Effects of Microbial Fertilizer Application on Soil Ecology in Saline–Alkali Fields
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description, Microbial Fertilizer, and Field Trial Design
- (1)
- Pre-treatment: In May, “Jian Kang” powder was mixed with the seeds at a ratio of 3 g per kilogram, resulting in a high concentration of a salt–alkaline-tolerant microbial community on the seed surface. After sowing, “Chumiao Bao” was administered by drip irrigation at an amount of 15–30 kg per hectare, establishing a medium-concentration microbial community in the soil around the seeds.
- (2)
- Mid-treatment: In mid-June, microbial liquid fertilizer (containing salt–alkaline-tolerant microbes at a concentration of 1 × 107 cfu·mL−1) was applied via drip irrigation at 150 kg per hectare along with 60 kg of compound fertilizer per hectare. In mid-July, microbial liquid fertilizer was applied at 300 kg per hectare, supplemented by 105 kg of compound fertilizer per hectare.
- (3)
- Post-treatment: At the beginning of August, 450 kg of microbial liquid fertilizer per hectare and 105 kg of compound fertilizer per hectare were applied using drip irrigation. In mid-August, 600 kg per hectare of microbial liquid fertilizer was applied, followed by 150 kg per hectare of compound fertilizer. Thus far, the field trials only used drip irrigation water, and the fertilization was completed.
2.2. Soil Samples
2.3. Soil Physicochemical Properties and Enzyme Activities
2.4. Soil DNA Extraction, PCR Amplification, and Sequencing
2.5. Library Construction
2.6. Statistical Analysis
3. Results
3.1. Effect of Microbial Fertilizer on Physicochemical Properties in Saline–Alkali Soil
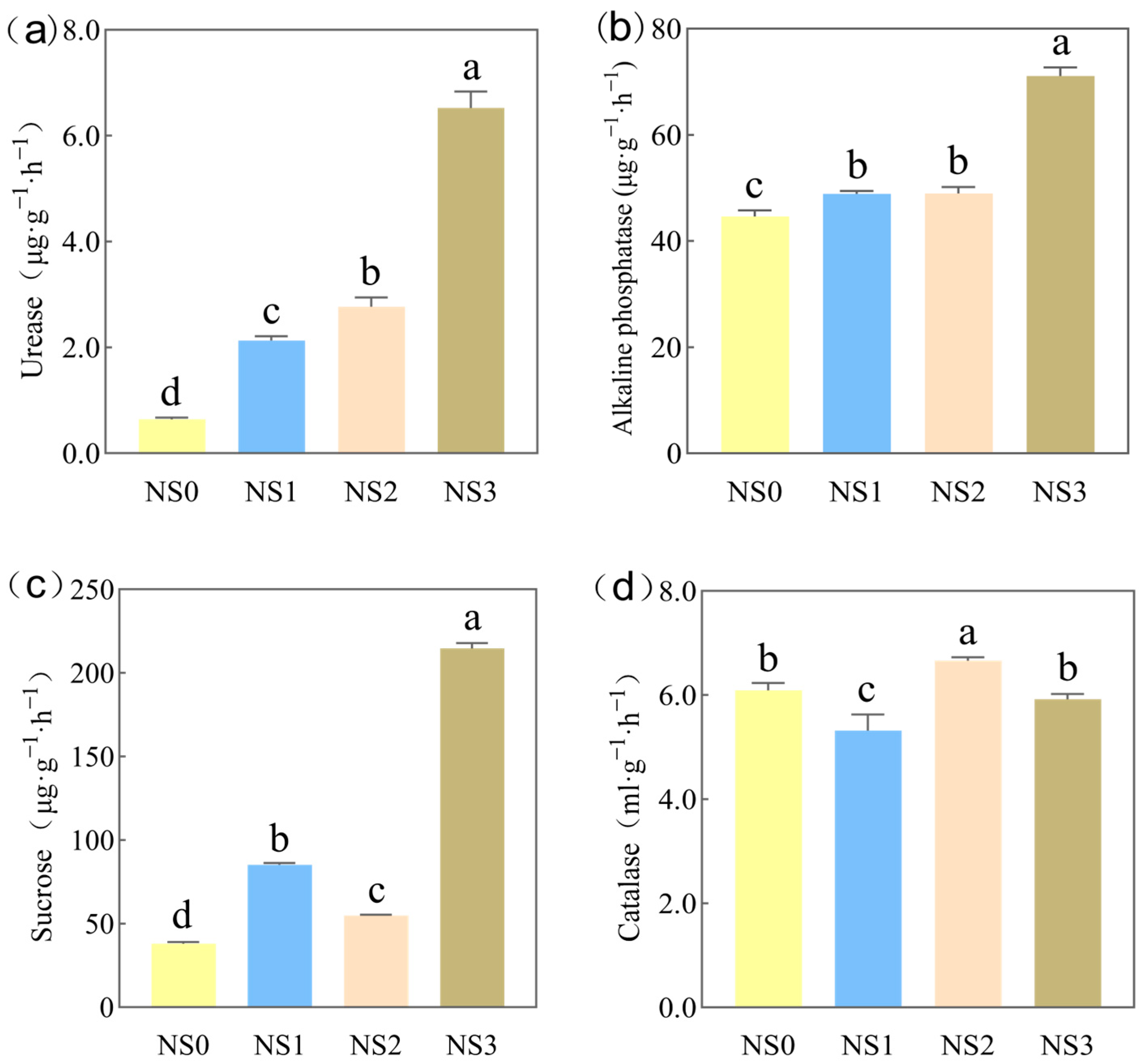
3.2. Effect of Microbial Fertilizer on Enzyme Activity in Saline–Alkali Soil
3.3. Alpha Diversity Analysis of Microbial Fertilizer on Soil Microbial Community in Saline–Alkali Soil
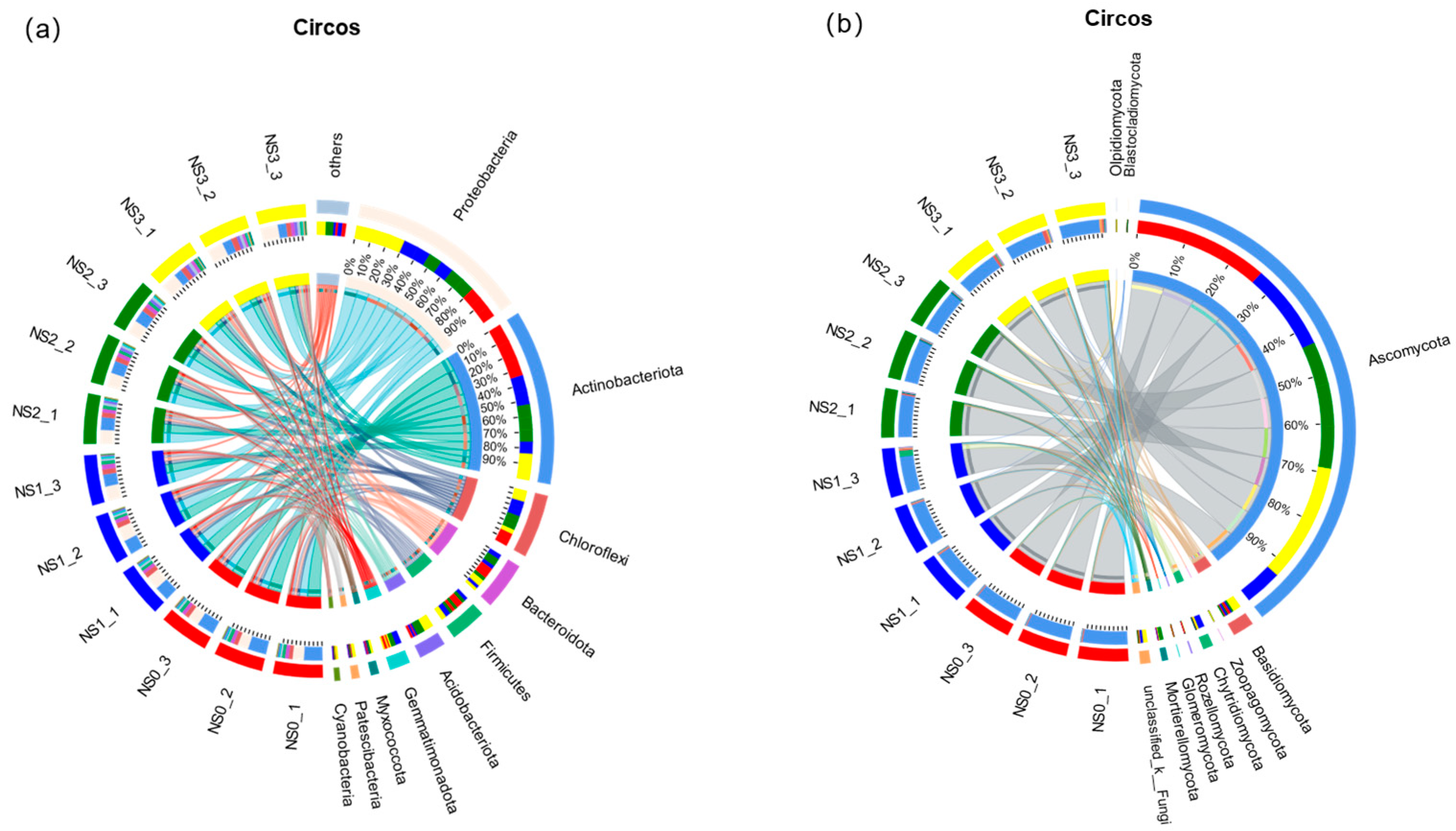
3.4. Analysis of Community Structure and Composition of Microbial Fertilizer in Saline–Alkali Soil
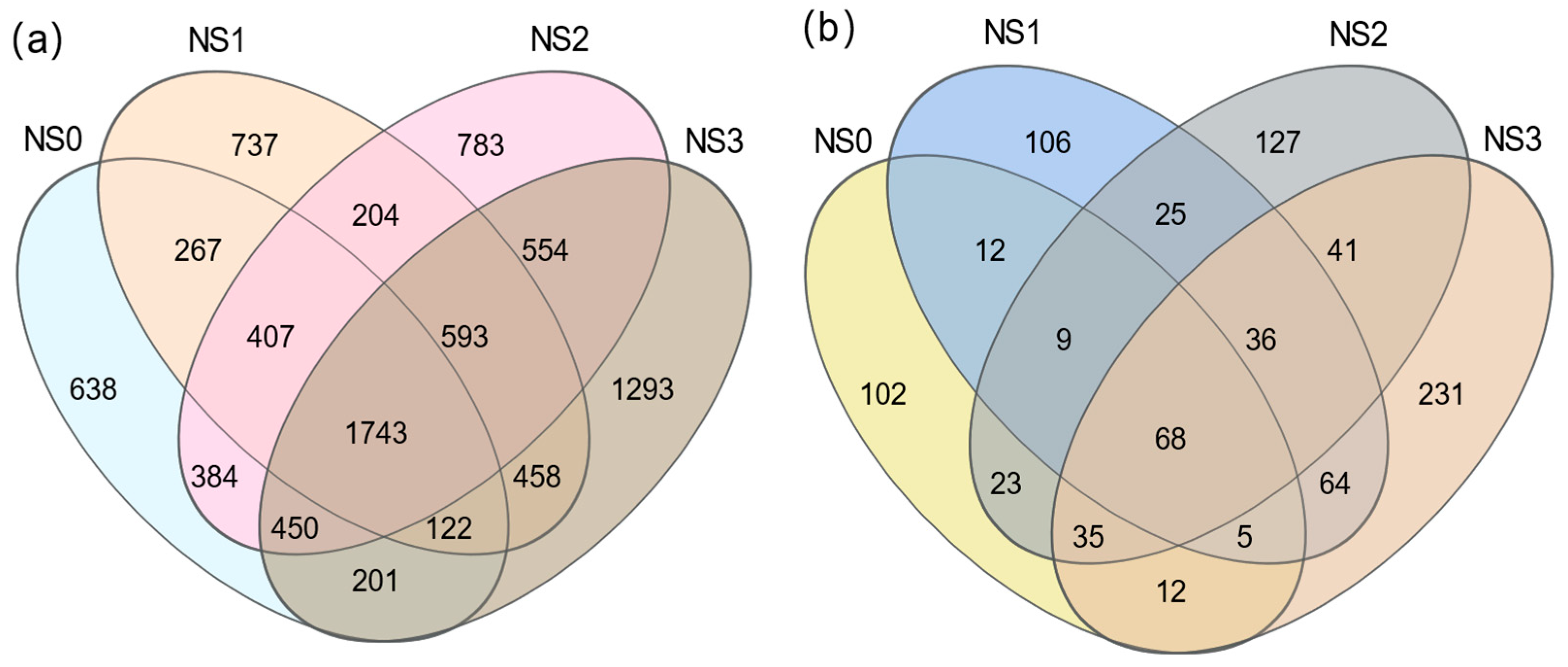
3.4.1. The Number of Soil Bacteria and Fungi at Different Periods
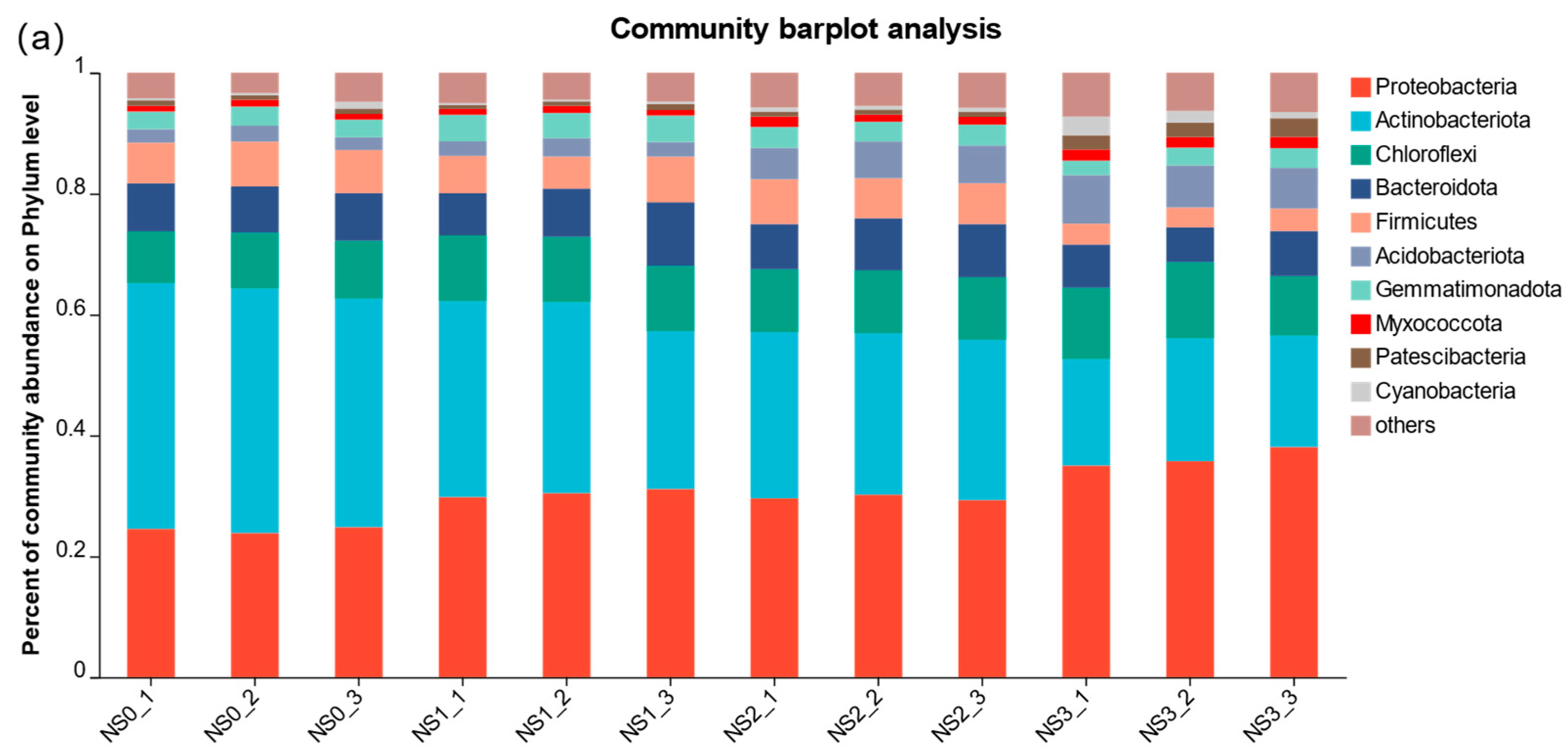
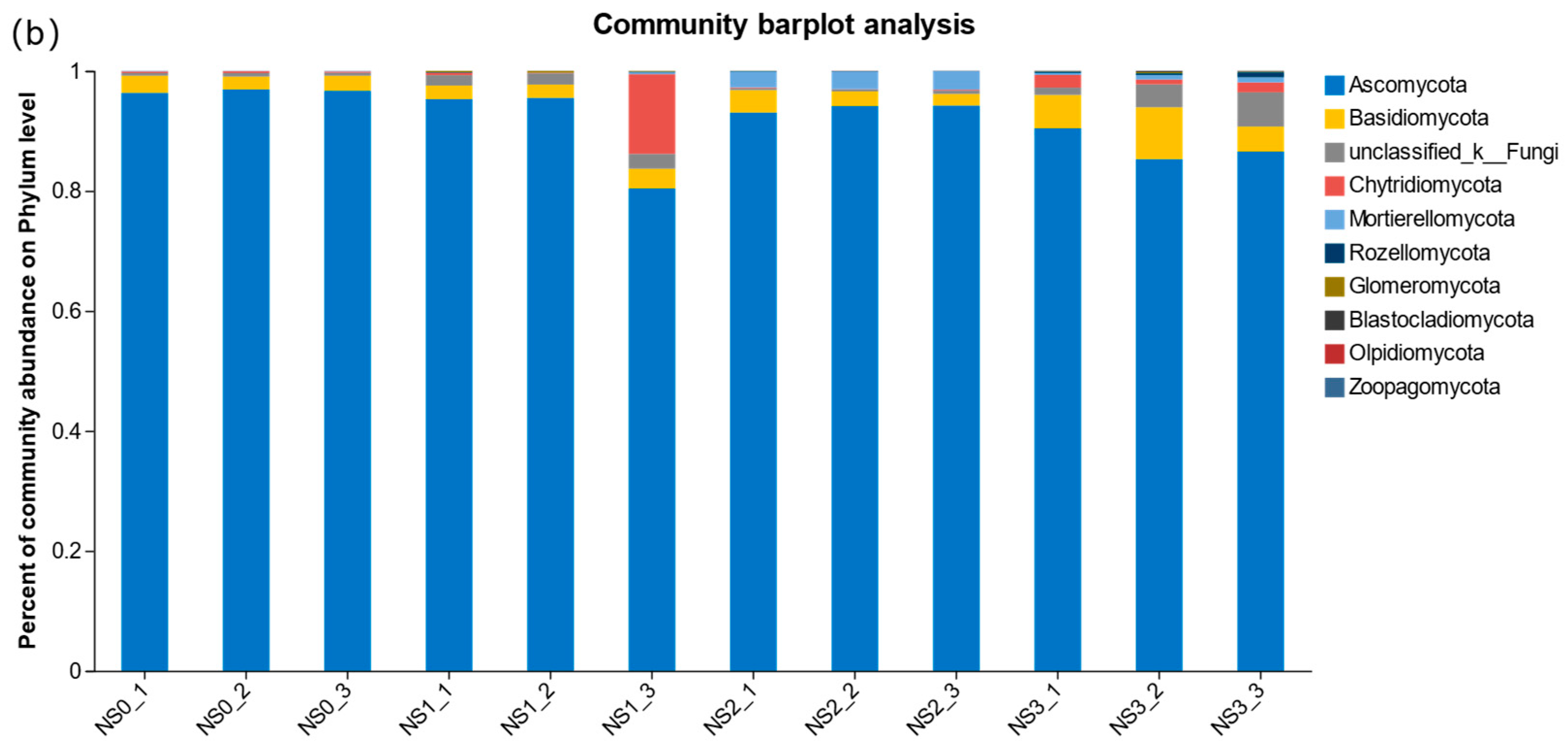
