Genetic Uniqueness and Pomological Diversity Among the Apple Accessions Maintained Within the Croatian National Clonal Germplasm Repository
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. SSR Genotyping
2.3. SNP Genotyping
2.4. Pomological Characterization
2.5. Statistical Analyses
2.6. Biostatistical Analyses
2.6.1. SSR Data Analysis
2.6.2. SNP Data Analysis
3. Results and Discussion
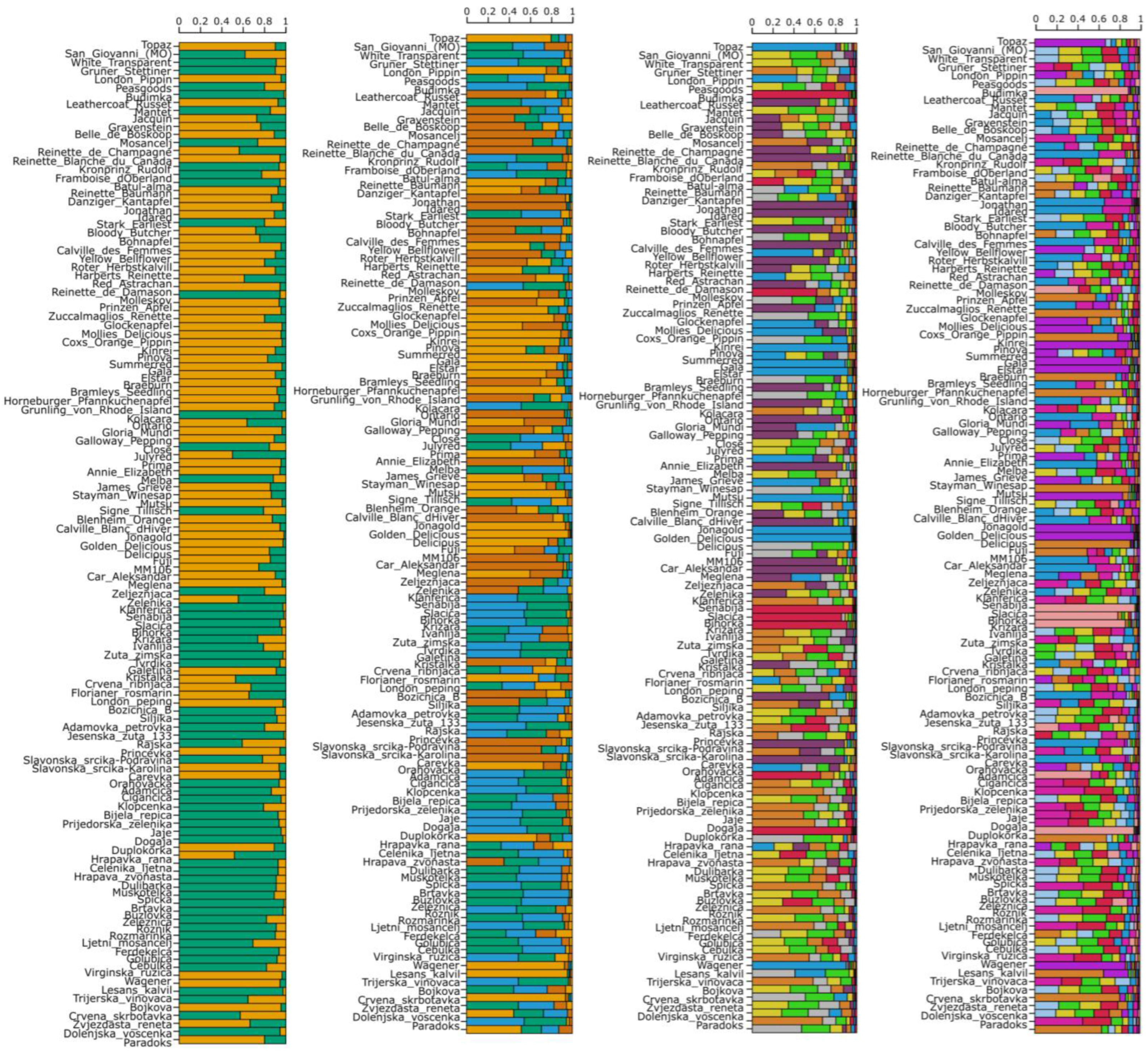
3.1. Identifying Redundancies Within the Collection
3.1.1. Genetic Identity
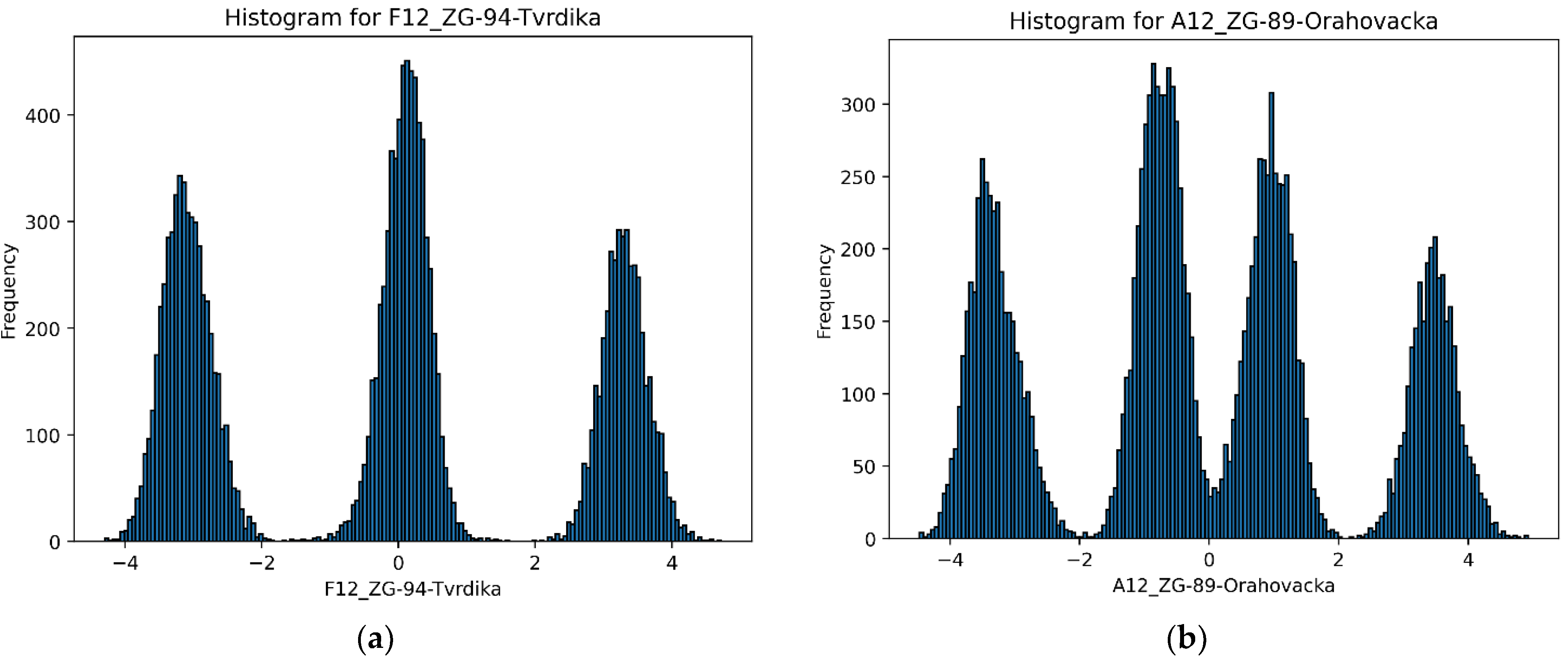
3.1.2. Ploidy Assessment Based on SSRs
3.1.3. SSR Polymorphism
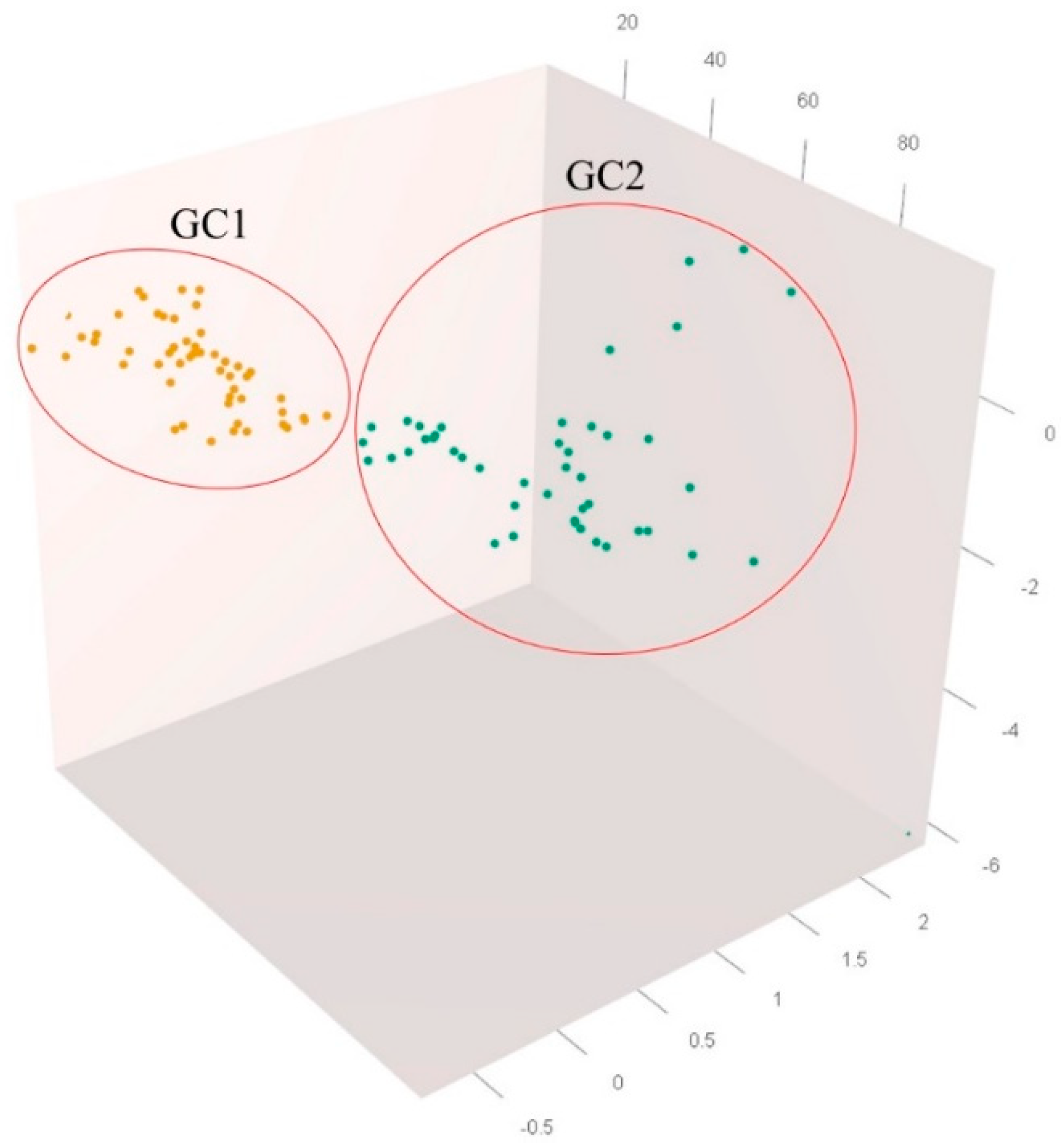
3.1.4. Genetic Structure
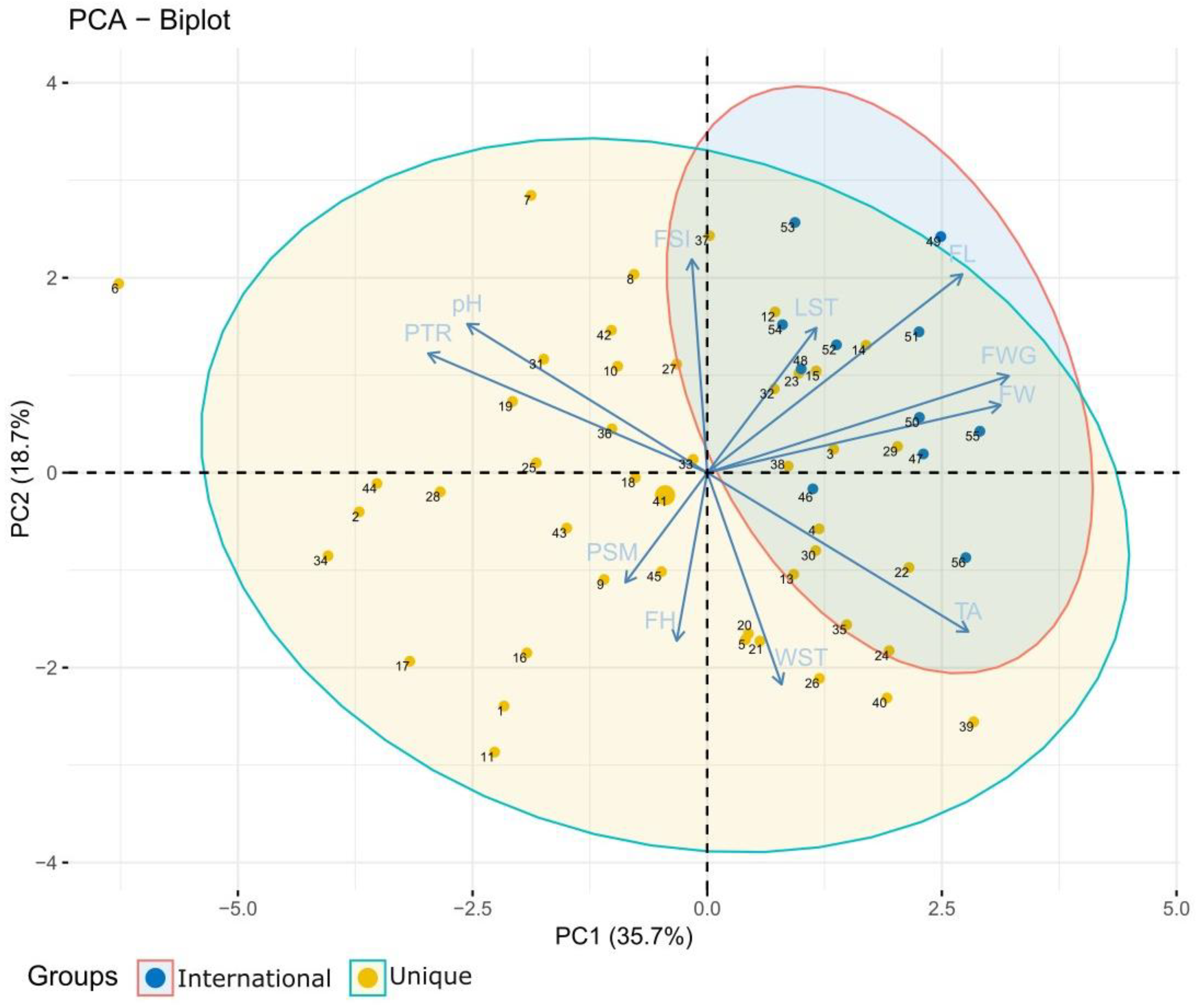
3.2. Pomological Characterization
3.3. Genetic Identity and Pedigree Reconstruction
3.3.1. Comparison of SSR and SNP Marker Systems
3.3.2. Ploidy Assessment Based on SNPs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Baric, S.; Storti, A.; Hofer, M.; Guerra, W.; Dalla Via, J. Molecular Genetic Identification of Apple Cultivars Based on Microsatellite DNA Analysis. I. The Database of 600 Validated Profiles. Erwerbs-Obstbau 2020, 62, 117–154. [Google Scholar] [CrossRef]
- Meland, M.; Aksic, M.F.; Frøynes, O.; Konjic, A.; Lasic, L.; Pojskic, N.; Gasi, F. Genetic Identity and Diversity of Apple Accessions within a Candidate Collection for the Norwegian National Clonal Germplasm Repository. Horticulturae 2022, 8, 630. [Google Scholar] [CrossRef]
- Ganopoulos, I.; Tourvas, N.; Xanthopoulou, A.; Aravanopoulos, F.A.; Avramidou, E.; Zambounis, A.; Tsaftaris, A.; Madesis, P.; Sotiropoulos, T.; Koutinas, N. Phenotypic and molecular characterization of apple (Malus × domestica Borkh.) genetic resources in Greece. Sci. Agric. 2018, 75, 509–518. [Google Scholar] [CrossRef]
- Čiček, D.; Vujević, P.; Mlinović, B.; Halapija Kazija, D.; Jelačić, T. Pomological characteristics of traditional cultivars of apples in the intensive breeding. In Proceedings of the 10th International Meeting Plant Breeding, Seed and Nursery Production, Sveti Martin na Muri, Croatia, 8–10 November 2017; Matotan, Z., Ed.; Croatian Society of Agronomists: Zagreb, Croatia, 2017; pp. 75–76. [Google Scholar]
- Jakobek, L.; Ištuk, J.; Buljeta, I.; Voća, S.; Žlabur, J.Š.; Babojelić, M.S. Traditional, Indigenous Apple Varieties, a Fruit with Potential for Beneficial Effects: Their Quality Traits and Bioactive Polyphenol Contents. Foods 2020, 9, 52. [Google Scholar] [CrossRef]
- Lončarić, A.; Skendrović Babojelić, M.; Kovač, T.; Šarkanj, B. Pomological Properties and Polyphenol Content of Conventional and Traditional Apple Cultivars from Croatia. Food Health Dis. Sci. Prof. J. Nutr. Diet. 2019, 8, 19–24. [Google Scholar]
- Gotal Skoko, A.; Skendrović Babojelić, M.; Šarkanj, B.; Flanjak, I.; Tomac, I.; Jozinović, A.; Babić, J.; Šubarić, D.; Sulyok, M.; Krska, R.; et al. Influence of Polyphenols on the Resistance of Traditional and Conventional Apple Varieties to Infection by Penicillium expansum during Cold Storage. Sustainability 2024, 16, 5019. [Google Scholar] [CrossRef]
