Sugarcane Genetic Diversity Study of Germplasm Bank and Assessment of a Core Collection †
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Extraction
2.3. DArT-Seq
2.4. Polymorphism Level in Genotyping Data
2.5. Genetic Structure Analysis
2.6. Core Collection
3. Results
3.1. Genetic Diversity
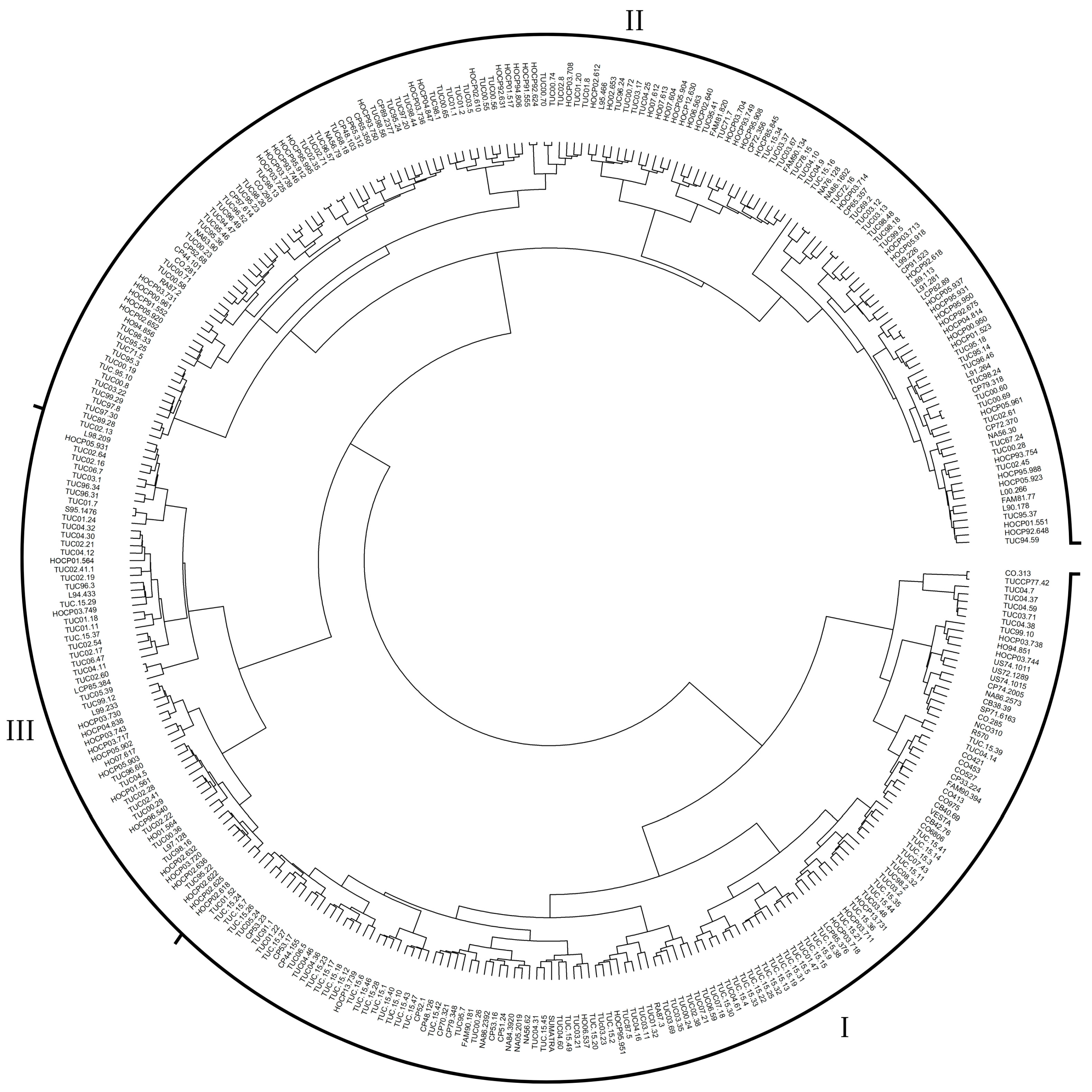
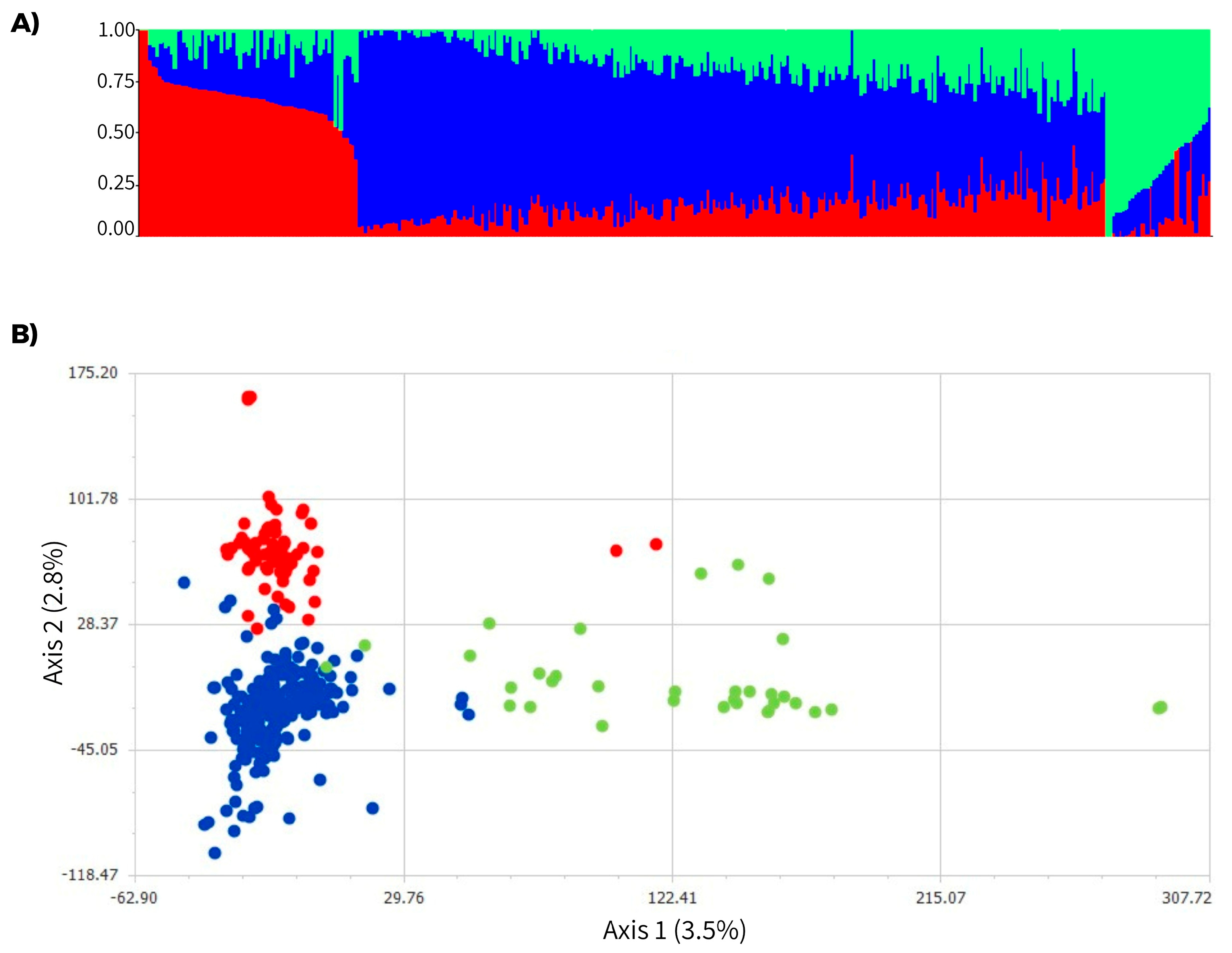
3.2. Genetic Structure Analysis
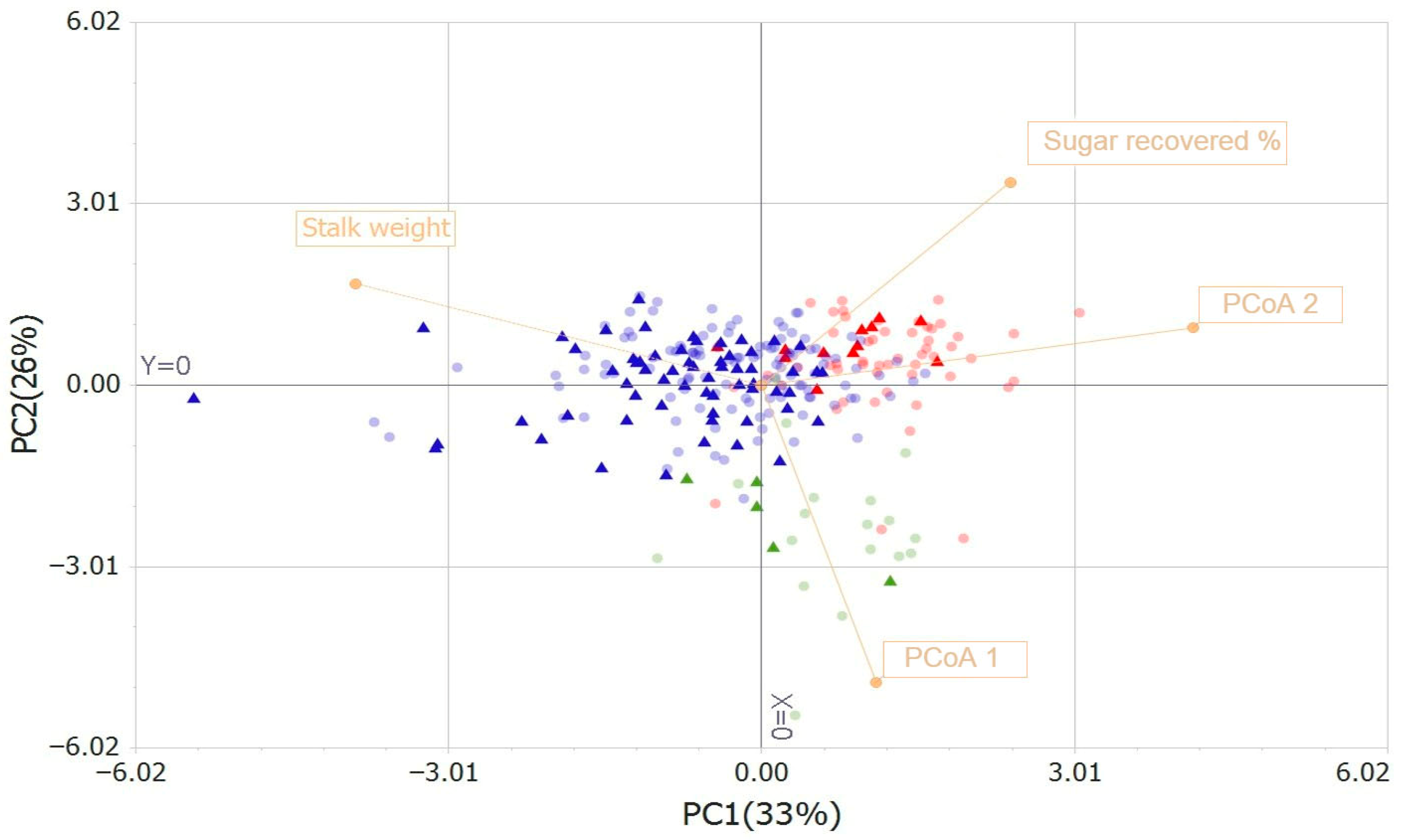
3.3. Core Collection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMOVA | analysis of molecular variance |
| DArT | Diversity Arrays Technology |
| Dj | discrimination power |
| EEAOC | Estación Experimental Agroindustrial Obispo Colombres |
| HE | expected heterozygosity |
| HO | observed heterozygosity |
| MAF | minor allele frequency |
| NA | null allele frequency |
| Ne | effective number of alleles |
| PCA | principal component analysis |
| PCoA | principal coordinate analysis |
| PIC | polymorphism information content |
| SNP | single-nucleotide polymorphism |
References
- Fickett, N.D.; Ebrahimi, L.; Parco, A.P.; Gutierrez, A.V.; Hale, A.L.; Pontif, M.J.; Todd, J.; Kimbeng, C.A.; Hoy, J.W.; Ayala-Silva, T.; et al. An enriched sugarcane diversity panel for utilization in genetic improvement of sugarcane. Sci. Rep. 2020, 10, 13390. [Google Scholar] [CrossRef] [PubMed]
- Crystian, D.; Messias dos Santos, J.; Veríssimo, G.; Souza, B.; Almeida, C. Genetic diversity trends in sugarcane germplasm: Analysis in the germplasm bank of the RB varieties. Crop Breed. Appl. Biotechnol. 2018, 18, 426–431. [Google Scholar] [CrossRef]
