Transcriptomic Analysis of Alfalfa Flowering and the Dual Roles of MsAP1 in Floral Organ Identity and Flowering Time
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatment
2.2. RNA-Seq Library Construction and Sequencing
2.3. Identification of DEGs and Gene Ontology (GO) Analysis
2.4. Reverse Transcription and Quantitative PCR (RT-QPCR)
2.5. Cloning and Construction and Plant Transformation
2.6. Sequence and Phylogenetic Analysis
2.7. Subcellular Localization and Transactive Activity Assay
2.8. Statistical Analysis
3. Results
3.1. Transcriptomic Analysis Revealed a Distinct Expression Pattern of Alfalfa Shoot Apex from Floral Buds
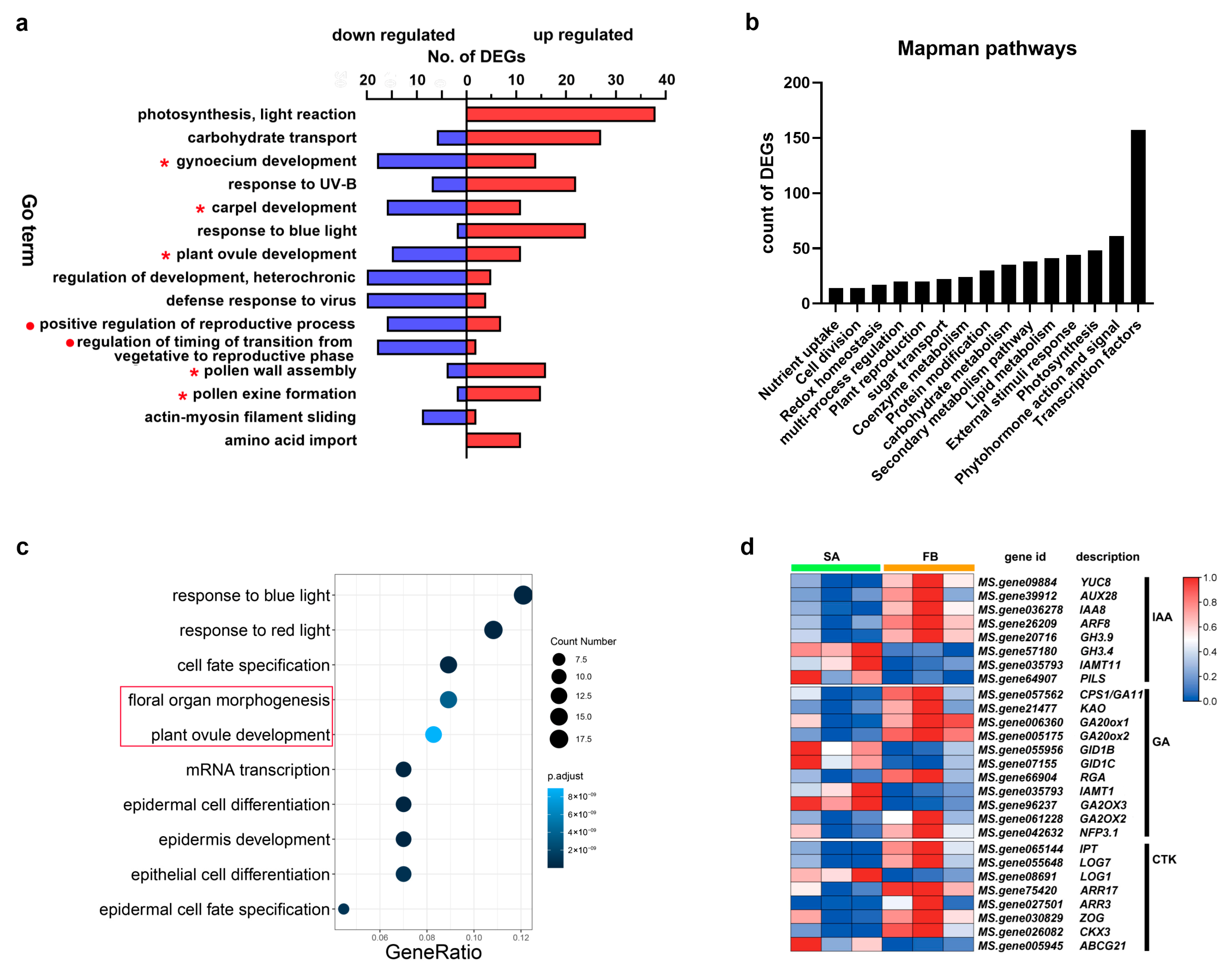
3.2. Genes Related to Reproduction Processes Were Enriched
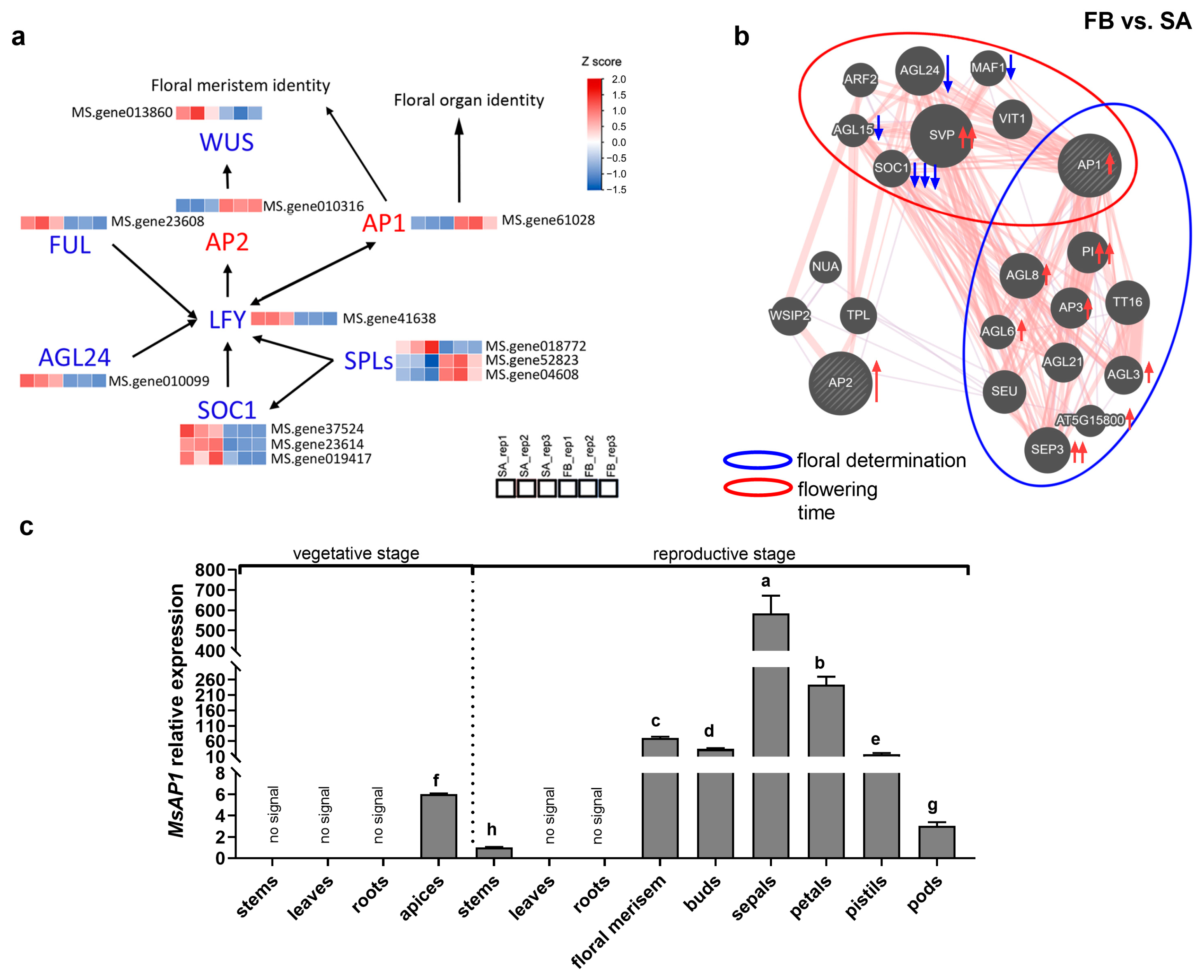
3.3. Components of Multiple Flowering-Regulation Pathways Were Involved in Alfalfa Phase Change
3.4. MsAP1 Might Be a Central Regulator of Alfalfa Flowering
3.5. MsAP1 Shares the Common Features of AP1
3.6. The Nuclear Protein MsAP1 Has Transcription Activity
3.7. Overexpression of MsAP1 in Arabidopsis Promoted Flowering and Altered Floral Structure
4. Discussion:
4.1. Alfalfa Flowering Consists of Multiple Regulatory Pathways
4.2. MsAP1 May Serve as a Candidate Gene for Flower Determination and Inflorescence Development
4.3. MsAP1 May Be Related to Alfalfa Pistil and Fruit Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Adhikari, L.; Makaju, S.O.; Missaoui, A.M. QTL mapping of flowering time and biomass yield in tetraploid alfalfa (Medicago sativa L.). BMC Plant Biol. 2019, 19, 359. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Han, L.; Pislariu, C.; Nakashima, J.; Fu, C.; Jiang, Q.; Quan, L.; Blancaflor, E.B.; Tang, Y.; Bouton, J.H.; et al. From model to crop: Functional analysis of a STAY-GREEN gene in the model legume Medicago truncatula and effective use of the gene for alfalfa improvement. Plant Physiol. 2011, 157, 1483–1496. [Google Scholar] [CrossRef] [PubMed]
- Wolabu, T.W.; Mahmood, K.; Jerez, I.T.; Cong, L.; Yun, J.; Udvardi, M.; Tadege, M.; Wang, Z.; Wen, J. Multiplex CRISPR/Cas9-mediated mutagenesis of alfalfa FLOWERING LOCUS Ta1 (MsFTa1) leads to delayed flowering time with improved forage biomass yield and quality. Plant Biotechnol. J. 2023, 21, 1383–1392. [Google Scholar] [CrossRef] [PubMed]