- Dolker, T.; Kumar, D.; Chandel, J.S.; Angmo, S.; Chaurasia, O.P.; Stobdan, T. Phenological and Pomological Characteristics of native Apple (Malus domestica Borkh.) cultivars of Trans Himalayan ladakh India. Def. Life Sci. J. 2021, 6, 64–69. [Google Scholar] [CrossRef]
- Skendrović Babojelić, M.; Korent, P.; Šindrak, Z.; Jemrić, T. Pomological properties and fruit quality of traditional apple varieties. Bull. Plant Prot. 2014, 37, 20–27. [Google Scholar]
- Božović, D.; Lazović, B.; Ercisli, S.; Adakalić, M.; Jaćimović, V.; Sezer, I.; Koc, A. Morphological characterization of autochthonous Apple Genetic Resources in Montenegro. Erwerbs-Obstbau 2015, 58, 93–102. [Google Scholar] [CrossRef]
- Arnal, A.; Lazaro, A.; Tardío, J. Morphological characterization of 23 Malus domestica Borkh. cultivars from central Spain. Plant Genet. Resour. 2022, 3, 22–37. [Google Scholar] [CrossRef]
- Karatas, N. Morphological, sensory and biochemical characteristics of summer apple genotypes. Braz. J. Biol. 2022, 82, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cice, D.; Ferrara, E.; Magri, A.; Adiletta, G.; Capriolo, G.; Rega, P.; Di Matteo, M.; Petriccione, M. Autochthonous Apple Cultivars from the Campania Region (Southern Italy): Bio-Agronomic and Qualitative Traits. Plants 2023, 12, 1160. [Google Scholar] [CrossRef] [PubMed]
- Nybom, H.; Weising, K. DNA-Based Identification of Clonally Propagated Cultivars. In Plant Breeding Reviews; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2010; Volume 34, pp. 221–295. [Google Scholar]
- Guilford, P.; Prakash, S.; Zhu, J.; Rikkerink, E.; Gardiner, S.; Bassett, H. Microsatellites in Malus × domestica (apple): Abundance, polymorphism and cultivar identification. Theor. Appl. Genet. 1997, 94, 249–254. [Google Scholar] [CrossRef]
- Pereira-Lorenzo, S.; Ramos-Cabrer, A.M.; Díaz-Hernández, M.B. Evaluation of genetic identity and variation of local apple cultivars (Malus × domestica Borkh.) from Spain using microsatellite markers. Genet. Resour. Crop. Evolut. 2007, 54, 405–420. [Google Scholar] [CrossRef]
- Pereira-Lorenzo, S.; Ramos-Cabrer, A.M.; González-Díaz, A.J.; Díaz-Hernández, M.B. Genetic assessment of local apple cultivars from La Palma, Spain, using simple sequence repeats (SSRs). Sci. Hortic. 2008, 117, 160–166. [Google Scholar] [CrossRef]
- Urrestarazu, J.; Miranda, C.; Santesteban, L.G.; Royo, J.B. Genetic diversity and structure of local apple cultivars from Northeastern Spain assessed by microsatellite markers. Tree Genet. Genomes 2012, 8, 1163–1180. [Google Scholar] [CrossRef]
- Garkava-Gustavsson, L.; Kolodinska Brantestam, A.; Sehic, J.; Nybom, H. Molecular characterisation of indigenous Swedish apple cultivars based on SSR and S-allele analysis. Hereditas 2008, 145, 99–112. [Google Scholar] [CrossRef]
- Gharghani, A.; Zamani, Z.; Talaie, A.; Oraguzie, N.C.; Fatahi, R.; Hajnajari, H.; Wiedow, C.; Gardiner, S.E. Genetic identity and relationships of Iranian apple (Malus × domestica Borkh.) cultivars and landraces, wild Malus species and representative old apple cultivars based on simple sequence repeat (SSR) marker analysis. Genet. Resour. Crop Evol. 2009, 56, 829–842. [Google Scholar] [CrossRef]
