Abstract
Spiroplasma citri, a bacterium from the class Mollicutes, is the causative agent of citrus stubborn disease, a serious threat to Moroccan citrus crops, with yield losses reaching 45%. Despite its long-standing presence since 1949 and regulations mandating disease-free citrus plants, data on S. citri in Morocco remain scarce. This study investigates the pathogenicity and symptom variability of Moroccan S. citri isolates using biological indexing and genetic mapping based on the Spiralin and P58 genes. Biological indexing through reverse inoculation revealed that seven out of ten isolates caused moderate to intense symptoms within 8 to 10 weeks, with symptom severity varying across citrus cultivars and regions. These findings suggest variations in pathogen titer. Molecular analysis showed that Moroccan isolates (27GH, 3GH, 8GH, 56MK, 16MK, 60MK, 2GLK, 13SS, and 30S1) exhibited complete (100%) sequence similarity with each other and the reference strain R2-A8. Furthermore, these isolates displayed a high degree of similarity (99.75%) to a Corsican isolate (U13995) and a 94% similarity to an Iranian isolate (KP666137). Analysis of the P58 gene confirmed a high level of homogeneity with Moroccan reference strain R8-A2, closely aligning (99.75%) with the American BR3-3X strain, and 98% similarity to isolates from Syria and Iran. This study lays a foundational insight into the molecular characterization of S. citri in Morocco and provides a groundwork for future research into managing citrus stubborn disease.
1. Introduction
The Green Morocco Plan has significantly boosted the country’s citrus sector, with production more than doubling from 1.3 million to over 2.6 million tonnes between 2003 and 2021 [1]. By 2021, Morocco had cultivated 135,000 hectares of citrus, exported 640,000 tonnes, and generated approximately MAD 336.74 million annually. Citrus farming is primarily concentrated in five regions, with Souss/Massa holding the largest share (32%). Key varieties include Clementine (35%), Maroc-Late (21%), and Navel (18%) [2].
Despite this growth, Morocco’s citrus crops face significant threats from abiotic and biotic stressors, including viruses and bacteria. Among these, Spiroplasma citri, the causative agent of Citrus Stubborn Disease (CSD), is a prominent graft-transmissible pathogen that severely impacts citrus production [3,4]. This wall-less, Gram-positive bacterium inhabits phloem sieve tubes [5] and is transmitted by leafhoppers, including Circulifer tenellus and Neoaliturus haematoceps [6].
Citrus Stubborn Disease (CSD) predominantly affects hot, semi-arid regions, including California, Arizona, and North Africa, as well as Mediterranean countries such as Morocco, Algeria, Tunisia, Libya, and Egypt [7,8,9]. First reported in Morocco in 1951, CSD has since been detected in commercial citrus fields across various production areas [10,11]. The complete nucleotide sequence of the circular chromosome and two plasmids of S. citri strain R8-A2T (Morocco-R8-A2 [ATCC 27556T]), originally isolated from diseased Citrus sinensis (L.) trees [12], was reported in 2017 [13]. However, limited studies have addressed the molecular and biological characterization of local isolates compared to those conducted in neighboring countries [8,14,15,16,17].
Given the scarcity of such data, this study aims to (1) detect and isolate S. citri strains from plant tissues in Moroccan citrus orchards, (2) characterize the biological properties of these isolates, and (3) perform molecular characterization targeting two genes: the spiralin gene and the P58 gene. The spiralin gene encodes a major membrane protein involved in insect transmission and pathogenicity, making it critical for understanding the disease’s epidemiology [18,19]. The P58 gene, which encodes an adhesin-like protein implicated in spiroplasma–insect interactions, is crucial for the bacterium’s ability to colonize and propagate within its insect vectors [20,21]. These genes were selected for their established roles in pathogenicity and vector transmission.
By investigating pathogen diversity and pathogenicity, this study seeks to refine diagnostic tools and contribute to developing more effective management strategies for CSD, ultimately fostering sustainable citrus production and reducing economic losses in Morocco.
2. Materials and Methods
In July 2020 and October 2021, surveys were conducted in the main Moroccan citrus-growing regions (Tadla, Loukkos, Gharb, Souss, Melouya, and Haouz) to collect samples exhibiting symptoms of stubborn disease [22]. Representative samples from these six regions were tested in this study.
2.1. Biological Indexing Assay
Biological indexing was performed to characterize 10 representative isolates of S. citri collected from six citrus-growing regions in Morocco: Tadla, Loukkos, Gharb, Souss, Melouya, and Haouz (Table 1). The inverse inoculation method was used, following the procedure described by Mannaa et al. [16]. This technique involves inoculating a cutting from the test sample with a bud from the Madame Vinous sweet orange indicator, which is then grafted onto a sour orange rootstock. The inoculated plants were maintained in a temperature-controlled greenhouse. To reduce humidity, the tops of the plastic bags covering the grafts were opened after ten days [23]. Twenty plants (two replicates per positive citrus sample) were tested alongside a positive control provided by the UCP (an agricultural organization specializing in certified citrus plant production). Negative controls were also included.

Table 1.
Analysis results of Moroccan S. citri isolates tested by isolation, PCR, and biological indexing.
