Functional Framework of Amino Acid Transporters in Quinoa: Genome-Wide Survey, Homology, and Stress Response
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of the AAT Gene Family in Quinoa
2.2. Chromosomal Localization, Duplication, and Selective Pressure Analysis
2.3. Phylogenetic Analyses, Conserved Motifs and Structure
2.4. Cis-Regulatory Element and AATs Expression in Quinoa
2.5. Expression of CqAAT Genes in Quinoa under Drought and Salt Stress
3. Results
3.1. Identification of AAT Gene Family Members in Quinoa
3.2. Chromosomal Localizations of CqAAT Genes and Gene Duplication Events
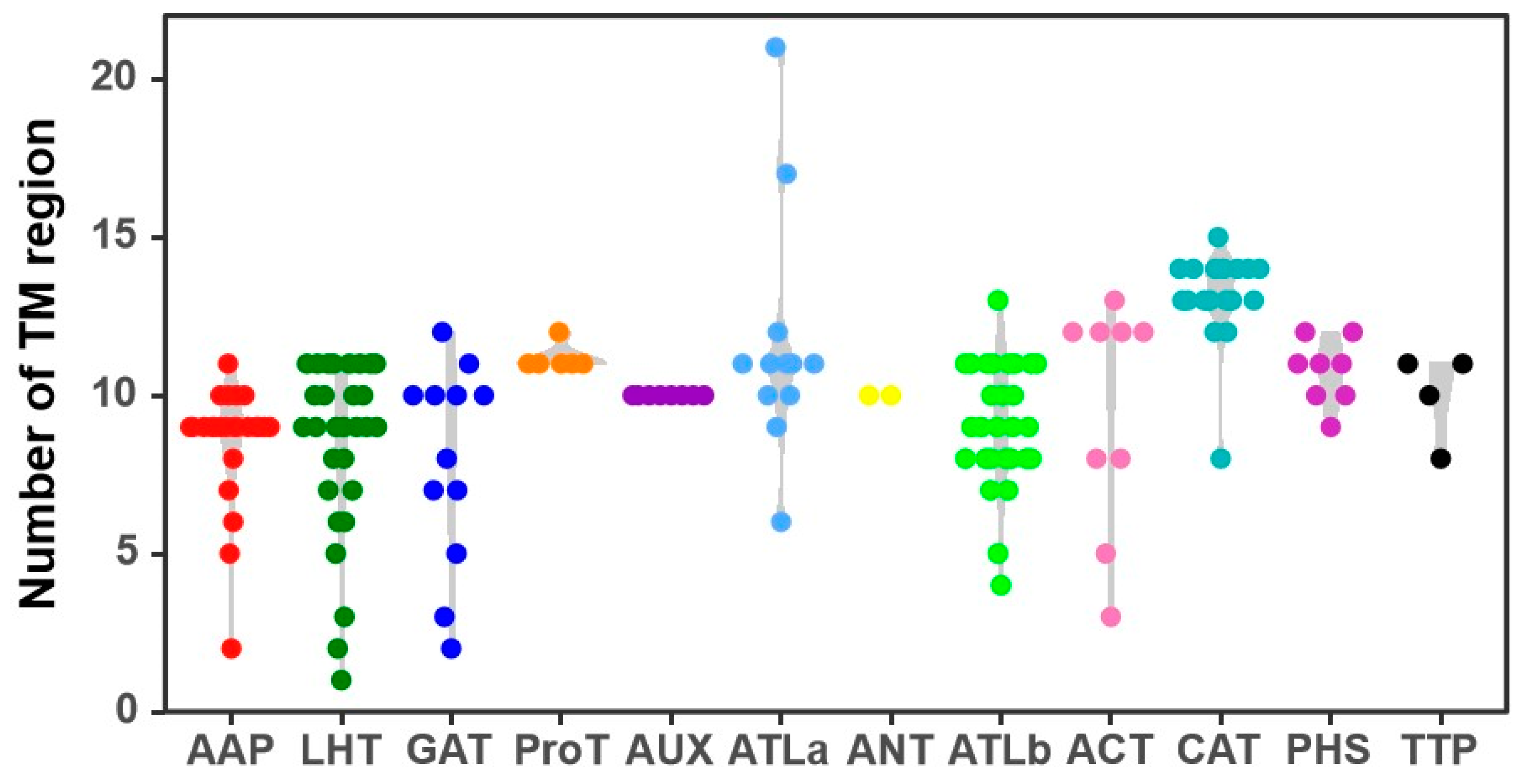
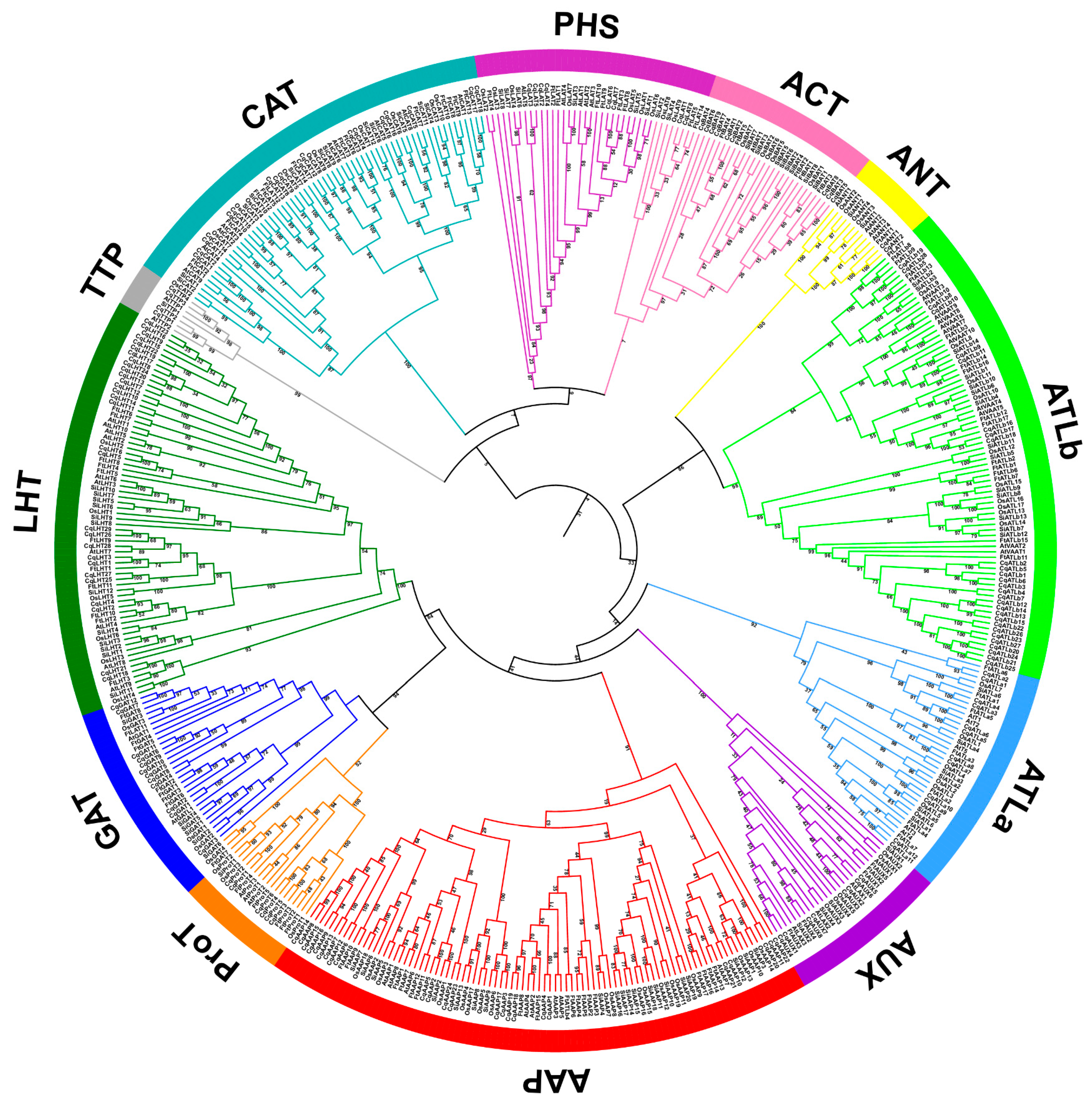
3.3. Phylogenetic Tree and Structures of CqAAT Genes
3.4. Cis-Regulatory Elements of CqAATs and Collinearity Analysis
3.5. Expression Patterns of CqAATs and Responses to Drought/Salt Stress
3.6. Functional Predictions for AAT Genes in Quinoa
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Choukr-Allah, R.; Rao, N.K.; Hirich, A.; Shahid, M.; Alshankiti, A.; Toderich, K.; Gill, S.; Butt, K.U. Quinoa for marginal environments: Toward future food and nutritional security in MENA and Central Asia Regions. Front. Plant Sci. 2016, 7, 346. [Google Scholar] [CrossRef] [PubMed]
- López-Marqués, R.L.; Nørrevang, A.F.; Ache, P.; Moog, M.; Visintainer, D.; Wendt, T.; Østerberg, J.T.; Dockter, C.; Jørgensen, M.E.; Salvador, A.T.; et al. Prospects for the accelerated improvement of the resilient crop quinoa. J. Exp. Bot. 2020, 71, 5333–5347. [Google Scholar] [CrossRef]
- Grimberg, A.; Saripella, G.V.; Repo-Carrasco Valencia, R.A.; Bengtsson, T.; Alandia, G.; Carlsson, A.S. Transcriptional regulation of quinoa seed quality: Identification of novel candidate genetic markers for increased protein content. Front. Plant Sci. 2022, 13, 816425. [Google Scholar] [CrossRef] [PubMed]
- Bastidas, E.G.; Roura, R.; Rizzolo, D.A.D.; Massanés, T.; Gomis, R. Quinoa (Chenopodium quinoa Willd.), from nutritional value to potential health benefits: An integrative review. J. Nutr. Food Sci. 2016, 6, 3. [Google Scholar] [CrossRef]
- Vega-Galvez, A.; Miranda, M.; Vergara, J.; Uribe, E.; Puente, L.; Martinez, E.A. Nutrition facts and functional potential of quinoa (Chenopodium quinoa Willd.), an ancient Andean grain: A review. J. Sci. Food Agric. 2010, 90, 2541–2547. [Google Scholar] [CrossRef] [PubMed]
- Rentsch, D.; Schmidt, S.; Tegeder, M. Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett. 2007, 581, 2281–2289. [Google Scholar] [CrossRef] [PubMed]
