Gosselin, N.; Burnette, M.; Demieville, J.; Hartling, S.; Lebauer, D.; Newcomb, M.; et al. Data-Driven Artificial Intelligence for Calibration of Hyperspectral Big Data. IEEE Trans. Geosci. Remote Sens. 2021, 60, 5510320. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Sagan, V.; Bhadra, S.; Nguyen, C.; Mockler, T.C.; Shakoor, N. A fully automated and fast approach for canopy cover estimation using super high-resolution remote sensing imagery. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2021, 3, 219–226. [Google Scholar] [CrossRef]
- Madec, S.; Irfan, K.; Velumani, K.; Baret, F.; David, E.; Daubige, G.; Samatan, L.B.; Serouart, M.; Smith, D.; James, C.; et al. VegAnn, Vegetation Annotation of multi-crop RGB images acquired under diverse conditions for segmentation. Sci. Data 2023, 10, 302. [Google Scholar] [CrossRef]
- Bolón-Canedo, V.; Sánchez-Maroño, N.; Alonso-Betanzos, A. Feature selection for high-dimensional data. Prog. Artif. Intell. 2016, 5, 65–75. [Google Scholar] [CrossRef]
- Dara, S.; Tumma, P. Feature Extraction by Using Deep Learning: A Survey. In Proceedings of the 2018 2nd International Conference on Electronics, Communication and Aerospace Technology (ICECA), Coimbatore, India, 29–31 March 2018; pp. 1795–1801. [Google Scholar]
- Mumuni, A.; Mumuni, F. Automated data processing and feature engineering for deep learning and big data applications: A survey. J. Inf. Intell. 2024, 3, 113–153. [Google Scholar] [CrossRef]
- Moreno, H.; Gómez, A.; Altares-López, S.; Ribeiro, A.; Andújar, D. Analysis of Stable Diffusion-derived fake weeds performance for training Convolutional Neural Networks. Comput. Electron. Agric. 2023, 214, 108324. [Google Scholar] [CrossRef]
- García-Navarrete, O.L.; Correa-Guimaraes, A.; Navas-Gracia, L.M. Application of Convolutional Neural Networks in Weed Detection and Identification: A Systematic Review. Agriculture 2024, 14, 568. [Google Scholar] [CrossRef]
- Castro-Valdecantos, P.; Apolo-Apolo, O.E.; Pérez-Ruiz, M.; Egea, G. Leaf area index estimations by deep learning models using RGB images and data fusion in maize. Precis. Agric. 2022, 23, 1949–1966. [Google Scholar] [CrossRef]
- Ford, J.; Sadgrove, E.; Paul, D. Developing an extreme learning machine based approach to weed segmentation in pastures. Smart Agric. Technol. 2023, 5, 100288. [Google Scholar] [CrossRef]
- Keerthinathan, P.; Amarasingam, N.; Kelly, J.E.; Mandel, N.; Dehaan, R.L.; Zheng, L.; Hamilton, G.; Gonzalez, F. African Lovegrass Segmentation with Artificial Intelligence Using UAS-Based Multispectral and Hyperspectral Imagery. Remote Sens. 2024, 16, 2363. [Google Scholar] [CrossRef]
- Okyere, F.G.; Cudjoe, D.; Sadeghi-Tehran, P.; Virlet, N.; Riche, A.B.; Castle, M.; Greche, L.; Mohareb, F.; Simms, D.; Mhada, M.; et al. Machine Learning Methods for Automatic Segmentation of Images of Field- and Glasshouse-Based Plants for High-Throughput Phenotyping. Plants 2023, 12, 2035. [Google Scholar] [CrossRef]
- Rebala, G.; Ravi, A.; Churiwala, S. Principal Component Analysis. In An Introduction to Machine Learning; Springer: Cham, Switzerland, 2019; pp. 141–152. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).