- van Treuren, R.; Kemp, H.; Ernsting, G.; Jongejans, B.; Houtman, H.; Visser, L. Microsatellite genotyping of apple (Malus × domestica Borkh.) genetic resources in the Netherlands: Application in collection management and variety identification. Genet. Resour. Crop Evol. 2010, 57, 853–865. [Google Scholar] [CrossRef]
- Mažeikienė, I.; Šikšnianienė, J.B.; Baniulis, D.; Gelvonauskienė, D.; Frercks, B.; Starkus, A.; Žebrauskienė, A.; Stanys, V. SSR analysis based on molecular characterisation of apple germplasm in Lithuania. Zemdirbyste 2019, 106, 159–166. [Google Scholar] [CrossRef]
- Butiuc-Keul, A.; Coste, A.; Farkas, A.; Cristea, V.; Isac, V.; Halmagyi, A. Molecular characterization of apple (Malus × domestica Borkh.) genotypes originating from three complementary conservation strategies. Turk. J. Agric. For. 2019, 43, 464–477. [Google Scholar] [CrossRef]
- Gaši, F.; Kurtović, M.; Šimon, S.; Pecina, M.; Pejić, I. Genetic and pomological analyses of apple germplasm in Bosnia and Herzegovina. Acta Hortic. 2010, 859, 111–116. [Google Scholar] [CrossRef]
- Gaši, F.; Šimon, S.; Pojskić, N.; Kurtović, M.; Pejic, I. Genetic assessment of apple germplasm in Bosnia and Herzegovina using microsatellite and morphologic markers. Sci. Hortic. 2010, 126, 164–171. [Google Scholar] [CrossRef]
- Gaši, F.; Šimon, S.; Pojskić, N.; Kurtović, M.; Pejić, I. Analysis of morphological variability in Bosnia and Herzegovina’s autochthonous apple germplasm. Int. J. Food Agric. Environ. 2011, 9, 444–448. [Google Scholar]
- Gaši, F.; Šimon, S.; Pojskić, N.; Kurtović, M.; Pejić, I.; Meland, M.; Kaiser, C. Evaluation of apple (Malus × domestica) genetic resources in Bosnia and Herzegovina using microsatellite markers. HortScience 2013, 48, 13–21. [Google Scholar] [CrossRef]
- Gaši, F.; Žulj Mihaljević, M.; Šimon, S.; Grahić, J.; Pojskić, N.; Kurtović, M.; Nikolić, D.; Pejić, I. Genetic structure of apple accessions maintained ex situ in Bosnia and Herzegovina examined by microsatellite markers. Genetika 2013, 45, 467–478. [Google Scholar] [CrossRef]
- Halapija Kazija, D.; Jelačić, T.; Vujević, P.; Milinović, B.; Čiček, D.; Biško, A.; Pejić, I.; Šimon, S.; Žulj Mihaljević, M.; Pecina, M.; et al. Plum germplasm in Croatia and neighboring countries assessed by microsatellites and DUS descriptors. Tree Genet. Genomes 2014, 10, 761–778. [Google Scholar] [CrossRef]
- Poljuha, D.; Kralj, I.; Krapac, M.; Klepo, T.; Strikić, F.; Radunić, M.; Benčić, Đ.; Marinović Peričević, M.; Žeravica, D.I.; Car, M.; et al. Genetic biodiversity of Croatian olives (Olea europaea L.) analyzed by SSR markers: A step toward the sustainable management of national genetic resources. In Proceedings of the 4th Congress of Croatian Geneticists with International Participation, Krk, Croatia, 26–29 September 2018; Croatian Genetic Society: Zagreb, Croatia, 2018; p. 59. [Google Scholar]
- Poljuha, D.; Kralj, I.; Krapac, M.; Klepo, T.; Radunić, M.; Strikić, F.; Weber, T.; Ercisli, S.; Baruca Arbeiter, A.; Hladnik, M.; et al. Analysis of genetic diversity in Croatian fig (Ficus carica L.) germplasm using SSR markers. Acta Hortic. 2021, 1310, 35–40. [Google Scholar] [CrossRef]