- Tegeder, M.; Offler, C.E.; Frommer, W.B.; Patrick, J.W. Amino acid transporters are localized to transfer cells of developing pea seeds. Plant Physiol. 2000, 122, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Tegeder, M. Transporters for amino acids in plant cells: Some functions and many unknowns. Curr. Opin. Plant Biol. 2012, 15, 315–321. [Google Scholar] [CrossRef]
- Williams, L.; Miller, A. Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 659–688. [Google Scholar] [CrossRef]
- Koch, W.; Kwart, M.; Laubner, M.; Heineke, D.; Stransky, H.; Frommer, W.B.; Tegeder, M. Reduced amino acid content in transgenic potato tubers due to antisense inhibition of the leaf H+/amino acid symporter StAAP1. Plant J. 2003, 33, 211–220. [Google Scholar] [CrossRef]
- Tegeder, M.; Rentsch, D. Uptake and partitioning of amino acids and peptides. Mol. Plant 2010, 3, 997–1011. [Google Scholar] [CrossRef] [PubMed]
- Rolletschek, H.; Hosein, F.; Miranda, M.; Heim, U.; Götz, K.P.; Schlereth, A.; Borisjuk, L.; Saalbach, I.; Wobus, U.; Weber, H. Ectopic expression of an amino acid transporter (VfAAP1) in seeds of Vicia narbonensis and pea increases storage proteins. Plant Physiol. 2005, 137, 1236–1249. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Foster, J.; Chen, J.; Voll, L.M.; Weber, A.P.; Tegeder, M. AAP1 transports uncharged amino acids into roots of Arabidopsis. Plant J. 2007, 50, 305–319. [Google Scholar] [CrossRef] [PubMed]
- Svennerstam, H.; Ganeteg, U.; Nasholm, T. Root uptake of cationic amino acids by Arabidopsis depends on functional expression of amino acid permease 5. New Phytol. 2008, 180, 620–630. [Google Scholar] [CrossRef]
- Wipf, D.; Loque, D.; Lalonde, S.; Frommer, W.B. Amino acid transporter inventory of the selaginella genome. Front. Plant Sci. 2012, 3, 36. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Ma, H.; Yu, L.; Wang, X.; Zhao, J. Genome-wide survey and expression analysis of amino acid transporter gene family in rice (Oryza sativa L.). PLoS ONE 2012, 7, e49210. [Google Scholar] [CrossRef]
- Hunt, E.; Gattolin, S.; Newbury, H.J.; Bale, J.S.; Tseng, H.M.; Barrett, D.A.; Pritchard, J. A mutation in amino acid permease AAP6 reduces the amino acid content of the Arabidopsis sieve elements but leaves aphid herbivores unaffected. J. Exp. Bot. 2010, 61, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Sanders, A.; Collier, R.; Trethewy, A.; Gould, G.; Sieker, R.; Tegeder, M. AAP1 regulates import of amino acids into developing Arabidopsis embryos. Plant J. 2009, 59, 540–552. [Google Scholar] [CrossRef]