To assess the transmission rate of S. citri, specific symptoms on the leaves of indicator plants were observed. The inoculated indicator plants with negative control plants showed no abnormalities. Evaluated parameters included the transmission rate and the time required for specific symptoms to appear, such as stunted shoots and chlorotic leaves resembling zinc deficiency [24].
2.2. Isolation and Growth of S. citri
Seventeen representative infected citrus samples collected from six distinct citrus-growing regions (Table 1) were used for the isolation and cultivation of S. citri. Three specific culture media, SP4 [25], C3-G [26], and R2 [27] were tested, with the R2 medium proving to be the most effective.
The composition of the R2 medium included PPLO Broth (7.5 g), glucose (50 g), phenol red (2 mg/mL, 5 mL), and distilled water (adjusted to 500 mL). After autoclaving at 121 °C for 20 min, 10% bovine serum was added as the final concentration.
Midribs of citrus leaves (3 g each) were excised, surface-cleaned, and diced with a sterile razor blade in 5 mL of R2 liquid medium. The mixture was left to stand for five minutes and then passed through a 0.45 μm filter membrane. Then, 1 mL of the filtered medium was transferred into culture tubes containing 10 mL of R2 medium [28]. The culture tubes were incubated at 30–32 °C for a period of 3 to 22 days following the initial isolation [27,29].
Cultures were monitored for a color change from red to yellow, indicating medium acidification and potential growth of S. citri. The time (in days) required for this color change was recorded. Subcultures were prepared by transferring 1 mL of the filtrate into 9 mL of fresh R2 medium, followed by 10-fold serial dilution to a final concentration of 1 × 10−⁷. Aliquots of 0.1 µL of each dilution were plated onto R2 solid medium containing 1.5% agar and incubated at 30 °C. Colony morphology was assessed, focusing on the characteristic “fried egg” appearance, as described in previous studies [30,31,32].
2.3. DNA Extraction and PCR Amplification
Pure colonies of S. citri obtained on solid media were suspended in 1 mL of R2 medium and incubated for 24 h. After centrifugation (5 min at 16,000 tr/min), the supernatant was discarded, and the resulting pellet underwent DNA extraction using the recommended protocol provided by the DNA extraction kit (Bioneer, Daejeon, Republic of Korea).
PCR amplification was performed according to the protocols described by Drais [8] and Sagouti et al. [22]. Two primer sets were used: “SC1-fw/SC1-rw”, targeting the Spiralin gene, and “P58-6f/P58-4r”, targeting the putative adhesion gene P58 [33].
The reaction mixture had a total volume of 25 µL, comprising 2 µL of template DNA, 0.5 µL of each primer (at 10 µM), 12.5 µL of HotGoldStar Mix (Eurogentec, Liège, Belgium), and 7.5 µL of PCR-grade ultrapure water. The amplification process included an initial step of Uracil-DNA Glycosylase (UDG) activation (50 °C for 2 min), then a denaturation step (95 °C for 2 min), followed by 40 cycles of 30 s at 95 °C, 30 s at 56 °C (P58-6f/P58-4r) and 58 °C (SC1-fw/SC1-rw), 90 s at 72 °C, and a final extension step of 5 min at 72 °C [33]. PCR products were then visualized by electrophoresis on a 1.5% agarose gel stained with Sybersafe. Electrophoresis was performed in a 1× TAE buffer and visualized under a UV transilluminator. Samples were considered positive if a band corresponding to the target size was observed.
2.4. Sequencing and Phylogenetic Analysis
The PCR products of the spiralin and P58 genes from our Spiroplasma citri isolates were sequenced by STABVIDA, a company based in Portugal. Sequences, along with those of related species from the GenBank database, were aligned using BIOEDIT version 7.0 software [34]. A nucleotide-level comparison was performed between the sequenced isolates and 20 different isolates retrieved from the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/ accessed on 22 January 2022) using MEGA 11 software (version 11.0.13), which estimates the evolutionary tree that best fits the observed DNA sequences. The maximum likelihood (ML) method was employed to estimate phylogenetic relationships, with branch support in the inferred tree evaluated through 1000 bootstrap replications.
3. Results
In all surveyed citrus-growing areas, symptoms consistent with Citrus Stubborn Disease (CSD) were observed (Table 1; Figure 1). These symptoms included general stunted tree growth and dense foliation on branches, alongside specific leaf abnormalities such as upright foliation, small leaves with short internodes, and cup-shaped leaves. Other notable signs included characteristic chlorosis and mottling, reduced fruit size, as well as off-season flowering and fruit setting (Figure 1).

Figure 1.
Chlorotic, upright, and cupped leaves (A); stunted plants and deformed fruits (B); bushy tree appearance (C); and over-season flowering (D).
3.1. Biological INDEXING
Biological indexing was evaluated based on the percentage of successful S. citri transmission and the time required for the first symptoms to appear on indicator plants (Figure 2). Transmission of S. citri was determined by observing characteristic symptoms in the indicator plants, including chlorosis, cup-shaped leaves with short internodes, and reduced leaf size. Negative control plants displayed no abnormalities.