3.4.2. Effects of the Horizontal Community Composition of Soil Bacteria and Fungi at Different Periods
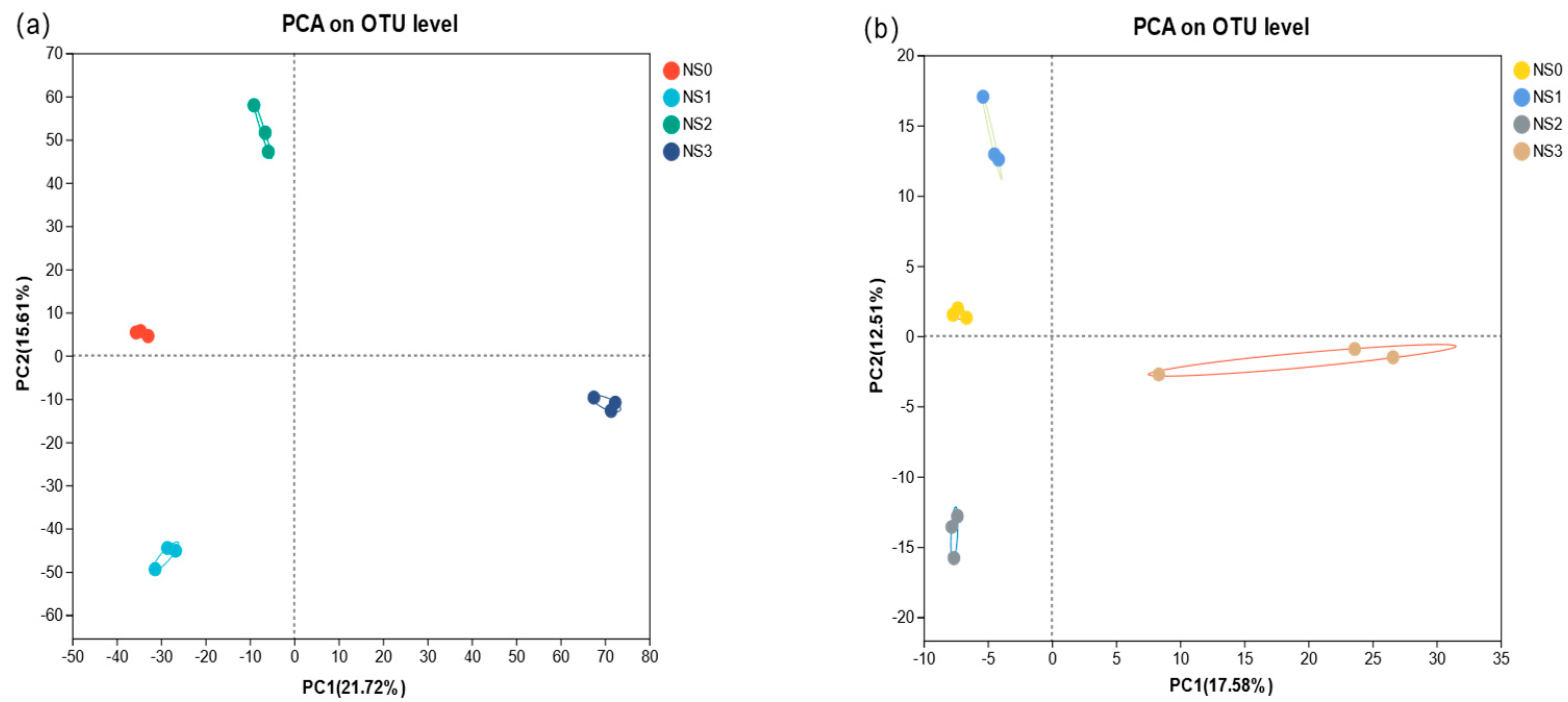
3.4.3. Relationship Between Soil Samples and Species at Different Periods
3.5. Difference Analysis of Microbial Fertilizer on Microbial Community Structure in Saline–Alkali Soil
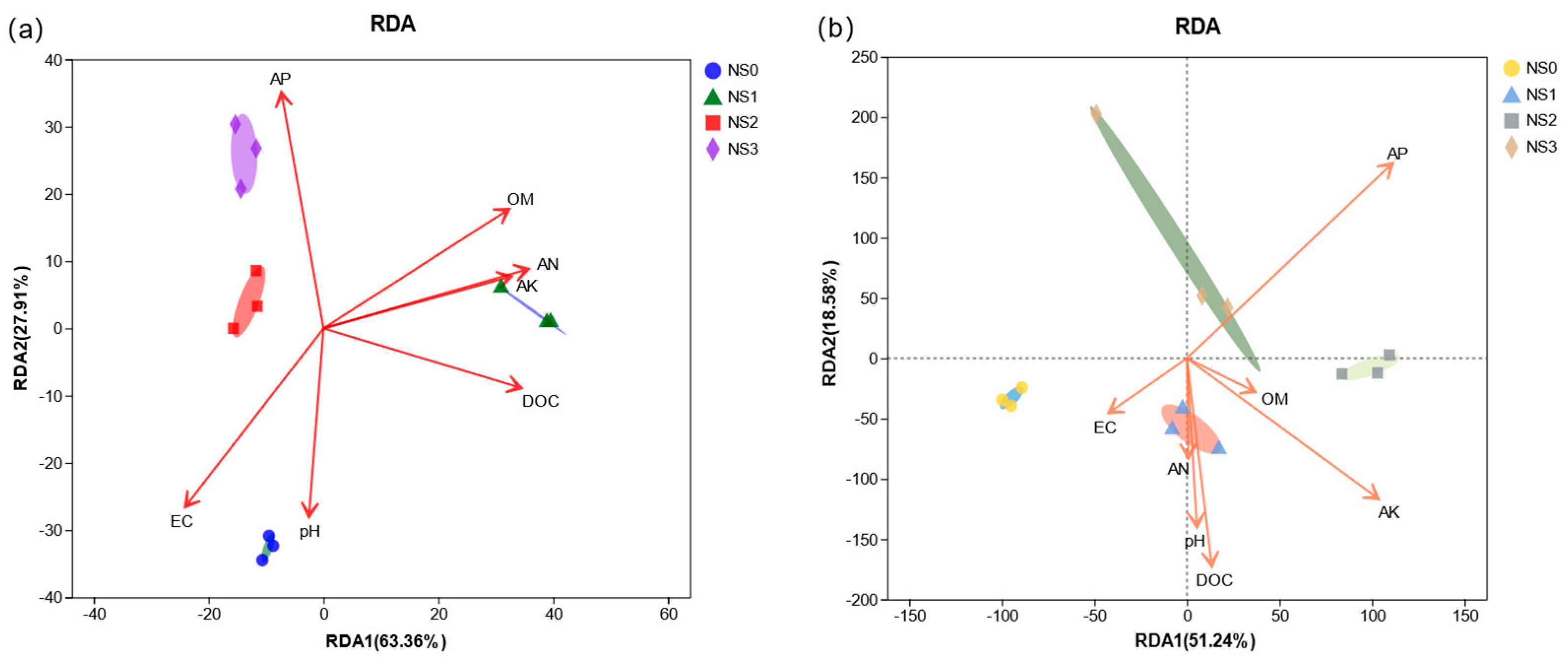
3.6. Correlation Analysis of Microbial Fertilizer on Physicochemical Properties and Microbial Communities in Saline–Alkali Soil
4. Discussion
4.1. Effects of Microbial Fertilizer on Physicochemical Properties and Enzyme Activities in Saline–Alkali Soil
4.2. Effects of Microbial Fertilizer on Microbial Diversity and Community Structure and Composition in Saline–Alkali Soil
4.3. Differences in Soil Microbial Community Structure and the Relationship to Soil Physicochemical Properties
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, Z.; Shang, H.; Han, F.; Zhang, M.; Li, Q.; Zhou, W. Improvement of nitrogen and phosphorus availability by Pseudoalteromonas sp. during salt-washing in saline-alkali soil. Appl. Soil Ecol. 2021, 168, 104117. [Google Scholar] [CrossRef]
- Shahbaz, M.; Ashraf, M. Improving salinity tolerance in cereals. Crit. Rev. Plant Sci. 2013, 32, 237–249. [Google Scholar] [CrossRef]
- Mathur, N.; Singh, J.; Bohra, S.; Vyas, A. Arbuscular mycorrhizal status of medicinal halophytes in saline areas of Indian Thar Desert. Int. J. Soil Sci. 2007, 2, 119–127. [Google Scholar]
- Yang, C.; Zhao, Y.; Cao, W.; Xing, M.; Xu, X.; Wang, Z.; Sun, J. Metagenomic analysis reveals antibiotic resistance genes and virulence factors in the saline-alkali soils from the Yellow River Delta, China. Environ. Res. 2022, 214, 113823. [Google Scholar] [CrossRef]
- Bao, X.; Chong, P.; He, C.; Wang, X.; Zhang, F. Mechanism on the promotion of host growth and enhancement of salt tolerance by Bacillaceae isolated from the rhizosphere of Reaumuria soongorica. Front. Microbiol. 2024, 31, 1408622. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, L.; Kellner, K. Restoring degraded patches in a semi-arid rangeland of South Africa. J. Arid. Environ. 2005, 61, 497–511. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, F.; Chen, Y.; Xu, T.; Cheng, Z.; Liang, J. Assessment of reclamation treatments of abandoned farmland in an arid region of China. Sustainability 2016, 8, 1183. [Google Scholar] [CrossRef]