- Okumoto, S.; Pilot, G. Amino acid export in plants: A missing link in nitrogen cycling. Mol. Plant 2011, 4, 453–463. [Google Scholar] [CrossRef]
- Ma, H.; Cao, X.; Shi, S.; Li, S.; Gao, J.; Ma, Y.; Zhao, Q.; Chen, Q. Genome-wide survey and expression analysis of the amino acid transporter superfamily in potato (Solanum tuberosum L.). Plant Physiol. Biochem. 2016, 107, 164–177. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, X.; Zheng, J.; Men, Y.; Zhang, Y.; Liu, L.; Han, Y.; Hou, S.; Sun, Z. Amino acid transporter (AAT) gene family in Tartary buckwheat (Fagopyrum tataricum L. Gaertn.): Characterization, expression analysis and functional prediction. Int. J. Biol. Macromol. 2022, 217, 330–344. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chai, Y.; Liu, J.; Zheng, J.; Zhao, Z.; Amo, A.; Cui, C.; Lu, Q.; Chen, L.; Hu, Y.G. Amino acid transporter (AAT) gene family in foxtail millet (Setaria italica L.): Widespread family expansion, functional differentiation, roles in quality formation and response to abiotic stresses. BMC Genom. 2021, 22, 519. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Yang, Y.; Chen, M. Genome-wide survey of the amino acid transporter gene family in wheat (Triticum aestivum L.): Identification, expression analysis and response to abiotic stress. Int. J. Biol. Macromol. 2020, 162, 1372–1387. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Yuan, H.Y.; Ren, R.; Zhao, S.Q.; Han, Y.P.; Zhou, Q.Y.; Ke, D.X.; Wang, Y.X.; Wang, L. Genome-wide identification, classification, and expression analysis of amino acid transporter gene family in Glycine max. Front. Plant Sci. 2016, 7, 515. [Google Scholar] [CrossRef] [PubMed]
- Fischer, W.N.; Kwart, M.; Hummel, S.; Frommer, W.B. Substrate specificity and expression profile of amino acid transporters (AAPs) in Arabidopsis. J. Biol. Chem. 1995, 270, 16315–16320. [Google Scholar] [CrossRef] [PubMed]
- Okumoto, S.; Schmidt, R.; Tegeder, M.; Fischer, W.N.; Rentsch, D.; Frommer, W.B.; Koch, W. High affinity amino acid transporters specifically expressed in xylem parenchyma and developing seeds of Arabidopsis. J. Biol. Chem. 2002, 277, 45338–45346. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; de Fay, E.; Fitz, M.; Wipf, D.; Blaudez, D.; Chalot, M. PtAAP11, a high affinity amino acid transporter specifically expressed in differentiating xylem cells of poplar. J. Exp. Bot. 2010, 61, 1671–1682. [Google Scholar] [CrossRef] [PubMed]