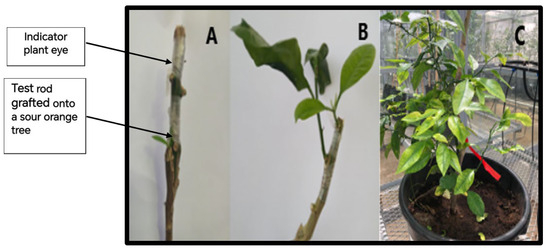
Figure 2.
Biological indexing: Reverse inoculation process of the C. sinensis “60MK” sample infected with S. citri on the “MadamVinous” indicator plant. (A) Grafting of an eye of an indicator plant grafted onto a test sample rod grafted in turn onto a sour orange plant; (B) emergence of leaves after 8 weeks; (C) symptoms of chlorosis and leaves with small cavities observed on the leaves of 60MK isolate after 12 weeks.
The results showed that severe, characteristic symptoms developed in five out of ten plants. The earliest signs of injury, such as leaf chlorosis, were observed eight weeks post-inoculation. Severe symptoms were associated with five isolates (4BM, 5TAD, 3GH, 16MK, and 60MK) and became significantly more pronounced after 12 weeks (Figure 2). In contrast, 3 out of 10 isolates exhibited moderate symptoms (2BLK, 6TAD, and 5GH), 1 out of 10 showed mild symptoms (27GH), and 1 isolate remained asymptomatic (13SS). This variation can be explained by the ability of the bacterium S. citri to thrive and multiply within citrus plants, gradually inducing characteristic symptoms over time (Table 1).
All isolates exhibiting severe symptoms tested positive through isolation, and qPCR analysis targeting the P58 adhesin gene further confirmed the presence of the bacterium in all plants, both symptomatic and asymptomatic. However, PCR targeting the spiralin gene yielded positive results only for isolates associated with moderate and severe symptoms (Table 1).
Differences in symptom onset and severity among isolates likely reflect variations in pathogen concentration within the host plant.
3.2. Isolation and Culturing of S. citri
A color change in the R2 medium, indicating the presence of S. citri, was recorded for 12 out of 17 samples 10–14 days post-inoculation (dpi). The absence of isolation in five samples could be attributed to low pathogen concentrations and uneven distribution within citrus plant tissues, as cited by Wang et al. [35]. In the second Subculture, color changes were observed after 3–5 days for all 12 samples (Figure 3). The presence of Spiroplasma was further confirmed by the characteristic “fried egg“-shaped colonies observed on R2 solid medium after 5–7 days of incubation at 30 °C. Optimal colony development was achieved at 30 °C with a dilution of 10−4 at five dpi. These colonies, characterized by their small size and clear appearance, were visible when observed directly on the surface of the plates (Figure 4). The isolated organism was identified as S. citri through PCR using P58 primers, according to the standard protocol [33].
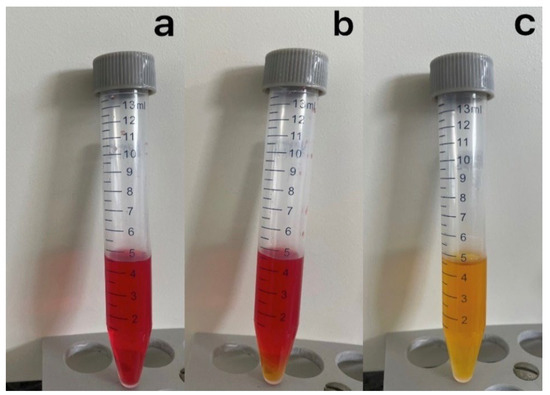
Figure 3.
Culture and isolation of S. citri on liquid medium R2. (a–c) The liquid media changed color following the presence of S. citri in the test sample. The change in color of the culture medium R2 from red to yellow indicates the presence and growth of S. citri in the medium.
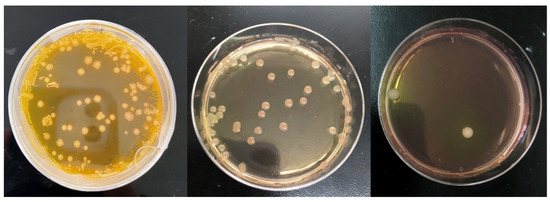
Figure 4.
The morphology of S. citri on solid medium R2 of different dilutions was examined using an optical microscope, revealing the presence of 10–15 dpi (days after inoculation) chip-shaped colonies at 30 °C.
3.3. Molecular Detection
PCR assays targeting the P58 gene produced positive results for all 17 isolates, with an amplification product of approximately 450 bp (Figure 5). However, PCR targeting the spiralin gene yielded positive results for only 9 of the 17 isolates, with the expected amplicon size of 336 bp (Figure 5).
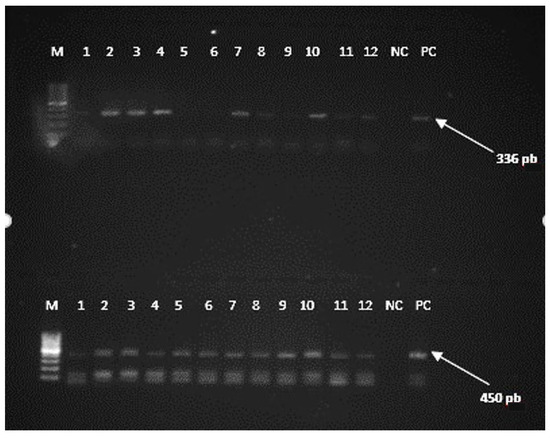
Figure 5.