- Healey, A.L.; Garsmeur, O.; Lovell, J.T.; Shengquiang, S.; Sreedasyam, A.; Jenkins, J.; Plott, C.B.; Piperidis, N.; Pompidor, N.; Llaca, V.; et al. The complex polyploid genome architecture of sugarcane. Nature 2024, 628, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Ostengo, S.; Serino, G.; Perera, M.F.; Racedo, J.; Mamaní González, S.Y.; Yañez Cornejo, F.; Cuenya, M.I. Sugarcane breeding, germplasm development and supporting genetic research in Argentina. Sugar Tech 2021, 24, 166–180. [Google Scholar] [CrossRef]
- Mohammadi, S.A.; Prasanna, B.M. Analysis of genetic diversity in crop plants. Salient statistical tools and considerations. Crop Sci. 2003, 43, 1235–1248. [Google Scholar] [CrossRef]
- de Oliveira, G.L.; de Souza, A.P.; de Oliveira, F.A.; Zucchi, M.I.; de Souza, L.M.; Moura, M.F. Genetic structure and molecular diversity of Brazilian grapevine germplasm: Management and use in breeding programs. PLoS ONE 2020, 15, e0240665. [Google Scholar] [CrossRef] [PubMed]
- Kanaka, K.K.; Sukhija, N.; Goli, R.C.; Singh, S.; Ganguly, I.; Dixit, S.; Dash, A.; Malik, A.A. On the concepts and measures of diversity in the genomics era. Curr. Plant Biol. 2023, 33, 100278. [Google Scholar] [CrossRef]
- Perera, M.F.; Ostengo, S.; Peña Malavera, A.N.; Balsalobre, T.W.A.; Onorato, G.D.; Noguera, A.S.; Hoffmann, H.P.; Carneiro, M.S. Genetic diversity and population structure of Saccharum hybrids. PLoS ONE 2023, 18, e0289504. [Google Scholar] [CrossRef] [PubMed]
- Yirgu, M.; Kebede, M.; Feyissa, T.; Lakew, B.; Woldeyohannes, A.B.; Fikere, M. Single nucleotide polymorphism (SNP) markers for genetic diversity and population structure study in Ethiopian barley (Hordeum vulgare L.) germplasm. BMC Genome 2024, 24, 7. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Crow, J.F. The number of alleles that can be maintained in a finite population. Genetics 1964, 49, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Medeiros, C.; Balsalobre, T.W.A.; Carneiro, M.S. Molecular diversity and genetic structure of Saccharum complex accessions. PLoS ONE 2020, 15, e0233211. [Google Scholar] [CrossRef] [PubMed]