- Freytes, S.N.; Canelo, M.; Cerdán, P.D. Regulation of Flowering Time: When and Where? Curr. Opin. Plant Biol. 2021, 63, 102049. [Google Scholar] [CrossRef]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef]
- Poethig, R.S.; Fouracre, J. Temporal regulation of vegetative phase change in plants. Dev. Cell 2024, 59, 4–19. [Google Scholar] [CrossRef]
- Bao, S.; Hua, C.; Shen, L.; Yu, H. New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 118–131. [Google Scholar] [CrossRef] [PubMed]
- Fukazawa, J.; Ohashi, Y.; Takahashi, R.; Nakai, K.; Takahashi, Y. DELLA degradation by gibberellin promotes flowering via GAF1-TPR-dependent repression of floral repressors in Arabidopsis. Plant Cell 2021, 33, 2258–2272. [Google Scholar] [CrossRef]
- Balouri, C.; Poulios, S.; Tsompani, D.; Spyropoulou, Z.; Ketikoglou, M.-C.; Kaldis, A.; Doonan, J.H.; Vlachonasios, K.E. Gibberellin Signaling through RGA Suppresses GCN5 Effects on Arabidopsis Developmental Stages. Int. J. Mol. Sci. 2024, 25, 6757. [Google Scholar] [CrossRef]
- Davis, S.J. Integrating hormones into the floral-transition pathway of Arabidopsis thaliana. Plant Cell Environ. 2009, 32, 1201–1210. [Google Scholar] [CrossRef]
- Izawa, T. What is going on with the hormonal control of flowering in plants? Plant J. 2021, 105, 431–445. [Google Scholar] [CrossRef]
- Mai, Y.X.; Wang, L.; Yang, H.Q. A gain-of-function mutation in IAA7/AXR2 confers late flowering under short-day light in Arabidopsis. J. Integr. Plant Biol. 2011, 53, 480–492. [Google Scholar] [CrossRef] [PubMed]
- Pauli, A.W.; Sorensen, E.L. Vegetative and reproductive responses of dwarf alfalfa plants to gibberellic acid. Crop Sci. 1961, 1, 269–271. [Google Scholar] [CrossRef]
- Ma, D.; Liu, B.; Ge, L.; Weng, Y.; Cao, X.; Liu, F.; Mao, P.; Ma, X. Identification and characterization of regulatory pathways involved in early flowering in the new leaves of alfalfa (Medicago sativa L.) by transcriptome analysis. BMC Plant Biol. 2021, 21, 8. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Chen, R. Auxin efflux transporter MtPIN10 regulates compound leaf and flower development in Medicago truncatula. Plant Signal Behav. 2011, 6, 1537–1544. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, C.D.; García-Gagliardi, P.; Antonietti, M.S.; Sánchez-Lamas, M.; Mancini, E.; Dezar, C.A.; Vazquez, M.; Watson, G.; Yanovsky, M.J.; Cerdán, P.D. Improvement of alfalfa forage quality and management through the down-regulation of MsFTa1. Plant Biotechnol. J. 2020, 18, 944–954. [Google Scholar] [CrossRef] [PubMed]