Amplification products of PCR tests targeting two genes of S. citri: the spiralin (Spi) gene using the primer set SC1-fw/SC1-rw (upper part) and the putative adhesion gene P58 using the primer set P58-6f/P58-4r (lower part). Lane M is the molecular weight marker, lanes 1–12 correspond to S. citri isolates from Moroccan citrus samples, lane NC is the negative control, and lane PC is the positive control. Amplicon sizes for the Spi and P58 genes were observed at expected sizes of approximately 336 bp and 450 pb, respectively.
Compared to the other tests, PCR using the P58 primers was found to be more sensitive than PCR targeting the spiralin gene.
3.4. Phylogenetic Sequencing Analysis
Sequence analysis of the putative adhesin gene and the spiralin gene revealed a similarity range of 98% to 100% identity across all Moroccan isolates when compared to reference sequences in GenBank. Sequence data for the Moroccan isolates were deposited at NCBI (Table 1).
Sequences from both genes were obtained from 17 of the selected isolates (9 out of 17 for the spiraline gene and 15 out of 17 for the putative adhesin P58 gene) and were used to construct a phylogenetic tree (Figure 6 and Figure 7). This analysis enabled us to classify our isolates into a single clade with various S. citri strains reported worldwide. The tree with the highest likelihood was chosen as the best-fit model, representing the most probable evolutionary relationships among the S. citri strains based on the sequence data. The analysis of spiraline gene sequences revealed a significant degree of similarity among S. citri isolates from various citrus-growing regions in Morocco. Specifically, Moroccan isolates (27GH, 3GH, 8GH, 56MK, 16MK, 60MK, 2GLK, 13SS, and 30S1) exhibited 100% similarity among themselves and with the Moroccan strain R2-A8, forming a distinct branch. Furthermore, these Moroccan isolates showed high similarity to other isolates, ranging from 99.75% similarity with the Corsican isolate (U13995) to 94% similarity with the Iranian isolate (KP666137).
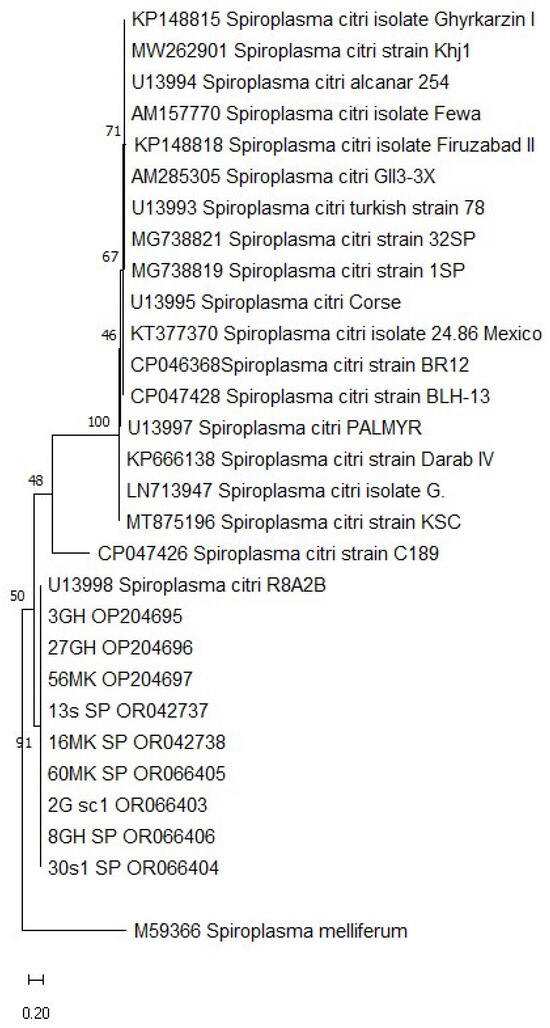
Figure 6.
Phylogenetic analysis of putative gene P58 in S. citri using Kimura 2-Parameter model and a bootstrap value of 1000 in MEGA 11 software.
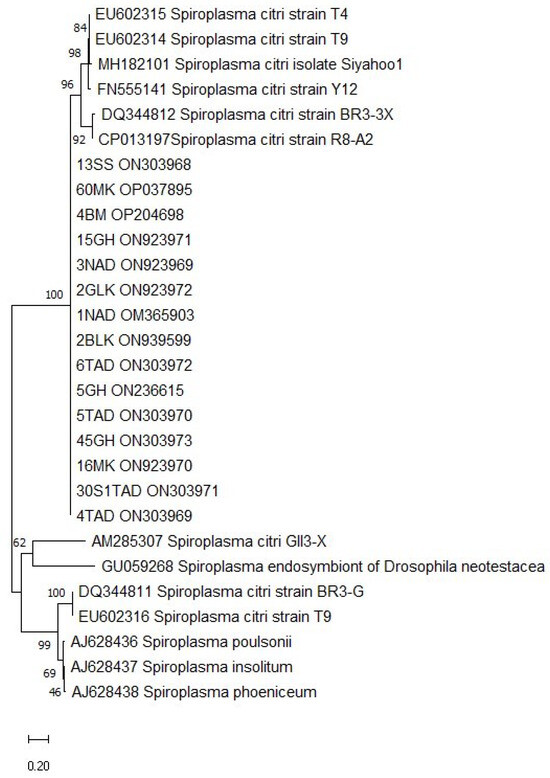
Figure 7.