- You, Y.; Wang, J.; Huang, X.; Tang, Z.; Liu, S.; Sun, O. Relating microbial community structure to functioning in forest soil organic carbon transformation and turnover. Ecol. Evol. 2014, 4, 633–647. [Google Scholar] [CrossRef]
- Zhang, G.; Bai, J.; Zhai, Y.; Jia, J.; Zhao, Q.; Wang, W.; Hu, X. Microbial diversity and functions in saline soils: A review from a biogeochemical perspective. J. Adv. Res. 2024, 59, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Budamagunta, V.; Shameem, N.; Irusappan, S.; Parray, J.A.; Thomas, M.; Marimuthu, S.; Kirubakaran, R.; Arul Jothi, K.N.; Sayyed, R.Z.; Show, P.L. Nanovesicle and extracellular polymeric substance synthesis from the remediation of heavy metal ions from soil. Environ. Res. 2023, 219, 114997. [Google Scholar] [CrossRef]
- Shan, S.; Wei, Z.; Cheng, W.; Du, D.; Zheng, D.; Ma, G. Biofertilizer based on halotolerant microorganisms promotes the growth of rice plants and alleviates the effects of saline stress. Front. Microbiol. 2023, 14, 1165631. [Google Scholar] [CrossRef] [PubMed]
- Ying, J.; Li, X.; Wang, N.; Lan, Z.; He, J.; Bai, B. Contrasting effects of nitrogen forms and soil pH on ammonia oxidizing microorganisms and their responses to long-term nitrogen fertilization in a typical steppe ecosystem. Soil Biol. Biochem. 2017, 107, 10–18. [Google Scholar] [CrossRef]
- Duan, C.; Fang, L.; Yang, C.; Chen, W.; Cui, Y.; Li, S. Reveal the response of enzyme activities to heavy metals through in situ zymography. Ecotoxicol. Environ. Saf. 2018, 156, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zhao, Q.; Chang, D.; Ndayisenga, F.; Yu, Z. Assessing the effect of physicochemical properties of saline and sodic soil on soil microbial communities. Agriculture 2022, 12, 782. [Google Scholar] [CrossRef]
- Guo, X.; Li, H.; Hu, Y. Effects of biochar on the diversity and community structure of soil fungi in intercropping system. Appl. Ecol. Environ. Res. 2019, 17, 8817–8834. [Google Scholar] [CrossRef]