- You, Q.; Yang, X.; Peng, Z.; Xu, L.; Wang, J. Development and Applications of a High Throughput Genotyping Tool for Polyploid Crops: Single Nucleotide Polymorphism (SNP) Array. Front. Plant Sci. 2018, 9, 104. [Google Scholar] [CrossRef]
- Chagné, D.; Crowhurst, R.N.; Troggio, M.; Davey, M.W.; Gilmore, B.; Lawley, C.; Vanderzande, S.; Hellens, R.P.; Kumar, S.; Cestaro, A.; et al. Genome-Wide SNP Detection, Validation, and Development of an 8K SNP Array for Apple. PLoS ONE 2012, 7, e31745. [Google Scholar] [CrossRef]
- Bianco, L.; Cestaro, A.; Sargent, D.J.; Banchi, E.; Derdak, S.; Di Guardo, M.; Salvi, S.; Jansen, J.; Viola, R.; Gut, I.; et al. Development and validation of a 20K single nucleotide polymorphism (SNP) whole genome genotyping array for apple (Malus × domestica Borkh). PLoS ONE 2014, 9, e110377. [Google Scholar] [CrossRef] [PubMed]
- Rymenants, M.; van de Weg, E.; Auwerkerken, A.; De Wit, I.; Czech, A.; Nijland, B.; Heuven, H.; De Storme, N.; Keulemans, W. Detection of QTL for apple fruit acidity and sweetness using sensorial evaluation in multiple pedigreed full-sib families. Tree Genet. Genomes 2020, 16, 71. [Google Scholar] [CrossRef]
- Bianco, L.; Cestaro, A.; Linsmith, G.; Muranty, H.; Denancé, C.; Théron, A.; Poncet, C.; Micheletti, D.; Kerschbamer, E.; Di Pierro, E.A.; et al. Development and validation of the Axiom(®) Apple480K SNP genotyping array. Plant Mol. Biol. 2016, 86, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Gilpin, L.; Røen, D.; Schubert, M.; Davik, J.; Rumpunen, K.; Gardli, K.A.; Hjeltnes, S.H.; Alsheikh, M. Genetic characterization of the Norwegian apple collection. Horticulturae 2023, 9, 575. [Google Scholar] [CrossRef]
- Howard, N.P.; Van de Weg, E.; Bedford, D.S.; Peace, C.P.; Vanderzande, S.; Clark, M.D.; Teh, S.L.; Cai, L.; Luby, J.J. Elucidation of the ‘Honeycrisp’ pedigree through haplotype analysis with a multi-family integrated SNP linkage map and a large apple (Malus x domestica) pedigree-connected SNP data set. Hortic. Res. 2017, 4, 17003. [Google Scholar] [CrossRef]
- Luby, J.J.; Howard, N.P.; Tillman, J.R.; Bedford, D.S. Extended pedigrees of apple cultivars from the University of Minnesota breeding program elucidated using SNP array markers. HortScience 2022, 57, 472–477. [Google Scholar] [CrossRef]
- Muranty, H.; Denancé, C.; Feugey, L.; Crépin, J.L.; Barbier, Y.; Tartarini, S.; Ordidge, M.; Troggio, M.; Lateur, M.; Nybom, H.; et al. Using whole-genome SNP data to reconstruct a large multi-generation pedigree in apple germplasm. BMC Plant Biol. 2020, 20, 2. [Google Scholar] [CrossRef]
- Chagné, D.; Kirk, C.; Whitworth, C.; Erasmuson, S.; Bicknell, R.; Sargent, D.J.; Kumar, S.; Troggio, M. Polyploid and aneuploid detection in apple using a single nucleotide polymorphism array. Tree Genet. Genomes 2015, 11, 94. [Google Scholar] [CrossRef]
- Urrestarazu, J.; Muranty, H.; Denancé, C.; Leforestier, D.; Ravon, E.; Guyader, A.; Guisnel, R.; Feugey, L.; Aubourg, S.; Celton, J.-M.; et al. Genome-Wide Association Mapping of Flowering and Ripening Periods in Apple. Front. Plant Sci. 2017, 8, 1923. [Google Scholar] [CrossRef]