Phylogenetic analysis of P58 sequences of S. citri strains using Kimura 2-Parameter model and a bootstrap value of 1000 in MEGA 11 software.
Comparative analysis of the putative adhesin gene P58 sequences among S. citri isolates from various citrus-growing regions in Morocco revealed a high degree of homogeneity with the Moroccan reference strain R8-A2 (CP013197). Additionally, these Moroccan isolates exhibited significant similarity to strains from other countries, sharing 99.75% similarity with the American strain BR3-3X (DQ344812), 98% with the Syrian isolate Y12 (FN55141), and 98% with the Iranian isolate (MH182101).
4. Discussion
Symptoms observed in the citrus orchard were characteristic of stubborn disease (Figure 1). Infected citrus trees exhibited small, chlorotic, spoon-shaped leaves and mottling. The fruits displayed abnormally small size, altered coloration, and lopsided, immature, acorn-shaped development. These symptoms are consistent with descriptions provided by several studies [4,14,17,36,37].
Haut du formulaire
Bas du formulaire
The isolation of various Moroccan isolates of S. citri was successfully achieved using R2 medium, detecting 12 out of 17 isolates. The presence of S. citri was indicated by a color change in the medium from red to yellow, as reported by Abd El-Fatah and Iftikhar [24,26], and confirmed by PCR. The R2 medium, known for its affordability and simplicity, proved highly effective in isolating S. citri from infected plant tissues, consistent with findings by several studies [27,38,39]. The characteristic “fried egg” colonies were observed, corroborated by El-Banna et al. [31] and Iftikhar et al. [24]. This method can enhance routine detection by increasing the titer of S. citri prior to molecular detection, mitigating low pathogen titer during colder periods.
Biological indexing was conducted through inverse inoculation, where 5 out of 10 isolates exhibited severe symptoms, 3 out of 10 showed moderate symptoms, 1 out of 10 displayed mild symptoms, and 1 isolate was symptomless, appearing 8 to 12 weeks post-inoculation. Indicator plants showed characteristic symptoms such as chlorotic and mottled leaves, consistent with the known effects of S. citri on citrus plants and previous research [9,16,24].
The bacterium S. citri thrives and multiplies within citrus plants, inducing characteristic symptoms. The severity of these symptoms is primarily influenced by bacterial titer rather than the virulence or genetic diversity of individual isolates [12,32]. This is supported by the high genetic similarity observed among Moroccan isolates in this study and by findings from Mannaa et al. [40], which attribute variability in symptom severity to differences in pathogen concentration within the bud sticks [16]. Regional variations in symptom severity may also reflect differences in pathogen titer at the time of inoculation.
All isolates with severe symptoms tested positive through isolation, and PCR targeting the P58 adhesin gene confirmed the presence of S. citri in both symptomatic and asymptomatic plants. In contrast, PCR targeting the spiralin gene produced positive results only for isolates causing moderate and severe symptoms, suggesting that the pathogen titer plays a critical role in symptom onset and severity.
The findings from the biological indexing of Moroccan isolates of Spiroplasma citri provide valuable contributions to advancing the application of this diagnostic method. Biological indexing offers multiple advantages: it enables the accurate identification of S. citri through characteristic symptom observation, reduces the likelihood of false positives, supports effective monitoring of disease progression, and deepens understanding of pathogen–plant interactions. These insights are essential for developing resistant citrus cultivars, optimizing disease management strategies, and mitigating the impact of S. citri on citrus production.
The variability in amplification efficiency between the P58 adhesin and spiralin genes can be attributed to their genetic architecture: the spiralin gene exists as a single copy in the genome of all S. citri strains, whereas the P58 adhesin gene likely occurs in multiple copies [19,20,33,41].
These findings underscore the importance of molecular testing for early detection, as S. citri can colonize plants before visible symptoms appear. Moreover, they provide a foundation for developing targeted diagnostic methods tailored to Moroccan isolates and for improving national strategies to manage S. citri infections effectively.
The analysis of spiraline gene sequences among S. citri isolates from various citrus-growing regions in Morocco revealed a very high level of genetic similarity. The Moroccan isolates (27GH, 3GH, 8GH, 56MK, 16MK, 60MK, 2GLK, 13SS, and 30S1) showed 100% sequence similarity with each other and with the Moroccan reference strain R2-A8, forming a distinct branch.
The observed genetic homogeneity among Moroccan S. citri isolates indicates a stable population with limited genetic variation. This stability may result from restricted pathogen movement or effective local agricultural practices that prevent the introduction of new variants. The high similarity (99.75%) with the Corsican isolate (U13995) suggests possible historical or recent interactions between Moroccan and Corsican citrus industries. In contrast, the lower similarity (94%) with the Iranian isolate (KP666137) points to more distant genetic relationships, likely due to geographical and ecological differences.