- Jeanbille, M.; Buée, M.; Bach, C.; Cébron, A.; Frey-Klett, P.; Turpault, M.P.; Uroz, S. Soil Parameters Drive the Structure, Diversity and Metabolic Potentials of the Bacterial Communities Across Temperate Beech Forest Soil Sequences. Microb. Ecol. 2016, 71, 482–493. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ren, W.; Yang, F.; Niu, L.; Li, Z.; Zhang, M. Effects of different tillage and residue retention measures on silage maize yield and quality and soil phosphorus in Karst areas. Agronomy 2023, 13, 2306. [Google Scholar] [CrossRef]
- Wu, Y.; Yu, Y. Soil carbon, nitrogen and phosphorus fractions and response to microorganisms and mineral elements in Zanthoxylum planispinum‘dintanensis’ plantations at different altitudes. Agronomy 2023, 13, 558. [Google Scholar] [CrossRef]
- Al-Jaloud, A.A.; Hussain, G.; Al-Saati, A.J.; Karimullah, S. Effect of wastewaters on plant growth and soil properties. Arid Soil Res. Rehabil. 1993, 7, 173–179. [Google Scholar] [CrossRef]
- Bhaduri, D.; Saha, A.; Desai, D.; Meena, H.N. Restoration of carbon and microbial activity in salt-induced soil by application of peanut shell biochar during short-term incubation study. Chemosphere 2016, 148, 86–98. [Google Scholar] [CrossRef]
- Jorge-Mardomingo, I.; Soler-Rovira, P.; Casermeiro, M.Á.; de la Cruz, M.T.; Polo, A. Seasonal changes in microbial activity in a semiarid soil after application of a high dose of different organic amendments. Geoderma 2013, 206, 40–48. [Google Scholar] [CrossRef]
- Tu, C.; Guan, F.; Sun, Y.; Guo, P.; Liu, Y.; Li, L.; Scheckel, K.G.; Luo, Y. Stabilizing effects on a Cd polluted coastal wetland soil using calcium polysulphide. Geoderma 2018, 332, 190–197. [Google Scholar] [CrossRef]
- Xie, H.; Yang, X.; Drury, C.; Yang, J.; Zhang, X. Predicting soil organic carbon and total nitrogen using mid- and near-infrared spectra for Brookston clay loam soil in Southwestern Ontario, Canada. Can. J. Soil Sci. 2011, 91, 53–63. [Google Scholar] [CrossRef]
- Xiong, W.; Zhao, Q.; Zhao, J.; Xun, W.; Li, R.; Zhang, R.; Wu, H.; Shen, Q. Different continuous cropping spans significantly affect microbial community membership and structure in a vanilla-grown soil as revealed by deep pyrosequencing. Microb. Ecol. 2015, 70, 209–218. [Google Scholar] [CrossRef]