- Hirner, B.; Fischer, W.N.; Rentsch, D.; Kwart, M.; Frommer, W.B. Developmental control of H+/amino acid permease gene expression during seed development of Arabidopsis. Plant J. 1998, 14, 535–544. [Google Scholar] [CrossRef]
- Zhang, L.; Tan, Q.; Lee, R.; Trethewy, A.; Lee, Y.H.; Tegeder, M. Altered xylem-phloem transfer of amino acids affects metabolism and leads to increased seed yield and oil content in Arabidopsis. Plant Cell 2010, 22, 3603–3620. [Google Scholar] [CrossRef]
- Swarup, R.; Kargul, J.; Marchant, A.; Zadik, D.; Rahman, A.; Mills, R.; Yemm, A.; May, S.; Williams, L.; Millner, P.; et al. Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 2004, 16, 3069–3083. [Google Scholar] [CrossRef]
- Swarup, K.; Benkova, E.; Swarup, R.; Casimiro, I.; Peret, B.; Yang, Y.; Parry, G.; Nielsen, E.; De Smet, I.; Vanneste, S.; et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 2008, 10, 946–954. [Google Scholar] [CrossRef] [PubMed]
- Hirner, A.; Ladwig, F.; Stransky, H.; Okumoto, S.; Keinath, M.; Harms, A.; Frommer, W.B.; Koch, W.J. Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell 2006, 18, 1931–1946. [Google Scholar] [CrossRef]
- Forsum, O.; Svennerstam, H.; Ganeteg, U.; Nasholm, T. Capacities and constraints of amino acid utilization in Arabidopsis. New Phytol. 2008, 179, 1058–1069. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Tegeder, M. Selective expression of a novel high-affinity transport system for acidic and neutral amino acids in the tapetum cells of Arabidopsis flowers. Plant J. 2004, 40, 60–74. [Google Scholar] [CrossRef]
- Ji, Y.; Huang, W.; Wu, B.; Fang, Z.; Wang, X. The amino acid transporter AAP1 mediates growth and grain yield by regulating neutral amino acid uptake and reallocation in Oryza sativa. J. Exp. Bot. 2020, 71, 4763–4777. [Google Scholar] [CrossRef]
- Yang, X.; Yang, G.; Wei, X.; Huang, W.; Fang, Z. OsAAP15, an amino acid transporter in response to nitrogen concentration, mediates panicle branching and grain yield in rice. Plant Sci. 2023, 330, 111640. [Google Scholar] [CrossRef]
- Su, Y.H.; Frommer, W.B.; Ludewig, U. Molecular and functional characterization of a family of amino acid transporters from Arabidopsis. Plant Physiol. 2004, 136, 3104–3113. [Google Scholar] [CrossRef] [PubMed]