- Jaudal, M.; Wen, J.; Mysore, K.S.; Putterill, J. Medicago PHYA promotes flowering, primary stem elongation and expression of flowering time genes in long days. BMC Plant Biol. 2020, 20, 329. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, J.; Liu, X.; Kong, Y.; Han, L. The Roles of the Pseudo-Response Regulators in Circadian Clock and Flowering Time in Medicago truncatula. Int. J. Mol. Sci. 2023, 24, 16834. [Google Scholar] [CrossRef] [PubMed]
- Andres, F.; Kinoshita, A.; Kalluri, N.; Fernández, V.; Falavigna, V.S.; Cruz, T.M.; Jang, S.; Chiba, Y.; Seo, M.; Mettler-Altmann, T.; et al. The sugar transporter SWEET10 acts downstream of FLOWERING LOCUS T during floral transition of Arabidopsis thaliana. BMC Plant Biol. 2020, 20, 53. [Google Scholar] [CrossRef]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef]
- Wigge, P.A.; Kim, M.C.; Jaeger, K.E.; Busch, W.; Schmid, M.; Lohmann, J.U.; Weigel, D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 2005, 309, 1056–1059. [Google Scholar] [CrossRef] [PubMed]
- Duk, M.A.; Gursky, V.V.; Samsonova, M.G.; Surkova, S.Y. Modeling the Flowering Activation Motif during Vernalization in Legumes: A Case Study of M. trancatula. Life 2024, 14, 26. [Google Scholar] [CrossRef]
- Mandel, M.A.; Yanofsky, M.F. A gene triggering flower formation in Arabidopsis. Nature 1995, 377, 522–524. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Tang, A.; Yu, J.; Cheng, T.; Wang, J.; Yang, W.; Pan, H.; Zhang, Q. RcAP1, a Homolog of APETALA1, is Associated with Flower Bud Differentiation and Floral Organ Morphogenesis in Rosa chinensis. Int. J. Mol. Sci. 2019, 20, 3557. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Mara, C. Regulatory mechanisms for floral homeotic gene expression. Semin. Cell Dev. Biol. 2010, 21, 80–86. [Google Scholar] [CrossRef] [PubMed]
- de Folter, S.; Immink, R.G.; Kieffer, M.; Pařenicová, L.; Henz, S.R.; Weigel, D.; Busscher, M.; Kooiker, M.; Colombo, L.; Kater, M.M.; et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 2005, 17, 1424–1433. [Google Scholar] [CrossRef]