- Liu, C.; Zhao, D.; Ma, W.; Guo, Y.; Wang, A.; Wang, Q.; Lee, D. Denitrifying sulfide removal process on high-salinity wastewaters in the presence of Halomonas sp. Appl. Microbiol. Biotechnol. 2016, 100, 1421–1426. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.; Tiedje, J. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Barberan, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Chen, Y.; Zhang, F. Effect of reclamation of abandoned salinized farmland on soil bacterial communities in arid northwest China. Sci. Total. Environ. 2018, 630, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shen, H.; He, X.; Thomas, B.W.; Lupwayi, N.Z.; Hao, X.; Thomas, M.C.; Shi, X. Fertilization shapes bacterial community structure by alteration of soil pH. Front. Microbiol. 2017, 8, 1325. [Google Scholar] [CrossRef]
- Wei, X.; Xie, B.; Wan, C.; Song, R.; Zhong, W.; Xin, S.; Song, K. Enhancing soil health and plant growth through microbial fertilizers: Mechanisms, benefits, and sustainable agricultural practices. Agronomy 2024, 14, 609. [Google Scholar] [CrossRef]
- Yin, R.; Deng, H.; Wang, H.; Zhang, B. Vegetation type affects soil enzyme activities and microbial functional diversity following re-vegetation of a severely eroded red soil in sub-tropical China. Catena 2014, 115, 96–103. [Google Scholar] [CrossRef]
- Peng, Y.; Zhang, H.; Lian, J.; Zhang, W.; Li, G.; Zhang, J. Combined application of organic fertilizer with microbial inoculum improved aggregate formation and salt leaching in a secondary salinized soil. Plants 2023, 12, 2945. [Google Scholar] [CrossRef]
- Yang, W.; Zhou, H.; Gu, J.; Liao, B.; Zhang, J.; Wu, P. Application of rapeseed residue increases soil organic matter, microbial biomass, and enzyme activity and mitigates cadmium pollution risk in paddy fields. Environ. Pollut. 2020, 264, 114681. [Google Scholar] [CrossRef]
- Frankenberger, W.T.; Dick, W.A. Relationships between enzyme activities and microbial growth and activity indices in soil. Soil Sci. Soc. Am. J. 1983, 47, 945–951. [Google Scholar] [CrossRef]