- Kohl, S.; Hollmann, J.; Blattner, F.R.; Radchuk, V.; Andersch, F.; Steuernagel, B.; Schmutzer, T.; Scholz, U.; Krupinska, K.; Weber, H.; et al. A putative role for amino acid permeases in sink-source communication of barley tissues uncovered by RNA-seq. BMC Plant Biol. 2012, 12, 154. [Google Scholar] [CrossRef]
- Yu, S.T.; Yu, H.B.; Yu, G.Q.; Zhao, L.R.; Sun, H.X.; Tang, Y.Y.; Wang, X.Z.; Wu, Q.; Sun, Q.X.; Wang, C.T. Isolation of differentially expressed genes from developing seeds of a high-protein peanut mutant and its wild type using genefishing TM Technology. In Adv Appl Biotechnol: Proceedings of the 2nd International Conference on Applied Biotechnology (ICAB 2014) 2015; Springer: Berlin/Heidelberg, Germany, 2015; Volume I, pp. 37–45. [Google Scholar] [CrossRef]
- Berg, J.A.; Hermans, F.W.K.; Beenders, F.; Abedinpour, H.; Vriezen, W.H.; Visser, R.G.F.; Bai, Y.; Schouten, H.J. The amino acid permease (AAP) genes CsAAP2A and SlAAP5A/B are required for oomycete susceptibility in cucumber and tomato. Mol. Plant Pathol. 2021, 22, 658–672. [Google Scholar] [CrossRef]
- Yao, X.; Li, H.; Nie, J.; Liu, H.; Guo, Y.; Lv, L.; Yang, Z.; Sui, X. Disruption of the amino acid transporter CsAAP2 inhibits auxin-mediated root development in cucumber. New Phytol. 2023, 239, 639–659. [Google Scholar] [CrossRef]
- Lehmann, S.; Gumy, C.; Blatter, E.; Boeffel, S.; Fricke, W.; Rentsch, D. In planta function of compatible solute transporters of the AtProT family. J. Exp. Bot. 2011, 62, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Ueda, A.; Shi, W.; Sanmiya, K.; Shono, M.; Takabe, T. Functional analysis of salt-inducible proline transporter of barley roots. Plant Cell Physiol. 2001, 42, 1282–1289. [Google Scholar] [CrossRef] [PubMed]
- Na, G.U.O.; Dong, X.U.E.; Zhang, W.; Zhao, J.M.; Xue, C.C.; Qiang, Y.A.N.; Xue, J.Y.; Wang, H.T.; Zhang, Y.M.; Han, X.I.N.G. Overexpression of GmProT1 and GmProT2 increases tolerance to drought and salt stresses in transgenic Arabidopsis. J. Integ Agric. 2016, 15, 1727–1743. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yue, H.; Feng, K.; Deng, P.; Song, W.; Nie, X. Genome-wide identification, phylogeny and expressional profiles of mitogen activated protein kinase kinase kinase (MAPKKK) gene family in bread wheat (Triticum aestivum L.). BMC Genom. 2016, 17, 668. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J.J.G. Proteomics, Bioinformatics, KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Lopez, A.; Chang, H.C.; Bush, D.R. Amino acid transporters in plants. Biochim. Biophys. Acta Biomembr. 2000, 1465, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.E.; Bick, J.A.; Neelam, A.; Weston, K.N.; Hall, J.L. Biochemical and molecular characterization of sucrose and amino acid carriers in Ricinus communis. J. Exp. Bot. 1996, 47, 1211–1216. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Ma, H.; DePamphilis, C.W. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef] [PubMed]