Additionally, notable similarity in spiralin sequences between Moroccan and American isolates has been previously reported by Drais [8], and high genetic similarity in spiralin gene sequences among Algerian isolates was also noted by Benazzouz et al. [14]. In addition, analysis of the putative adhesin gene P58 sequences revealed that Moroccan isolates share a high degree of homogeneity with the Moroccan reference strain R8-A2 (CP013197), suggesting that the P58 gene is highly conserved among Moroccan isolates. These isolates also exhibited 99.75% similarity with the American strain BR3-3X, 98% with the Syrian isolate Y12, and 98% with the Iranian isolate (MH182101).
This genetic stability, coupled with the high conservation of key virulence genes such as P58 and spiralin, offers an opportunity to develop precise diagnostic and management strategies tailored to Moroccan citrus farming. Targeting these conserved genes could enhance early detection and control efforts, reducing the risk of disease spread across orchards. Furthermore, understanding the potential pathways of pathogen introduction, as indicated by similarities with Corsican and American isolates, can inform biosecurity measures aimed at preventing new variants from entering Moroccan citrus orchards.
5. Conclusions
This study represents a significant advancement in the molecular characterization of Spiroplasma citri in Morocco, marking the first comprehensive mapping of S. citri variants in the region. The findings reveal low genetic diversity among isolates, suggesting a common origin and highlighting the role of global trade in the spread of the pathogen. This work lays a solid foundation for future research and practical advancements in S. citri detection and management.
A key contribution of this study is the genetic sequence data, which forms the foundation for the design and development of a highly specific and sensitive molecular diagnostic method that can enhance the management of citrus stubborn disease (CSD) through early detection and targeted interventions.
To strengthen CSD management, it is essential to integrate molecular findings with practical agricultural practices. The widespread presence of genetically similar isolates emphasizes the need for region-specific strategies, such as the use of certified pathogen-free planting materials, stricter quarantine measures, and the promotion of resistant citrus varieties.
Future research should focus on identifying citrus cultivars with resistance or tolerance to S. citri, facilitating the development of more resilient crops. Additionally, integrated control strategies, including biological control, insect vector management, and resistant rootstocks, should be explored. Expanded epidemiological studies are crucial to understanding the role of vectors in pathogen transmission and defining disease dynamics. Further genetic analysis, including studies of the 16S rRNA gene, will deepen our understanding of S. citri diversity and pathogenicity. Collectively, these efforts are vital for developing sustainable control strategies to mitigate the impact of CSD on Morocco’s citrus industry.
Author Contributions
Conceptualization, T.S. and R.L.; methodology, T.S., N.R. and R.L.; software, T.S., I.L. and R.L.; validation, Z.B., R.L. and A.T.; formal analysis, T.S.; investigation, R.L.; data curation, T.S. and R.L.; writing—original draft preparation, T.S. and I.L.; writing—review and editing, I.L., Z.B., M.E.J., E.A.B. and R.L.; supervision, R.L., E.A.B. and N.R.; project administration, R.L. and S.A.; funding acquisition, R.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by The Faculty of Science and Technology of Mohammadia (FSTM) and the Department of Plant Protection, Ecole Nationale d’Agriculture de Meknes.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- FNH. Agrumiculture: La Production Devrait S’Inscrire en Hausse de 14%. Finances News. 3 December 2021. Available online: https://fnh.ma/article/alaune/agrumiculture-la-production-devrait-s-inscrire-en-hausse-de-14 (accessed on 1 April 2024).
- Ezrari, S.; Radouane, N.; Tahiri, A.; El Housni, Z.; Mokrini, F.; Özer, G.; Lazraq, A.; Belabess, Z.; Amiri, S.; Lahlali, R. Dry Root Rot Disease, an Emerging Threat to Citrus Industry Worldwide under Climate Change: A Review. Physiol. Mol. Plant Pathol. 2022, 117, 101753. [Google Scholar] [CrossRef]