- Yang, D.; Tang, L.; Cui, Y.; Chen, J.; Liu, L.; Guo, C. Saline-alkali stress reduces soil bacterial community diversity and soil enzyme activities. Ecotoxicology 2022, 31, 1356–1368. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Zhu, L.; Wang, J.; Wang, J.; Xie, H.; Lv, D. Enzymatic activities and microbial biomass in black soil as affected by azoxystrobin. Environ. Earth Sci. 2015, 74, 1353–1361. [Google Scholar] [CrossRef]
- Cruz, C.; Cardoso, P.; Santos, J.; Matos, D.; Sá, C.; Figueira, E. Application of plant growth-promoting bacteria from cape verde to increase maize tolerance to salinity. Antioxidants 2023, 12, 488. [Google Scholar] [CrossRef]
- Pineda, A.; Kaplan, I.; Bezemer, T.M. Steering soil microbiomes to suppress aboveground insect pests. Trends Plant Sci. 2017, 22, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Araujo, R.; Dunlap, C.; Franco, C.M.M. Analogous wheat root rhizosphere microbial successions in field and greenhouse trials in the presence of biocontrol agents Paenibacillus peoriae SP9 and Streptomyces fulvissimus FU14. Mol. Plant Pathol. 2020, 21, 622–635. [Google Scholar] [CrossRef]
- Wang, F.; Wang, Q.; Adams, C.A.; Sun, Y.; Zhang, S. Effects of microplastics on soil properties: Current knowledge and future perspectives. J. Hazard. Mater. 2022, 424, 127531. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, A.; Ueda, Y.; Takase, H.; Yazaki, K. Pyrosequencing assessment of rhizosphere fungal communities from a soybean field. Can. J. Microbiol. 2014, 60, 687–690. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ding, C.; Zhang, T.; Wang, X. Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 2014, 72, 11–18. [Google Scholar] [CrossRef]
- Buyer, J.S.; Roberts, D.P.; Russek-Cohen, E. Soil and plant effects on microbial community structure. Can. J. Microbiol. 2002, 48, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Nunan, N.; Daniell, T.J.; Singh, B.K.; Papert, A.; McNicol, J.W.; Prosser, J.I. Links between plant and rhizoplane bacterial communities in grassland soils, characterized using molecular techniques. Appl. Environ. Microbiol. 2005, 71, 6784–6792. [Google Scholar] [CrossRef]
- Li, X.; Lu, Q.; Li, D.; Wang, D.; Ren, X.; Yan, J.; Ahmed, T.; Li, B. Effects of different microbial fertilizer on growth and rhizosphere soil properties of corn in newly reclaimed land. Plants 2022, 11, 1978. [Google Scholar] [CrossRef]