- Li, W.H.; Gojobori, T.; Nei, M. Pseudogenes as a paradigm of neutral evolution. Nature 1981, 292, 237–239. [Google Scholar] [CrossRef] [PubMed]
- Die, J.V.; Castro, P.; Millán, T.; Gil, J. Segmental and tandem duplications driving the recent NBS-LRR gene expansion in the asparagus genome. Genes 2018, 9, 568. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yu, H.; Cao, P.B.; Fawal, N.; Mathé, C.; Azar, S.; Cassan-Wang, H.; Myburg, A.A.; Grima-Pettenati, J.; Marque, C.; et al. Explosive tandem and segmental duplications of multigenic families in Eucalyptus grandis. Genome Biol. Evol. 2015, 7, 1068–1081. [Google Scholar] [CrossRef]
- Schilling, S.; Kennedy, A.; Pan, S.; Jermiin, L.S.; Melzer, R. Genome-wide analysis of MIKC-type MADS-box genes in wheat: Pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 2020, 225, 511–529. [Google Scholar] [CrossRef]
- Liu, Y.; Patra, B.; Pattanaik, S.; Wang, Y.; Yuan, L. GATA and phytochrome interacting factor transcription factors regulate light-induced vindoline biosynthesis in Catharanthus roseus. Plant Physiol. 2019, 180, 1336–1350. [Google Scholar] [CrossRef]
- Tarczynski, M.C.; Jensen, R.G.; Bohnert, H.J.J.S. Stress protection of transgenic tobacco by production of the osmolyte mannitol. Science 1993, 259, 508–510. [Google Scholar] [CrossRef] [PubMed]
- Rentsch, D.; Hirner, B.; Schmelzer, E.; Frommer, W.B. Salt stress-induced proline transporters and salt stress-repressed broad specificity amino acid permeases identified by suppression of a yeast amino acid permease-targeting mutant. Plant Cell 1996, 8, 1437–1446. [Google Scholar] [CrossRef] [PubMed]
- Dinkeloo, K.; Boyd, S.; Pilot, G. Update on amino acid transporter functions and on possible amino acid sensing mechanisms in plants. Semin. Cell Dev. Biol. 2018, 74, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Dhatterwal, P.; Mehrotra, S.; Miller, A.J.; Mehrotra, R. Promoter profiling of Arabidopsis amino acid transporters: Clues for improving crops. Plant Mol. Biol. 2021, 107, 451–475. [Google Scholar] [CrossRef]





| Eudicots | Monocots | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Quino a | Tartary Buckwheat | Arabidopsis | Potato | Foxtail Millet | Maize | Rice | Wheat | ||