- Mubeen, M.; Arshad, H.M.I.; Iftikhar, Y.; Irfan Ullah, M.; Bilqees, I. Bio-Chemical Characterization of Xanthomonas Axonopodis Pv. Citri: A Gram Negative Bacterium Causing Citrus Canker. Int. J. Sci. Nat. 2015, 6, 151–154. [Google Scholar]
- Timmer, L.W.; Garnsey, S.M.; Graham, J.H. Front Matter. In Compendium of Citrus Diseases, 2nd ed.; Timmer, L.W., Garnsey, S.M., Graham, J.H., Eds.; Diseases and Pests Compendium Series; The American Phytopathological Society: St. Paul, MN, USA, 2000; pp. i–iv. ISBN 978-0-89054-585-0. [Google Scholar]
- Bové, J.M.; Renaudin, J.; Saillard, C.; Foissac, X.; Garnier, M. Spiroplasma citri, a Plant Pathogenic Mollicute: Relationships with Its Two Hosts, the Plant and the Leafhopper Vector. Annu. Rev. Phytopathol. 2003, 41, 483–500. [Google Scholar] [CrossRef]
- Liu, H.-Y.; Gumpf, D.J.; Oldfield, G.N.; Calavan, E.C. The Relationship of Spiroplasma citri and Circulifer Tenellus. Phytopathology 1983, 73, 585–590. [Google Scholar] [CrossRef]
- Bové, J.M. Virus and Virus-Like Diseases of Citrus in the Near East Region; FAO: Rome, Italy, 1995. [Google Scholar]
- Drais, M.I. Development of New Tools for On-Site Detection of Spiroplasma citri, the Causal Agent of Citrus Stubborn Disease; Università degli studi della Tuscia-Viterbo: Viterbo, Italy, 2018. [Google Scholar]
- Nejat, N.; Vadamalai, G.; Dickinson, M. Spiroplasma Citri: A Wide Host Range Phytopathogen. Plant Pathol. J. 2011, 10, 46–56. [Google Scholar] [CrossRef][Green Version]
- Afechtal, M. Present Status of Virus and Virus-like Diseases of Citrus in Morocco. IOBC/WPRS Bull. 2018, 132, 215–220. [Google Scholar]
- Sagouti, T.; Belabess, Z.; Rhallabi, N.; Barka, E.A.; Tahiri, A.; Lahlali, R. Citrus Stubborn Disease: Current Insights on an Enigmatic Problem Prevailing in Citrus Orchards. Microorganisms 2022, 10, 183. [Google Scholar] [CrossRef]
- Saglio, P.; Lhospital, M.; Lafleche, D. Spiroplasma citri Gen. and Sp. n.: A Mycoplasma like Organism Associated with “Stubborn” Disease of Citrus. Int. J. Syst. Bacteriol. 1973, 23, 191–204. [Google Scholar] [CrossRef]
- Davis, R.E.; Shao, J.; Zhao, Y.; Gasparich, G.E.; Gaynor, B.J.; Donofrio, N. Complete Genome Sequence of Spiroplasma citri Strain R8-A2T, Causal Agent of Stubborn Disease in Citrus Species. Genome Announc. 2017, 5, 10–1128. [Google Scholar] [CrossRef]
- Benazzouz, K.; Belkahla, H.; Dubrana, M.-P.; Lehad, A. Occurrence of Spiroplasma citri in Algerian Citrus Orchards. Preprint 2023. [Google Scholar] [CrossRef]
- Khanchezar, A.; Izadpanah, K.; Taghavi, M.; Béven, L. Genetic Diversity of Strains of Spiroplasma citri Isolated in Southern Iran. Eur. J. Plant Pathol. 2022, 163, 381–392. [Google Scholar] [CrossRef]
- Mannaa, M.; D’Onghia, A.M.; Djelouah, K.; Cavallo, G.; Valentini, F. Improvement of Detection Methods and Further Characterization of Spiroplasma citri, the Causal Agent of Citrus Stubborn Disease in Egypt. Am. J. Plant Sci. 2013, 4, 245–249. [Google Scholar] [CrossRef]
- Çağlar, B.K.; Satar, G.Ü.L.; Baloğlu, S.; DRAİS, M.İ.; Djelouah, K. Detection of Spiroplasma citri from Citrus Trees in Turkey by Molecular Techniques. Mediterr. Agric. Sci. 2020, 33, 37–42. [Google Scholar] [CrossRef]
- Wróblewski, H.; Johansson, K.-E.; Hjérten, S. Purification and Characterization of Spiralin, the Main Protein of the Spiroplasma citri Membrane. Biochim. Biophys. Acta (BBA)-Biomembr. 1977, 465, 275–289. [Google Scholar] [CrossRef]
- Foissac, X.; Saillard, C.; Gandar, J.; Zreik, L.; Bove, J.M. Spiralin Polymorphism in Strains of Spiroplasma citri Is Not Due to Differences in Posttranslational Palmitoylation. J. Bacteriol. 1996, 178, 2934–2940. [Google Scholar] [CrossRef]
- Comer, J.; Fletcher, J.; Davis, R.E.; Melcher, U. Evolution of the Spiroplasma P58 Multigene Family. Biochem. Genet. 2007, 45, 25–32. [Google Scholar] [CrossRef]
- Duret, S.; Berho, N.; Danet, J.-L.; Garnier, M.; Renaudin, J. Spiralin Is Not Essential for Helicity, Motility, or Pathogenicity but Is Required for Efficient Transmission of Spiroplasma citri by Its Leafhopper Vector Circulifer Haematoceps. Appl. Environ. Microbiol. 2003, 69, 6225–6234. [Google Scholar] [CrossRef]
- Sagouti, T.; Rhallabi, N.; Polizzi, G.; Tahiri, A.; Belabess, Z.; Barka, E.A.; Lahlali, R. Comparison of Serological and Molecular Methods for Detection of Spiroplasma citri in Moroccan Citrus-Growing Areas. Plants 2023, 12, 667. [Google Scholar] [CrossRef]