- Diez, O.; Zossi, S.; Chavanne, E.R.; Cárdenas, G. Calidad industrial de las cañas de azúcar de maduración temprana LCP85-384 y LCP85-376 en Tucumán. Análisis de sus principales constituyentes físico-químicos. RIAT 2000, 77, 39–48. [Google Scholar]
- Suman, A.; Ali, K.; Arro, J.; Parco, A.S.; Kimbeng, C.A.; Baisakh, N. Molecular diversity among members of the Saccharum complex assessed using TRAP markers based on lignin-related genes. Bioenergy Res. 2012, 5, 197–205. [Google Scholar] [CrossRef]
- Mahadevaiah, C.; Appunu, C.; Aitken, K.; Suresha, G.S.; Vignesh, P.; Swamy, H.K.M.; Valarmathi, R.; Hemaprabha, G.; Alagarasan, G.; Ram, B. Genomic selection in sugarcane: Current status and future prospects. Front. Plant Sci. 2021, 12, 708233. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Luo, Z.; Todd, J.; Sood, S.; Wang, J. Genome-wide association study of multiple yield traits in a diversity panel of polyploid sugarcane (Saccharum spp.). Plant Genome. 2020, 13, e20006. [Google Scholar] [CrossRef] [PubMed]
- Melchinger, A.E. Use of RFLP markers for analyses of genetic relationships among breeding materials and prediction of hybrid performance. In International Crop Science I; Buxton, D.R., Shibles, R., Forsberg, R.A., Blad, B.L., Asay, K.H., Paulsen, G.M., Wilson, R.F., Eds.; Crop Science Society of America: Madison, WI, USA, 1993; pp. 621–628. [Google Scholar]
- Glynn, N.C.; McCorkle, K.; Comstock, J.C. Diversity among mainland USA sugarcane cultivars examined by SSR genotyping. J. Am. Soc. Sugar Cane Technol. 2009, 29, 36–52. [Google Scholar]
- Tazeb, A.; Haileselassie, T.; Tesfaye, K. Molecular characterization of introduced sugarcane genotypes in Ethiopia using inter simple sequence repeat (ISSR) molecular markers. Afr. J. Biotechnol. 2017, 16, 434–449. [Google Scholar]