| AAAP | AAP | 24 (15.00%) | 17 (16.35%) | 8 (12.70%) | 8 (11.11%) | 20 (21.27%) | 24 (22.43%) | 19 (22.35%) | 66 (22.30%) |
| LHT | 29 (18.13%) | 11 (10.58%) | 10 (15.87%) | 11 (15.28%) | 12 (12.77%) | 15 (14.42%) | 6 (7.05%) | 24 (8.11%) | |
| GAT | 12 (7.50%) | 8 (7.69%) | 2 (3.17%) | 3 (4.17%) | 6 (6.38%) | 2 (1.92%) | 4 (4.71%) | 14 (4.73%) | |
| ProT | 6 (3.75%) | 5 (4.81%) | 3 (4.76%) | 4 (6.35%) | 1 (1.06%) | 2 (1.92%) | 3 (3.53%) | 9 (3.04%) | |
| AUX | 8 (5.00%) | 5 (4.81%) | 4 (6.35%) | 5 (7.94%) | 4 (4.25%) | 6 (5.60%) | 5 (5.88%) | 15 (5.07%) | |
| ATLa | 12 (7.50%) | 7 (6.73%) | 5 (7.94%) | 8 (11.11%) | 6 (6.38%) | 7 (6.54%) | 7 (8.24%) | 18 (6.08%) | |
| ANT | 2 (1.25%) | 1 (0.96%) | 4 (6.34%) | 5 (7.94%) | 2 (2.13%) | 3 (2.8%) | 4 (4.71%) | 18 (6.08%) | |
| ATLb | 28 (17.50%) | 17 (16.35%) | 10 (15.87%) | 8 (11.11%) | 14 (14.89%) | 17 (15.89%) | 10 (11.76%) | 40 (13.51%) | |
| APC | ACT | 9 (5.63%) | 5 (4.81%) | 1 (1.59%) | 1 (1.39%) | 8 (8.51%) | 7 (6.54%) | 7 (8.24%) | 21 (7.09%) |
| CAT | 18 (11.25%) | 14 (13.46%) | 9 (14.29%) | 9 (12.5%) | 12 (12.77%) | 14 (13.08%) | 11 (12.94%) | 31 (10.47%) | |
| PHS | 8 (5.00%) | 14 (13.46%) | 5 (7.94%) | 8 (11.11%) | 8 (8.51%) | 7 (6.54%) | 9 (10.59%) | 31 (10.47%) | |
| TTP | 4 (2.50%) | 0 (0.00%) | 2 (3.17%) | 2 (2.78%) | 1 (1.06%) | 3 (2.80%) | 0 (0.00%) | 9 (3.04%) | |
| Total | 160 | 104 | 63 | 72 | 94 | 107 | 85 | 296 |
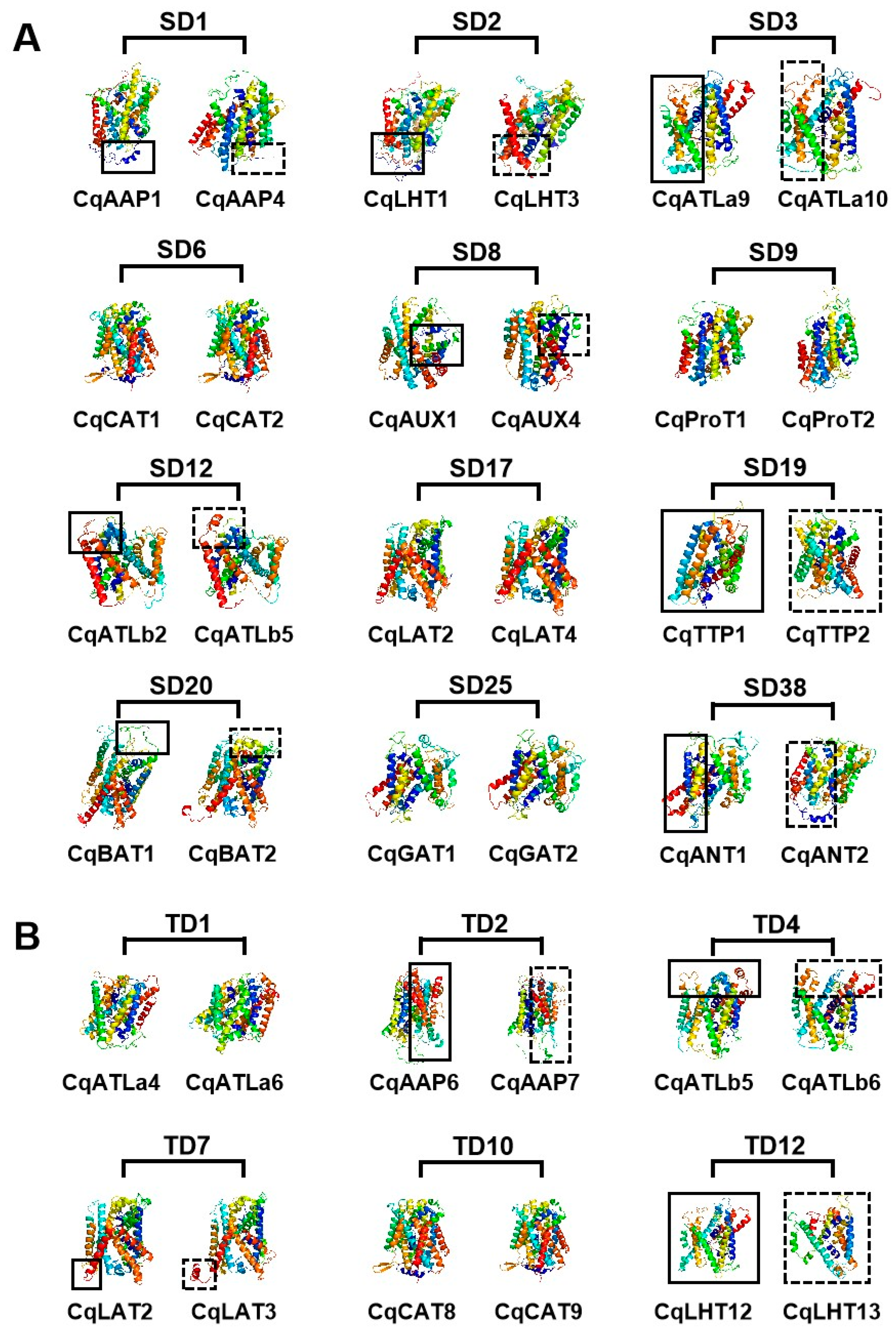
| Type of Duplication | Subfamily | No. of Duplicated Gene Groups | Gene and Protein Structure Variation | Range of Ka/Ks Values | Duplicated Gene Groups | ||
|---|---|---|---|---|---|---|---|
| No. of TM Variation | No. of Gene Structure Variation | No. of Conserved Motif Variation | |||||
| Tandem duplication | AAP | 7 | 2 | 3 | 3 | 0.17–0.38 | TD2, 5, 6, 9 |
| ATLa | 2 | 1 | 1 | 0 | 0.12–0.16 | TD1, 3 | |
| ATLb | 5 | 3 | 3 | 3 | 0.19–0.42 | TD4, 11, 14, 15, 17 | |
| LHT | 5 | 4 | 4 | 4 | 0.10–0.26 | TD12, 13, 16, 18 | |
| GAT | 4 | 4 | 4 | 4 | 0.13–0.37 | TD19, 20 | |
| CAT | 1 | 0 | 0 | 0 | 0.35 | TD10 | |
| PHS | 2 | 2 | 0 | 2 | 0.12–0.14 | TD7, 8 | |
| Sum | 26 | 16 | 15 | 16 | 0.10–0.42 | ||
| Segmental duplication | AAP | 6 | 4 | 1 | 2 | 0.09–0.19 | SD1, 4, 7, 14, 21, 41 |
| ANT | 1 | 0 | 0 | 1 | 0.11 | SD38 | |
| ATLa | 3 | 1 | 1 | 1 | 0.11–0.32 | SD3, 11, 13 | |
| ATLb | 8 | 7 | 3 | 4 | 0.17–0.70 | SD12, 15, 16, 31, 32, 39, 42, 43 | |