- D’Onghia, A.M. Updated Regulation on Certification of Citrus Propagating Material in the Mediterranean Region. In Citrus Tristeza Virus and Toxoptera Citricidus: A Serious Threat to the Mediterranean Citrus Industry; CIHEAM: Bari, Italy, 2009; pp. 189–210. [Google Scholar]
- Iftikhar, Y.; Bakhtawar, F.; Hussain, I.; Sajid, A.; Mubeen, M.; Zeshan, M.A.; Sohail, M.A.; Fatima, N.; Umer, M.; Iqbal, S. Detection of Spiroplasma citri Causing Citrus Stubborn Disease in Sargodha, Pakistan. Int. J. Bot. Stud. 2020, 5, 481–485. [Google Scholar]
- Raju, B.C.; Chen, T.A. Growth, Morphology and Ultrastructural Studies of a Spiroplasma Associated with Bermudagrass Showing Witches’-Broom Symptoms/Wachstum, Morphologie Und Ultrastruktur von Spiroplasma in Hexenbesenkrankem Bermudagras. Z. Pflanzenkrankh. Pflanzenschutz/J. Plant Dis. Prot. 1980, 87, 37–45. [Google Scholar]
- Abd El-Fatah, W.; Egiza, A.O.; Youssef, S.A.; Shalaby, A.A. Isolation and Identification of Spiroplasma citri Associated with Citrus Stubborn Disease in Egypt. Int. J. Adv. Res. Biol. Sci. 2016, 3, 223–231. [Google Scholar]
- Moulder, R.W.; French, F.E.; Chang, C.J. Simplified Media for Spiroplasmas Associated with Tabanid Flies. Can. J. Microbiol. 2002, 48, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.J. Nutrition and Cultivation of Spiroplasmas. In The Mycoplasmas; Whitcomb, R.F., Tully, J.G., Eds.; Elsevier: Amsterdam, The Netherlands, 1989; Volume 5. [Google Scholar]
- Bi, K.; Huang, H.; Gu, W.; Wang, J.; Wang, W. Phylogenetic Analysis of Spiroplasmas from Three Freshwater Crustaceans (Eriocheir sinensis, Procambarus clarkia and Penaeus vannamei) in China. J. Invertebr. Pathol. 2008, 99, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Sidaros, S.A.; El-Kewey, S.A.; Omar, A.F. Detection of Spiroplasma citri the Causal Organism of Citrus Stubborn Disease. In Proceedings of the 9th Congres of the Phytopathology, Egyptian Phytopathology Society, Giza, Egypt, 8–10 May 2000; pp. 19–33. [Google Scholar]
- El-Banna, O.-H.M.; Kheder, A.A.; Ali, M.A.H.; Sayed, A.M.; Haseeb, G.M. Biological and Molecular Characterization of Different Egyptian Isolates of Spiroplasma citri. Plant Arch. 2020, 20, 6457–6469. [Google Scholar]
- Mello, A.F.S.; Yokomi, R.K.; Melcher, U.; Chen, J.C.; Fletcher, J. Citrus Stubborn Severity Is Associated with Spiroplasma citri Titer But Not with Bacterial Genotype. Plant Dis. 2010, 94, 75–82. [Google Scholar] [CrossRef]
- Yokomi, R.K.; Mello, A.F.S.; Saponari, M.; Fletcher, J. Polymerase Chain Reaction-Based Detection of Spiroplasma citri Associated with Citrus Stubborn Disease. Plant Dis. 2008, 92, 253–260. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Wang, X.; Doddapaneni, H.; Chen, J.; Yokomi, R.K. Improved Real-Time PCR Diagnosis of Citrus Stubborn Disease by Targeting Prophage Genes of Spiroplasma citri. Plant Dis. 2015, 99, 149–154. [Google Scholar] [CrossRef]
- Mello, A.F.S.; Yokomi, R.K.; Payton, M.E.; Fletcher, J. Effect of Citrus Stubborn Disease on Navel Orange Production in a Commercial Orchard in California. J. Plant Pathol. 2010, 92, 429–438. [Google Scholar]
- Sagouti, T.; Rhallabi, N.; Tahiri, A.; Belabess, Z.; Radouane, N.; Lahlali, R. Development and Application of an Immunocapture Real-Time PCR for the Detection of Spiroplasma citri, the Causal Agent of Citrus Stubborn Disease. J. Plant Dis. Prot. 2024, 131, 1537–1547. [Google Scholar] [CrossRef]
- Bennani, B. Contribution à L’Étude Génétique et Elaboration de Diagnostics Immunologique et Mo-Léculaire Pour la Detection de Deux Phytopathogènes D’Agrumes: Spiroplasma Citri et Citrus Variegation Virus; Faculty of Sciences Ain Chok: Casablanca, Morocco, 2002. [Google Scholar]
- Chang, C.J.; Zheng, B. Isolation of Spiroplasma citri from Flowers and Seeds Collected from Infected Periwinkles. Plant Dis. 1999, 83, 60–61. [Google Scholar] [CrossRef] [PubMed]
- Mannai, S.; Boughalleb-M’Hamdi, N. In Vitro and in Vivo Effects of Some Chemical Fungicides against Pythium Ultimum and Phytophthora Citrophthora Associated with Peach Seedlings Decline. Nov. Res. Microbiol. J. 2021, 5, 1431–1446. [Google Scholar]
- Ye, F.; Melcher, U.; Fletcher, J. Molecular Characterization of a Gene Encoding a Membrane Protein of Spiroplasma citri. Gene 1997, 189, 95–100. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).