- Manechini, J.R.V.; Costa, J.B.; Pereira, B.T.; Carlini-Garcia, L.; Xavier, M.A.; Landell, M.G.A.; Rossini Pinto, L. Unraveling the genetic structure of Brazilian commercial sugarcane cultivars through microsatellite markers. PLoS ONE 2018, 23, e0195623. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. Evolution and the Genetics of Populations. Volume 4: Variability within and among Natural Populations; University of Chicago Press: Chicago, IL, USA, 1978. [Google Scholar]
- Perera, M.F.; Peña Malavera, A.N.; Noguera, A.S.; Racedo, J.; Ostengo, S. Sugarcane genetic diversity study of EEAOC´s germplasm bank and assessment of a core collection. In Proceedings of the ISSCT XXXII Centennial Congress 2025, Cali, Colombia, 24–28 August 2025; Volume 32, pp. 180–186. [Google Scholar]

| Variable | Mean | S.D. | Min | Max |
|---|---|---|---|---|
| Ne | 1.54 | 0.40 | 1.00 | 2.00 |
| NA | 0.38 | 0.22 | 0.00 | 0.90 |
| HO | 0.28 | 0.33 | 0.00 | 1.00 |
| HE | 0.30 | 0.20 | 0.00 | 0.50 |
| PIC | 0.22 | 0.15 | 0.00 | 0.38 |
| Dj | 0.28 | 0.20 | 0.00 | 0.50 |
| Size (%) | Number of Genotypes | Number of Alleles | Captured Diversity | Average SR% | Average SW |
|---|---|---|---|---|---|
| 10 | 35 | 137,040 | 95.78% | 11.08 ± 1.34 | 0.76 ± 0.22 |
| 20 | 70 | 139,013 | 97.16% | 11.02 ± 1.24 | 0.73 ± 0.20 |
| 30 | 104 | 140,554 | 98.24% | 11.13 ± 1.16 | 0.72 ± 0.18 |
| 100 | 348 | 143,063 | 100% | 11.43 ± 1.15 | 0.67 ± 0.19 |
| Genotypes | Core (%) | Genotypes | Core (%) | Genotypes | Core (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 20 | 30 | 10 | 20 | 30 | 10 | 20 | 30 | |||
| CB 38-39 | x | HOCP 95-951 | x | x | x b | TUC 06-47 | x | x | |||