- McCarthy, E.W.; Mohamed, A.; Litt, A. Functional Divergence of APETALA1 and FRUITFULL is due to Changes in both Regulation and Coding Sequence. Front. Plant Sci. 2015, 6, 1076. [Google Scholar] [CrossRef]
- Li, C.; Chen, L.; Fan, X.; Qi, W.; Ma, J.; Tian, T.; Zhou, T.; Ma, L.; Chen, F. MawuAP1 promotes flowering and fruit development in the basal angiosperm Magnolia wufengensis (Magnoliaceae). Tree Physiol. 2020, 40, 1247–1259. [Google Scholar] [CrossRef]
- Theissen, G.; Becker, A.; Di Rosa, A.; Kanno, A.; Kim, J.T.; Münster, T.; Winter, K.-U.; Saedler, H. A short history of MADS-box genes in plants. Plant Mol. Biol. 2000, 42, 115–149. [Google Scholar] [CrossRef]
- Pabón-Mora, N.; Ambrose, B.A.; Litt, A. Poppy APETALA1/FRUITFULL orthologs control flowering time, branching, perianth identity, and fruit development. Plant Physiol. 2012, 158, 1685–1704. [Google Scholar] [CrossRef]
- Sun, Y.; Fan, Z.; Li, X.; Li, J.; Yin, H. The APETALA1 and FRUITFUL homologs in Camellia japonica and their roles in double flower domestication. Mol. Breed. 2014, 33, 821–834. [Google Scholar] [CrossRef]
- Chen, L.; Nan, H.; Kong, L.; Yue, L.; Yang, H.; Zhao, Q.; Fang, C.; Li, H.; Cheng, Q.; Lu, S.; et al. Soybean AP1 homologs control flowering time and plant height. J. Integr. Plant Biol. 2020, 62, 1868–1879. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zeng, Y.; Yang, Y.; Huang, L.; Tang, B.; Zhang, H.; Hao, F.; Liu, W.; Li, Y.; Liu, Y.; et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 2020, 11, 2494. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Logemann, E.; Birkenbihl, R.P.; Ülker, B.; Somssich, I.E. An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods 2006, 2, 16. [Google Scholar] [CrossRef]
- Jiang, X.; Cui, H.; Wang, Z.; Kang, J.; Yang, Q.; Guo, C. Genome-Wide Analysis of the Lateral Organ Boundaries Domain (LBD) Members in Alfalfa and the Involvement of MsLBD48 in Nitrogen Assimilation. Int. J. Mol. Sci. 2023, 24, 4644. [Google Scholar] [CrossRef] [PubMed]