- Coupel-Ledru, A.; Pallas, B.; Delalande, M.; Segura, V.; Guitton, B.; Muranty, H.; Durel, C.-E.; Regnard, J.-L.; Costes, E. Tree architecture, light interception and water-use related traits are controlled by different genomic regions in an apple tree core collection. New Phytol. 2022, 234, 209–226. [Google Scholar] [CrossRef]
- García-Fernández, B.; Dolcet-Sanjuan, R.; Micheletti, D.; Antón-Díaz, M.J.; Solsona, C.; Fernández, M.; Abad, X.; Dapena, E. Susceptibility evaluation to fire blight and genome-wide associations within a collection of asturian apple accessions. Plants 2023, 12, 4068. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Deng, C.H.; Molloy, C.; Kirk, C.; Plunkett, B.; Lin-Wang, K.; Allan, A.; Espley, R. Extreme-phenotype GWAS unravels a complex nexus between apple (Malus domestica) red-flesh colour and internal flesh browning. Fruit Res. 2022, 2, 1–14. [Google Scholar] [CrossRef]
- Skytte af Sätra, J.; Troggio, M.; Odilbekov, F.; Sehic, J.; Mattisson, H.; Hjalmarsson, I.; Ingvarsson, P.K.; Garkava-Gustavsson, L. Genetic Status of the Swedish Central collection of heirloom apple cultivars. Sci. Hortic. 2020, 272, 109599. [Google Scholar] [CrossRef]
- Larsen, B.; Gardner, K.; Pedersen, C.; Ørgaard, M.; Migicovsky, Z.; Myles, S.; Toldam-Andersen, T.B. Population structure, relatedness and ploidy levels in an apple gene bank revealed through genotyping-by-sequencing. PLoS ONE 2018, 13, e0201889. [Google Scholar] [CrossRef]
- Konjić, A.; Kurtović, M.; Grahić, J.; Pojskić, N.; Kalajdžić, A.; Gaši, F. Using High-Density SNP Array to Investigate Genetic Relationships and Structure of Apple Germplasm in Bosnia and Herzegovina. Horticulturae 2023, 9, 527. [Google Scholar] [CrossRef]
- Đurić, G.; Skytte af Sätra, J.; Gaši, F.; Konjić, A.; Flachowsky, H.; Howard, N.P.; Zeljković, M.K.; Garkava-Gustavsson, L. Genetic diversity of apple heirloom germplasm in Bosnia and Herzegovina, as revealed by SNP markers. Tree Genet. Genomes 2024, 20, 28. [Google Scholar] [CrossRef]
- Edge Garza, D.A.; Rowland, T.V.; Haendiges, S.; Peace, C. A high-throughput and cost-efficient DNA extraction protocol for the tree fruit crops of apple, sweet cherry, and peach relying on silica beads during tissue sampling. Mol. Breed. 2014, 34, 2225–2228. [Google Scholar] [CrossRef]
- R Studio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 12 May 2022).
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses, R Package Version 1.0.7. 2020. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 4 April 2024).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 4 April 2024)ISBN 978-3-319-24277-4.
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. dplyr: A Grammar of Data Manipulation, R Package Version 1.1.4. 2023. Available online: https://dplyr.tidyverse.org (accessed on 4 April 2024).
- Auguie, B. gridExtra: Miscellaneous Functions for “Grid” Graphics, Version 2.0.0. 2015. Available online: http://CRAN.R-project.org/package=gridExtra (accessed on 4 April 2024).
- Iannone, R.; Cheng, J.; Schloerke, B.; Hughes, E.; Lauer, A.; Seo, J.; Brevoort, K. gt: Easily Create Presentation-Ready Display Tables, R Package Version 0.10.1.9000. 2024. Available online: https://gt.rstudio.com (accessed on 4 April 2024).