| CB 42-76 | x | x b | HOCP 95-995 | x | TUC 07-18 | x | x | ||||
| CO 413 | x | x | x b | HOCP 95-988 | x | TUC 07-21 | x | x b | |||
| CO 453 | x | x b | L 00-266 | x | TUC 68-18 | x | x | x b | |||
| CO 527 | x | x | x b | L 94-433 | x | TUC 94-47 | x b | ||||
| CO 6806 | x | x | x b | L 98-209 | x a | TUC 94-59 | x | ||||
| CO 290 | x | LCP 85-376 | x | x | TUC 95-7 | x | x | x b | |||
| CP 33-224 | x | x | x b | NA 05-2019 | x | x | x b | TUC 95-23 | x | ||
| CP 44-101 | x | NA 84-3920 | x | x b | TUC 95-24 | x | x | ||||
| CP 44-155 | x | x | NA 86-2392 | x b | TUC 95-36 | x | |||||
| CP 48-126 | x | x | x | NA 86-2573 | x | x | TUC 95-46 | x | x | ||
| CP 51-24 | x | x | x | R 570 | x | x | x b | TUC 96-3 | x | ||
| CP 52-1 | x | x | x | SUMATRA | x | x | x | TUC 96-34 | x | x | |
| CP 53-16 | x b | TUC 00-19 | x | x | TUC 96-46 | x | |||||
| CP 53-17 | x | x | TUC 00-23 | x | TUC 96-49 | x | |||||
| CP 53-23 | x | TUC 00-26 | x | x | TUC 96-52 | x | |||||
| CP 65-350 | x | TUC 01-2 | x | TUC 97-20 | x | x | x | ||||
| CP 70-321 | x | x | x | TUC 01-11 | x a | TUC 98-48 | x | ||||
| CP 74-2005 | x | x | TUC 00-71 | x | TUC 99-10 | x | x | ||||
| CP 89-2377 | x | TUC 01-22 | x | x | x | TUC 15-2 | x | ||||
| FAM 81-77 | x b | TUC 01-47 | x | TUC 15-5 | x | x | |||||
| FAM 90-181 | x | x b | TUC 02-13 | x | TUC 15-7 | x | |||||
| FAM 90-394 | x | x | x b | TUC 02-16 | x | TUC 15-12 | x | x | |||
| HO 07-613 | x | TUC 02-17 | x | x a | TUC 15-20 | x | x | ||||
| HO 07-604 | x | x | TUC 02-22 | x a | TUC 15-21 | x | x | ||||
| HO 07-612 | x | x | TUC 02-41 | x | TUC 15-22 | x | x | ||||
| HO 94-851 | x | x | x | TUC 02-54 | x | x | TUC 15-23 | x | |||
| HOCP 00-961 | x | TUC 02-71 | x | TUC 15-24 | x | x | |||||
| HOCP 01-517 | x | x | TUC 03-11 | x | TUC 15-27 | x | |||||
| HOCP 03-704 | x | TUC 03-23 | x | TUC 15-29 | x | x | |||||