- Hung, F.Y.; Chen, F.F.; Li, C.; Chen, C.; Chen, J.H.; Cui, Y.; Wu, K. The LDL1/2-HDA6 Histone Modification Complex Interacts With TOC1 and Regulates the Core Circadian Clock Components in Arabidopsis. Front. Plant Sci. 2019, 10, 233. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Zhuo, R.; Wang, H.; Xu, J.; Jin, K.; Huang, B.; Qiao, G. A comprehensive analysis of the floral transition in ma bamboo (Dendrocalamus latiflorus) reveals the roles of DlFTs involved in flowering. Tree Physiol. 2022, 42, 1899–1911. [Google Scholar] [CrossRef]
- Gao, X.; Wang, L.; Zhang, H.; Zhu, B.; Lv, G.; Xiao, J. Transcriptome analysis and identification of genes associated with floral transition and fruit development in rabbiteye blueberry (Vaccinium ashei). PLoS ONE 2021, 16, e0259119. [Google Scholar] [CrossRef]
- Guo, X.; Yu, C.; Luo, L.; Wan, H.; Zhen, N.; Li, Y.; Cheng, T.; Wang, J.; Pan, H.; Zhang, Q. Developmental transcriptome analysis of floral transition in Rosa odorata var. gigantea. Plant Mol. Biol. 2018, 97, 113–130. [Google Scholar] [CrossRef]
- Guo, X.; Yu, C.; Luo, L.; Wan, H.; Zhen, N.; Xu, T.; Tan, J.; Pan, H.; Zhang, Q. Transcriptome of the floral transition in Rosa chinensis ‘Old Blush’. BMC Genom. 2017, 18, 199. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Melzer, R.; Theissen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Litt, A.; Irish, V.F. Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: Implications for the evolution of floral development. Genetics 2003, 165, 821–833. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Huang, F.; Liu, H.; Yang, S.; Yu, D. An APETALA1-like gene of soybean regulates flowering time and specifies floral organs. J. Plant Physiol. 2011, 168, 2251–2259. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yan, G.; Wen, Z.; An, Y.-Q.; Singer, S.D.; Liu, Z. Two tobacco AP1-like gene promoters drive highly specific, tightly regulated and unique expression patterns during floral transition, initiation and development. Planta 2014, 239, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Goslin, K.; Zheng, B.; Serrano-Mislata, A.; Rae, L.; Ryan, P.T.; Kwaśniewska, K.; Thomson, B.; Ó’Maoiléidigh, D.S.; Madueño, F.; Wellmer, F.; et al. Transcription Factor Interplay between LEAFY and APETALA1/CAULIFLOWER during Floral Initiation. Plant Physiol. 2017, 174, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yang, R.; Shen, S.; Zhao, J. Molecular mechanism of fl owering time regulation in Brassica rapa: Similarities and differences with Arabidopsis. Hortic. Plant J. 2024, 10, 615–628. [Google Scholar] [CrossRef]