- Drost, H.G. Philentropy: Information Theory and Distance Quantification with R. J. Open Source Softw. 2018, 3, 765. Available online: https://joss.theoj.org/papers/10.21105/joss.00765 (accessed on 4 April 2024). [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Hardy, O.J.; Vekemans, X. SPAGeDi: A versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2022, 2, 618–620. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; Von Holdt, B.M. Structure harvester: A Website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
- Ramasamy, R.K.; Ramasamy, S.; Bindroo, B.B.; Naik, V.G. STRUCTURE PLOT: A program for drawing elegant STRUCTURE bar plots in user friendly interface. SpringerPlus 2014, 3, 431. [Google Scholar] [CrossRef] [PubMed]
- Carey, V.J. gee: Generalized Estimation Equation Solver, R Package Version 4.13–20. 2019. Available online: https://CRAN.R-project.org/package=gee (accessed on 4 April 2024).
- Hahsler, M.; Chelluboina, S.; Hornik, K.; Buchta, C. The arules R-Package ecosystem: Analyzing interesting patterns from large transaction datasets. JMLR 2011, 12, 1977–1981. [Google Scholar]
- Chasalow, S. combinat: Routines for Combinatorics, R Package Version 0.0–8. 2015. Available online: https://CRAN.R-project.org/package=combinat (accessed on 4 April 2024).
- Thioulouse, J.; Dray, S.; Dufour, A.; Siberchicot, A.; Jombart, T.; Pavoine, S. Multivariate Analysis of Ecological Data with ade4; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar] [CrossRef]
- Murdoch, D.; Adler, D. Rgl: 3D Visualization Using OpenGL, R Package Version 0.108.3. 2021. Available online: https://CRAN.R-project.org/package=rgl (accessed on 4 April 2024).
- Ligges, U.; Mächler, M. Scatterplot3d—An R package for visualizing multivariate data. J. Stat. Softw. 2003, 8, 1–20. [Google Scholar] [CrossRef]
- Clark, L.; Jasieniuk, M. Polysat: An R package for polyploid microsatellite analysis. Mol. Ecol. Resour. 2011, 11, 562–566. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Oksanen, J.; Guillaume Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, R Package Version 2.5–7. 2020. Available online: https://cran.r-project.org/package=vegan (accessed on 4 April 2024).
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Denancé, C.; Muranty, H.; Durel, C.-E. MUNQ—Malus UNiQue Genotype Code for Grouping Apple Accessions Corresponding to a Unique Genotypic Profile. 2020. [CrossRef]
- Howard, N.P.; Peace, C.; Silverstein, K.A.T.; Poets, A.; Luby, J.J.; Vanderzande, S.; Durel, C.-E.; Muranty, H.; Denancé, C.; van de Weg, E. The use of shared haplotype length information for pedigree reconstruction in asexually propagated outbreeding crops, demonstrated for apple and sweet cherry. Hortic. Res. 2021, 8, 1–13. [Google Scholar] [CrossRef]
- Konjić, A.; Uzunović, M.; Gaši, F.; Grahić, J.; Kanlić, K.; Bogunić, F.; Howard, N.P. axiomFP.py: A Software for Ploidy and Call Quality Assessment of Axiom SNP Array Data. 2024; submitted. [Google Scholar]
- Urrestarazu, J.; Denancé, C.; Ravon, E.; Guyader, A.; Guisnel, R.; Feugey, L.; Poncet, C.; Lateur, M.; Houben, P.; Ordidge, M.; et al. Analysis of the genetic diversity and structure across a wide range of germplasm reveals prominent gene flow in apple at the European level. BMC Plant Biol. 2016, 16, 130. [Google Scholar] [CrossRef]
- Liang, W.; Dondini, L.; De Franceschi, P.; Paris, R.; Sansavini, S.; Tartarini, S. Genetic diversity, population structure and construction of a core collection of apple cultivars from Italian germplasm. Plant Mol. Biol. Rep. 2015, 33, 458–473. [Google Scholar] [CrossRef]
- Lassois, L.; Denancé, C.; Ravon, E.; Guyader, A.; Guisnel, R.; Hibrand-Saint-Oyant, L.; Poncet, C.; Lasserre-Zuber, P.; Feugey, L.; Durel, C.E. Genetic diversity, population structure, parentage analysis, and construction of core collections in the French apple germplasm based on SSR markers. Plant Mol. Biol. Rep. 2016, 34, 827–844. [Google Scholar] [CrossRef]
- Gaši, F.; Kanlić, K.; Stroil, B.K.; Pojskić, N.; Asdal, Å.; Rasmussen, M.; Kaiser, C.; Meland, M. Redundancies and Genetic Structure among ex situ Apple Collections in Norway Examined with Microsatellite Markers. HortScience 2016, 51, 1458–1462. [Google Scholar] [CrossRef]
- Marconi, G.; Ferradini, N.; Russi, L.; Concezzi, L.; Veronesi, F.; Albertini, E. Genetic Characterization of the Apple Germplasm Collection in Central Italy: The Value of Local Varieties. Front. Plant Sci. 2018, 9, 1460. [Google Scholar] [CrossRef]
- Testolin, R.; Foria, S.; Baccichet, I.; Messina, R.; Danuso, F.; Losa, A.; Scarbolo, E.; Stocco, M.; Cipriani, G. Genotyping Apple (Malus × domestica Borkh.) heirloom germplasm collected and maintained by the Regional Administration of friuli Venezia Giulia (Italy). Sci. Hortic. 2019, 252, 229–237. [Google Scholar] [CrossRef]