| HOCP 03-711 | x | TUC 03-67 | x b | TUC 15-36 | x | ||||||
| HOCP 03-731 | x | x | x | TUC 04-9 | x | TUC 15-37 | x | x | |||
| HOCP 03-736 | x | x | TUC 03-71 | x | TUC 15-39 | x | |||||
| HOCP 03-738 | x | x | TUC 04-11 | x | x | x | TUC 15-40 | x | x | ||
| HOCP 03-739 | x | x b | TUC 04-30 | x | TUC 15-42 | x | x | x | |||
| HOCP 03-744 | x | x | x b | TUC 04-31 | x | x | TUC 15-43 | x | |||
| HOCP 03-749 | x | TUC 04-38 | x | x a | TUC 15-45 | x | x | x | |||
| HOCP 04-847 | x | TUC 04-60 | x | x | x | TUC 15-49 | x | x | x | ||
| HOCP 05-920 | x | TUC 04-61 | x | US 74-1011 | x | x | x | ||||
| HOCP 93-750 | x | TUC 05-24 | x | x | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perera, M.F.; Peña Malavera, A.N.; Henriquez, D.D.; Noguera, A.S.; Racedo, J.; Ostengo, S. Sugarcane Genetic Diversity Study of Germplasm Bank and Assessment of a Core Collection. Agronomy 2025, 15, 2638. https://doi.org/10.3390/agronomy15112638
Perera MF, Peña Malavera AN, Henriquez DD, Noguera AS, Racedo J, Ostengo S. Sugarcane Genetic Diversity Study of Germplasm Bank and Assessment of a Core Collection. Agronomy. 2025; 15(11):2638. https://doi.org/10.3390/agronomy15112638
Chicago/Turabian StylePerera, Maria Francisca, Andrea Natalia Peña Malavera, Diego Daniel Henriquez, Aldo Sergio Noguera, Josefina Racedo, and Santiago Ostengo. 2025. "Sugarcane Genetic Diversity Study of Germplasm Bank and Assessment of a Core Collection" Agronomy 15, no. 11: 2638. https://doi.org/10.3390/agronomy15112638
APA StylePerera, M. F., Peña Malavera, A. N., Henriquez, D. D., Noguera, A. S., Racedo, J., & Ostengo, S. (2025). Sugarcane Genetic Diversity Study of Germplasm Bank and Assessment of a Core Collection. Agronomy, 15(11), 2638. https://doi.org/10.3390/agronomy15112638