- Reyes, P.d.L.; Serrano-Bueno, G.; Romero-Campero, F.J.; Gao, H.; Romero, J.M.; Valverde, F. CONSTANS alters the circadian clock in Arabidopsis thaliana. Mol. Plant 2024, in press. [Google Scholar]
- Cho, L.H.; Yoon, J.; An, G. The control of flowering time by environmental factors. Plant J. 2017, 90, 708–719. [Google Scholar] [CrossRef]
- Liu, L.; Zheng, Y.; Feng, S.; Yu, L.; Li, Y.; Zong, Y.; Chen, W.; Liao, F.; Yang, L.; Guo, W. Transcriptomic and Physiological Analysis Reveals the Responses to Auxin and Abscisic Acid Accumulation During Vaccinium corymbosum Flower Bud and Fruit Development. Front. Plant Sci. 2022, 13, 818233. [Google Scholar] [CrossRef]
- Irish, V.F.; Sussex, I.M. Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 1990, 2, 741–753. [Google Scholar] [PubMed]
- Elo, A.; Lemmetyinen, J.; Turunen, M.L.; Tikka, L.; Sopanen, T. Three MADS-box genes similar to APETALA1 and FRUITFULL from silver birch (Betula pendula). Physiol. Plant. 2001, 112, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, X.; Yan, G.; Zhou, Y.; Zhang, K. Over-expression of the PaAP1 gene from sweet cherry (Prunus avium L.) causes early flowering in Arabidopsis thaliana. J. Plant Physiol. 2013, 170, 315–320. [Google Scholar] [CrossRef]
- Wang, C.; Liu, J.; Xie, X.; Wang, J.; Ma, Q.; Chen, P.; Yang, D.; Ma, X.; Hao, F.; Su, J. GhAP1-D3 positively regulates flowering time and early maturity with no yield and fiber quality penalties in upland cotton. J. Integr. Plant Biol. 2023, 65, 985–1002. [Google Scholar] [CrossRef]
- Hu, T.; Li, X.; Du, L.; Manuela, D.; Xu, M. LEAFY and APETALA1 down-regulate ZINC FINGER PROTEIN 1 and 8 to release their repression on class B and C floral homeotic genes. Proc. Natl. Acad. Sci. USA 2023, 120, e2221181120. [Google Scholar] [CrossRef]
- Shannon, S. A Developmental and Genetic Analysis of tfl1, a Regulator of Inflorescence Meristem Identity in Arabidopsis. Ph.D. Thesis, University of Oregon, Eugene, OR, USA, 1993. [Google Scholar]
- Eswaramoorthy, V.; Kandasamy, T.; Thiyagarajan, K.; Chockalingam, V.; Jegadeesan, S.; Natesan, S.; Adhimoolam, K.; Prabhakaran, J.; Singh, R.; Muthurajan, R. Characterization of terminal flowering cowpea (Vigna unguiculata L. Walp.) mutants obtained by induced mutagenesis digs out the loss-of-function of phosphatidylethanolamine-binding protein. PLoS ONE 2023, 18, e0295509. [Google Scholar] [CrossRef]