- Hou, Y.; Zeng, W.; Hou, M.; Wang, Z.; Luo, Y.; Lei, G.; Zhou, B.; Huang, J. Responses of the soil microbial community to salinity stress in maize fields. Biology 2021, 10, 1114. [Google Scholar] [CrossRef]
- Costa-Broseta, Á.; Castillo, M.; León, J. Nitrite reductase 1 is a target of nitric oxide-mediated post-translational modifications and controls nitrogen flux and growth in arabidopsis. Int. J. Mol. Sci. 2020, 21, 7270. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jiang, N.; Condron, L.M.; Dunfield, K.E.; Chen, Z.; Wang, J.; Chen, L. Impact of long-term phosphorus fertilizer inputs on bacterial phoD gene community in a maize field, Northeast China. Sci. Total Environ. 2019, 669, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
| Property | Liquid Organic Fertilizer | Microbial Fertilizer |
|---|---|---|
| Ingredients | Biogas slurry, a small amount of amino acids and humic acid. | Functional strain (effective viable bacteria count ≥ 1 × 107 cfu·mL−1), biogas slurry as carrier, a small amount of amino acids and humic acid. |
| pH | 7.1 | 6.5 |
| OM (g·kg−1) | 24.5 | 28.6 |
| DOC (g·kg−1) | 14.2 | 16.5 |
| N + P2O5 + K2O (g·kg−1) | ≥26.0 | ≥18.0 |
| Property | NS0 | NS1 | NS2 | NS3 |
|---|---|---|---|---|
| pH | 8.62 ± 0.1 a | 8.27 ± 0.03 b | 8.6 ± 0.13 a | 7.87 ± 0.04 c |
| EC (µs·cm−1) | 1269.33 ± 13.5 a | 500.67 ± 25.32 d | 1086 ± 54 b | 655.33 ± 29.26 c |
| OM (g·kg−1) | 2.61 ± 0.01 d | 4.6 ± 0.03 a | 2.96 ± 0.09 c | 3.51 ± 0.12 b |
| DOC (mg·kg−1) | 47.07 ± 1.64 b | 72.59 ± 0.05 a | 47.95 ± 0.19 b | 33.52 ± 1.02 c |
| AN (mg·kg−1) | 9.53 ± 0.25 c | 19.37 ± 0.21 a | 9.33 ± 0.5 c | 11.73 ± 0.47 b |
| AP (mg·kg−1) | 6.6 ± 0.26 c | 8.03 ± 0.23 b | 8.5 ± 0.3 b | 10.23 ± 0.25 a |
| AK (mg·kg−1) | 59.88 ± 0.33 d | 137.79 ± 0.78 a | 95.95 ± 2.34 b | 66.99 ± 0.96 c |
| K+ (mg·kg−1) | 10.62 ± 0.17 c | 32.17 ± 0.08 a | 13.04 ± 1.09 b | 10.11 ± 0.51 c |
| Na+ (mg·kg−1) | 2551.91 ± 28.07 b | 5249.87 ± 14.55 a | 982.68 ± 1.87 c | 325.97 ± 8.92 d |
| Ca2+ (mg·kg−1) | 83.33 ± 0.21 c | 438.95 ± 0.81 a | 40.39 ± 0.64 d | 99.17 ± 0.27 b |
| Mg2+ (mg·kg−1) | 61.65 ± 0.43 a | 41.24 ± 0.26 b | 21.24 ± 0.25 c | 41.47 ± 0.17 b |
| SO42− (mg·kg−1) | 122.4 ± 4.62 a | 104.03 ± 4.13 b | 66.37 ± 6.68 c | 29 ± 2.42 d |