- Pereira-Lorenzo, S.; Ramos-Cabrer, A.M.; Ferreira, V.; Díaz-Hernández, M.B.; Carnide, V.; Pinto-Carnide, O.; Rodrigues, R.; Velázquez Barrera, M.E.; Rios-Mesa, D.; Ascasíbar-Errasti, J.; et al. Genetic diversity and core collection of Malus × domestica in northwestern Spain, Portugal and the Canary Islands by SSRs. Sci. Hortic. 2018, 240, 49–56. [Google Scholar] [CrossRef]
- BakIr, M.; Dumanoglu, H.; Aygun, A.; Erdogan, V.; Efe Dost, S.; Gülsen, O.; Serdar, U.; Kalkisim, O.; Bastas, K. Genetic diversity and population structure of apple germplasm from Eastern Black Sea region of Turkey by SSRs. Sci. Hortic. 2022, 294, 110793. [Google Scholar] [CrossRef]
- Kanlić, K.; Kalamujić-Stroil, B.; Grahić, J.; Asdal, Å.; Meland, M.; Kurtović, M.; Gaši, F. Influence of selection pressure on the frequency of triploid genotypes among different traditional apple germplasms. Work. Fac. Agric. Food Sci. Univ. Sarajevo 2016, 66, 287–290. [Google Scholar]
- Venison, E.P.; Litthauer, S.; Laws, P.; Denancé, C.; Fernández-Fernández, F.; Durel, C.E.; Ordidge, M. Microsatellite markers as a tool for active germplasm management and bridging the gap between national and local collections of apple. Genet. Resour. Crop Evol. 2022, 69, 1817–1832. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Babić, D. Italians in Požega-Slavonia County between assimilation and identity preservation. Work. Inst. Sci. Artist. Work Požega 2018, 7, 177–196. [Google Scholar] [CrossRef]
- Korban, S.S.; Wannarat, W.; Rayburn, C.M.; Tatum, T.C.; Rayburn, A.L. Genome Size and Nucleotypic Variation in Malus Germplasm. Genome 2009, 52, 148–155. [Google Scholar] [CrossRef]


| Accession Name | Identification Result | Method of Identification | |
|---|---|---|---|
| Duplicates | ‘Bijela zimska rebrača’ | ‘Leathercoat Russet’ | SSR |
| ‘Bisera’ | ‘Mantet’ | SSR | |
| ‘Carska reneta’ | ‘Bloody Butcher’ | SSR | |
| ‘Cox’s Orange 1’ | ‘Leathercoat Russet’ | SSR | |
| ‘Crvena caklenka’ | ‘Jonatan’ | SSR | |
| ‘Crvena kožara’ | ‘Crveni Boskop’ | SSR | |
| ‘Crvena zimska’ | ‘Jonatan’ | SSR | |
| ‘Crvenka’ | ‘Danciška rebrača’ | SSR | |
| ‘Dulibe velika’ | ‘Bietigheimer’, ‘San Giovanni’ | SSR | |
| ‘Francuska kožara’ | ‘Crveni Boskop’ | SSR | |
| ‘Gospojinska’ | ‘Grafenštajn’ | SSR | |
| ‘Imperica’ | ‘Jonathan’ | SSR | |
| ‘Jakobovka’ | ‘Bietigheimer’, ‘San Giovanni’ | SSR | |
| ‘Ječmenika 151’ | ‘Mantet’ | SSR | |
| ‘Jesenska žuta 103’ | ‘Peasgood’ | SSR | |
| ‘Kanada B’ | ‘Kanada’ | SSR | |
| ‘Kraljevčica kasna’ | ‘Grafenštajn’ | SSR | |
| ‘Lojzendorf’ | ‘Roter Herbstkalvill’ | SSR | |
| ‘Mađarica’ | ‘Calville des Femmes’ | SSR | |
| ‘Mirišljava Budići’ | ‘Danziger Kantapfel’ | SSR | |
| ‘Parkerov peping’ | ‘Leathercoat Russet’ | SSR | |
| ‘Petrovača žuta’ | ‘Bjeličnik’ | SSR | |
| ‘Prsnika’ | ‘Lijepocvjetka’ | SSR | |
| ‘Rebrača Brinje’ | ‘London peping’ | SSR | |
| ‘Slatka srčika’ | ‘Crveni Boskop’ | SSR | |
| ‘Slavonska srčika’ | ‘Crveni Boskop’ | SSR | |
| ‘Slavonska srčika Veić’ | ‘Zeleni štetinec’ | SSR | |
| ‘Slavonska srčika-Milić’ | ‘Zeleni štetinec’ | SSR | |
| ‘Slovenka’ | ‘Carević Rudolf’ | SSR | |
| ‘Srčika 1’ | ‘Zeleni štetinec’ | SSR | |
| ‘Srčika 2’ | ‘Zeleni štetinec’ | SSR | |
| ‘Stara šara’ | ‘Bietigheimer’, ‘San Giovanni’ | SSR | |
| ‘Starkova’ | ‘Starkova najranija’ | SSR | |
| ‘Svijetla musulja’ | ‘Leathercoat Russet’ | SSR | |
| ‘Šampanjka B’ | ‘Šampanjka’ | SSR | |
| ‘Šarulja’ | ‘Baumanova reneta’ | SSR | |
| ‘Winston’ | ‘Leathercoat Russet’ | SSR | |
| ‘Zelenika 35’ | ‘Krupnara’ | SSR | |
| ‘Zelenika 109’ | ‘Krupnara’ | SSR | |
| ‘Žuta jesenska’ | ‘Bloody Butcher’ | SSR | |
| Synonyms | ‘Bijela ribnjača’ | ‘Batulenka’, ‘Turkova’ | SSR |
| ‘Car Aleksandar’ | ‘Kardinal’ | SSR | |
| ‘Carevka’ | ‘Oberdikova reneta’ | SSR/SNP | |
| ‘Crvena djevojačka’ | ‘Framboise’, ‘Malinovača’ | SSR | |
| ‘Div jabuka’ | ‘Calville des Femmes’, ‘Mađarica’ | SSR | |
| ‘Gospojinka B’ | ‘Bloody Butcher’ | SSR | |
| ‘Funtača’ | ‘Bramley’s Seedling’ | SSR | |
| ‘Irmgard’ | ‘Horneburger Pfannkuchen’ | SSR | |
| ‘Kablarka’ | ‘Galloway Pippin’ | SSR | |
| ‘Kamenica’ | ‘Jacquin’, ‘Muškatnica’ | SSR | |
| ‘Kristalka’ | ‘Schoner von Nordhausen’ | SSR/SNP | |
| ‘Krvavka’ | ‘Danziger Kantapfel’ | SSR | |
| ‘Lopatičanka’ | ‘Molleskov’ | SSR | |
| ‘Lovrenčovka’ | ‘Grafenštajn’ | SSR | |
| ‘Lubeničarka’ | ‘Bloody Ploughman’ | SSR | |
| ‘Meglena’ | ‘Dugata’ | SSR/SNP | |
| ‘Pisanika ranka’ | ‘Close’ | SSR | |
| ‘Prinčevka’ | ‘Conique’ | SSR/SNP | |
| ‘Siva jesenska’ | ‘Learthercoat Russet’ | SSR | |
| ‘Slatka’ | ‘Klanferica’ | SSR | |
| ‘Umačka’ | ‘Kinrei’ | SSR | |
| ‘Vrtna’ | ‘Signe Tillisch’ | SSR | |
| ‘Zeleni štetinec’ | ‘Srčika’ | SSR | |
| ‘Zelenika’ | ‘Rhode Island Greening’ | SSR | |
| ‘Žuta Brinje’ | ‘Blajnhajmska reneta’ | SSR | |