- Huijser, P.; Klein, J.; Lönnig, W.; Meijer, H.; Saedler, H.; Sommer, H. Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J. 1992, 11, 1239–1249. [Google Scholar] [CrossRef]
- Theissen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001, 4, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Benlloch, R.; d’Erfurth, I.; Ferrandiz, C.; Cosson, V.; Beltrán, J.P.; Canas, L.A.; Kondorosi, A.; Madueno, F.; Ratet, P. Isolation of mtpim proves Tnt1 a useful reverse genetics tool in Medicago truncatula and uncovers new aspects of AP1-like functions in legumes. Plant Physiol. 2006, 142, 972–983. [Google Scholar] [CrossRef]
- Kotoda, N.; Wada, M.; Kusaba, S.; Kano-Murakami, Y.; Masuda, T.; Soejima, J. Overexpression of MdMADS5, an APETALA1-like gene of apple, causes early flowering in transgenic Arabidopsis. Plant Sci. 2002, 162, 679–687. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Chen, X.; Zeng, L.; Su, Y.; Liu, Z. Characterization of Two AGAMOUS-like Genes and Their Promoters from the Cymbidium faberi (Orchidaceae). Plants 2023, 12, 2740. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, X.; Cui, H.; Wang, Z.; Long, R.; Yang, Q.; Kang, J. Transcriptomic Analysis of Alfalfa Flowering and the Dual Roles of MsAP1 in Floral Organ Identity and Flowering Time. Agronomy 2024, 14, 1741. https://doi.org/10.3390/agronomy14081741
Jiang X, Cui H, Wang Z, Long R, Yang Q, Kang J. Transcriptomic Analysis of Alfalfa Flowering and the Dual Roles of MsAP1 in Floral Organ Identity and Flowering Time. Agronomy. 2024; 14(8):1741. https://doi.org/10.3390/agronomy14081741
Chicago/Turabian StyleJiang, Xu, Huiting Cui, Zhen Wang, Ruicai Long, Qingchuan Yang, and Junmei Kang. 2024. "Transcriptomic Analysis of Alfalfa Flowering and the Dual Roles of MsAP1 in Floral Organ Identity and Flowering Time" Agronomy 14, no. 8: 1741. https://doi.org/10.3390/agronomy14081741
APA StyleJiang, X., Cui, H., Wang, Z., Long, R., Yang, Q., & Kang, J. (2024). Transcriptomic Analysis of Alfalfa Flowering and the Dual Roles of MsAP1 in Floral Organ Identity and Flowering Time. Agronomy, 14(8), 1741. https://doi.org/10.3390/agronomy14081741