| CO32− (mg·kg−1) | 0 | 0 | 0 | 0 |
| HCO3− (mg·kg−1) | 311.67 ± 31.12 a | 297.1 ± 21.38 a | 326.13 ± 6.12 a | 240.87 ± 6.1 b |
| Cl− (mg·kg−1) | 2330.57 ± 35.05 b | 3936.47 ± 11.07 a | 711.87 ± 15.54 c | 444.97 ± 6.31 d |
| Genus | Property | NS0 | NS1 | NS2 | NS3 |
|---|---|---|---|---|---|
| Bacteria | Shannon | 6.07 ± 0.05 c | 6.25 ± 0.05 b | 6.63 ± 0.06 a | 6.73 ± 0.07 a |
| Simpson | 0.01 ± 0 a | 0.01 ± 0 a | 0.01 ± 0 a | 0.01 ± 0 a | |
| Ace | 3276.64 ± 17.93 d | 3595.35 ± 27.38 c | 4082.68 ± 164.01 b | 4286.01 ± 31.36 a | |
| Chao1 | 3244.13 ± 21.97 d | 3523.77 ± 15.44 c | 4013.57 ± 148.15 b | 4198.77 ± 30.92 a | |
| Sobs | 2841.33 ± 34.31 d | 3115 ± 36.35 c | 3534 ± 107.68 b | 3785.67 ± 54.31 a | |
| Coverage | 0.99 ± 0 a | 0.99 ± 0 a | 0.99 ± 0 a | 0.99 ± 0 a | |
| Fungi | Shannon | 3.25 ± 0.07 c | 3.86 ± 0.02 a | 3.41 ± 0.05 bc | 3.72 ± 0.37 ab |
| Simpson | 0.07 ± 0.01 a | 0.04 ± 0 a | 0.06 ± 0 a | 0.06 ± 0.04 a | |
| Ace | 156.69 ± 9.66 d | 186.38 ± 4.94 c | 223.48 ± 18.08 b | 310.75 ± 20.16 a | |
| Chao1 | 156.5 ± 9.53 d | 186.44 ± 5.59 c | 223.46 ± 19.21 b | 312.98 ± 18.1 a | |
| Sobs | 156 ± 10.39 d | 184.33 ± 4.73 c | 220.33 ± 17.39 b | 303.33 ± 17.21 a | |
| Coverage | 1 ± 0 a | 1 ± 0 a | 1 ± 0 a | 1 ± 0 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, X.; Zhang, X.; Yang, G.; Wang, Y.; Liu, Q.; Song, J. Effects of Microbial Fertilizer Application on Soil Ecology in Saline–Alkali Fields. Agronomy 2025, 15, 14. https://doi.org/10.3390/agronomy15010014
Tian X, Zhang X, Yang G, Wang Y, Liu Q, Song J. Effects of Microbial Fertilizer Application on Soil Ecology in Saline–Alkali Fields. Agronomy. 2025; 15(1):14. https://doi.org/10.3390/agronomy15010014
Chicago/Turabian StyleTian, Xingguo, Xiu Zhang, Guoping Yang, Yu Wang, Qianru Liu, and Jingjing Song. 2025. "Effects of Microbial Fertilizer Application on Soil Ecology in Saline–Alkali Fields" Agronomy 15, no. 1: 14. https://doi.org/10.3390/agronomy15010014
APA StyleTian, X., Zhang, X., Yang, G., Wang, Y., Liu, Q., & Song, J. (2025). Effects of Microbial Fertilizer Application on Soil Ecology in Saline–Alkali Fields. Agronomy, 15(1), 14. https://doi.org/10.3390/agronomy15010014