| ‘Željeznjača’ | Majdofija’ | SSR | |
| Mislabeling of accessions | ‘Blatnjača’ | ‘MM 106’ | SSR/SNP |
| ‘Galetina’ | ‘Enterprise’ | SSR/SNP | |
| ‘Grofova’ | ‘Mutsu’ | SSR | |
| ‘Harbertova reneta ‘ | ‘Annie Elizabeth’ | SSR | |
| ‘Ječmenika’ | ‘Julyred’ | SSR | |
| ‘Kasler reneta’ | ‘Harbertova reneta’ | SSR | |
| ‘Kraljevača’ | ‘Prima’ | SSR | |
| ‘Krivopeteljka’ | ‘Bobovec’ | SSR | |
| ‘Ljetni špincl’ | ‘Mollie’s Delicious’ | SSR | |
| ‘Slična Božićnici’ | ‘Idared’ | SSR | |
| ‘Šarlamovski’ | ‘Budimka’ | SSR | |
| ‘Zelenika 77’ | ‘MM 106’ | SSR | |
| ‘Žrnovnička jabuka’ | ‘Mašanka’ | SSR |
| All (n = 125) | Accessions (n = 114) | Reference (n = 11) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Locus | No. of Alleles | No. of Effective Alleles | Gene Diversity He | No. of Alleles | No. of Effective Alleles | Gene Diversity He | No. of Alleles | No. of Effective Alleles | Gene Diversity He |
| CH04c07 | 18 | 7.98 | 0.87 | 18 | 7.76 | 0.87 | 7 | 8.1 | 0.88 |
| CH01h10 | 11 | 3.28 | 0.69 | 11 | 3.23 | 0.69 | 4 | 3.48 | 0.72 |
| CH01h01 | 15 | 6.67 | 0.85 | 15 | 6.88 | 0.85 | 6 | 4.83 | 0.80 |
| Hi02c07 | 11 | 4.49 | 0.78 | 11 | 4.56 | 0.78 | 5 | 3.82 | 0.74 |
| CH01f02 | 26 | 8.3 | 0.88 | 25 | 8.55 | 0.88 | 7 | 6.69 | 0.85 |
| CH01f03b | 13 | 4.68 | 0.79 | 13 | 4.54 | 0.78 | 8 | 6.7 | 0.85 |
| GD12 | 14 | 4.33 | 0.77 | 14 | 4.34 | 0.77 | 4 | 3.63 | 0.73 |
| GD147 | 16 | 6.59 | 0.85 | 16 | 6.9 | 0.86 | 6 | 3.44 | 0.71 |
| CH04e05 | 17 | 4.34 | 0.77 | 17 | 4.46 | 0.78 | 7 | 2.83 | 0.65 |
| CH02d08 | 16 | 7.3 | 0.86 | 15 | 7.2 | 0.86 | 7 | 8.28 | 0.88 |
| CH02c11 | 16 | 10.6 | 0.91 | 16 | 10.93 | 0.91 | 9 | 5.44 | 0.82 |
| CH02c09 | 11 | 7.61 | 0.87 | 11 | 7.8 | 0.87 | 6 | 4.9 | 0.80 |
| Mean | 15.33 | 6.35 | 0.82 | 15.17 | 6.43 | 0.83 | 6.33 | 5.18 | 0.79 |
| Accession Name | Group |
|---|---|
| ‘Adamčica’ | 1 |
| ‘Adamovka petrovka’ | 2 |
| ‘Blatnjača’ | 5 |
| ‘Carevka’ | 6 |
| ‘Cigančica’ | 7 |
| ‘Crvena ribnjača’ | 8 |
| ‘Čebulka’ | 9 |
| ‘Čelenika ljetna’ | 10 |
| ‘Dogaja’ | 11 |
| ‘Galetina’ | 16 |
| ‘Golubica’ | 17 |
| ‘Grinštantin’ | 18 |
| ‘Hrapava zvonasta’ | 19 |
| ‘Hrapavka rana’ | 20 |
| ‘Ivanlija’ | 21 |
| ‘Kristalka’ | 25 |
| ‘Križara’ | 26 |
| ‘Ljetni mošancelj’ | 28 |
| ‘Muškotelka’ | 31 |
| ‘Prijedorska zelenika’ | 33 |
| ‘Prinčevka’ | 34 |
| ‘Rožmarinka’ | 36 |
| ‘Senabija’ | 39 |
| ‘Slačica’ | 40 |
| ‘Slavonska srčika-Karolina’ | 41 |
| ‘Slavonska srčika-Podravina’ | 42 |
| ‘Slična Božićnici’ | 43 |
| ‘Šiljika’ | 44 |
| ‘Špička’ | 45 |
| ‘Braeburn’ | 49 |
| PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | |
|---|---|---|---|---|---|---|---|
| FWG | −0.44 | −0.19 | 0.18 | −0.24 | −0.18 | −0.06 | 0.11 |
| FL | −0.37 | −0.38 | 0.05 | −0.29 | 0.19 | 0.03 | 0.05 |
| FW | −0.43 | −0.13 | 0.29 | −0.2 | −0.25 | −0.05 | 0.13 |
| FSI | 0.02 | −0.41 | −0.34 | −0.21 | 0.67 | 0.15 | −0.1 |
| LST | −0.16 | −0.28 | −0.51 | 0.08 | −0.44 | −0.18 | −0.53 |
| WST | −0.11 | 0.41 | 0.39 | −0.32 | 0.25 | 0.07 | −0.61 |
| FH | 0.04 | 0.33 | −0.31 | −0.49 | 0.06 | −0.71 | 0.1 |
| PSM | 0.12 | 0.21 | −0.33 | −0.54 | −0.29 | 0.59 | 0.15 |
| TA | −0.38 | 0.31 | −0.25 | 0.12 | −0.02 | 0.29 | −0.12 |
| PTR | 0.41 | −0.23 | 0.16 | −0.28 | −0.19 | 0.02 | 0.19 |
| pH | 0.35 | −0.29 | 0.24 | −0.19 | −0.21 | 0.01 | −0.47 |
| Eigen value | 3.93 | 2.06 | 1.49 | 1.32 | 0.88 | 0.6 | 0.45 |
| Component variance (%) | 35.73 | 18.75 | 13.54 | 13.03 | 8.02 | 5.46 | 4.05 |
| Cumulative variance (%) | 35.73 | 54.47 | 68.01 | 80.04 | 88.06 | 93.52 | 97.57 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Čiček, D.; Konjić, A.; Skendrović Babojelić, M.; Vujević, P.; Šimon, S.; Gaši, F. Genetic Uniqueness and Pomological Diversity Among the Apple Accessions Maintained Within the Croatian National Clonal Germplasm Repository. Agronomy 2025, 15, 113. https://doi.org/10.3390/agronomy15010113
Čiček D, Konjić A, Skendrović Babojelić M, Vujević P, Šimon S, Gaši F. Genetic Uniqueness and Pomological Diversity Among the Apple Accessions Maintained Within the Croatian National Clonal Germplasm Repository. Agronomy. 2025; 15(1):113. https://doi.org/10.3390/agronomy15010113
Chicago/Turabian StyleČiček, Danijel, Almira Konjić, Martina Skendrović Babojelić, Predrag Vujević, Silvio Šimon, and Fuad Gaši. 2025. "Genetic Uniqueness and Pomological Diversity Among the Apple Accessions Maintained Within the Croatian National Clonal Germplasm Repository" Agronomy 15, no. 1: 113. https://doi.org/10.3390/agronomy15010113
APA StyleČiček, D., Konjić, A., Skendrović Babojelić, M., Vujević, P., Šimon, S., & Gaši, F. (2025). Genetic Uniqueness and Pomological Diversity Among the Apple Accessions Maintained Within the Croatian National Clonal Germplasm Repository. Agronomy, 15(1), 113. https://doi.org/10.3390/agronomy15010113