| AUX | 3 | 0 | 1 | 1 | 0–0.20 | SD8, 10, 44 | |
| LHT | 9 | 4 | 3 | 4 | 0.05–0.34 | SD2, 5, 18, 29, 30, 35, 36, 37, 45 | |
| ProT | 2 | 0 | 0 | 2 | 0.08–0.25 | SD9, 40 | |
| GAT | 3 | 3 | 2 | 2 | 0.20–0.48 | SD25, 46, 47 | |
| ACT | 3 | 2 | 2 | 3 | 0.11–0.39 | SD20, 33, 34 | |
| CAT | 5 | 2 | 3 | 4 | 0.09–0.25 | SD6. 22, 24, 27, 28 | |
| PHS | 2 | 0 | 1 | 1 | 0.12–0.16 | SD17, 23 | |
| TTP | 2 | 1 | 0 | 1 | 0.31–0.59 | SD19, 26 | |
| Sum | 47 | 24 | 17 | 26 | 0–0.70 | ||
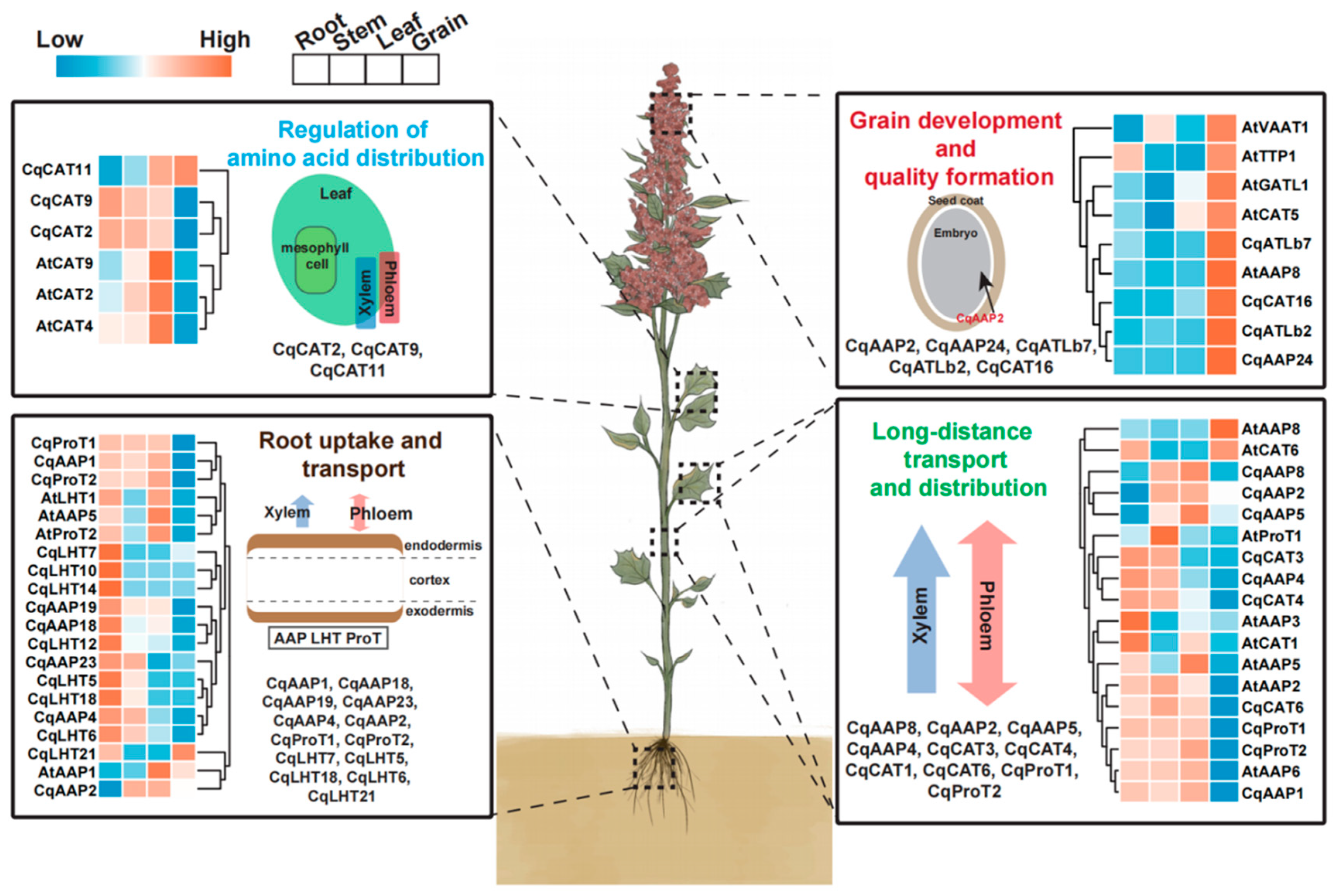
| Subfamilies | AATs in Quinoa | Expression Organization | Functional Prediction |
|---|---|---|---|
| AAP | CqAAP15, 24 | G | Play a role in the formation of grain quality |
| CqAAP21, 3 | L | Play a role in the transfer and distribution of amino acids from stem to leaf | |
| CqAAP6, 12, 13, 19 | R | Participate in the uptake of amino acids by roots | |
| CqAAP10, 20 | L, G | Transfer and distribution of amino acids from leaf to grain | |
| CqAAP4, 7, 9, 16, 22, 23 | R, S | Amino acid uptake in root and transport in stem | |
| CqAAP2, 5, 8, 11 | S, L | Transfer and distribution of amino acids from stem to leaf | |
| CqAAP1, 17, 18 | R, S, L | Uptake of amino acids in root, transfer and distribution from stem to leaf | |
| GAT | CqGAT6, 8, 11 | G | Play a role in grain development |
| CqGAT1, 9 | R | Play a role in amino acid uptake in root | |
| CqGAT10 | S | Long-distance transport of amino acids in stem | |
| CqGAT2, 7, 12 | R, S | Amino acid uptake in root and long-distance transport of GABA | |
| CqGAT3, 5 | R, S, L | Amino acid uptake in root; long-distance transport in the stem; distribution of amino acids in leaf | |
| ProT | CqProT4 | S | Transport of proline in the stem |
| CqProT3, 5 | S, L | Proline transport in the stem; distribution in stem and leaf | |
| CqProT1, 2, 6 | R, S, L | Proline uptake in roots; long-distance transport in the stem; distribution in stem and leaf | |
| LHT | CqLHT7, 10, 12, 14, 15, 16, 22, 27, 28 | R | Participate in amino acid uptake in root |
| CqLHT8, 9, 26 | S | Long-distance transport of amino acids in stem | |
| CqLHT21 | R, G | Amino acid uptake in roots; transfer in storage tissue | |
| CqLHT1, 2, 4, 5, 6, 18, 20, 25, 29 | R, S | Amino acid uptake in root; transportation in root and stem | |
| CqLHT17 | S, L | Transport of amino acids in root and stem | |
| CqLHT3 | R, S, L | Uptake of amino acids in root; long-distance transport from stem to leaf | |
| AUX | CqAUX1, 4 | R | Uptake and transport of amino acids in root |
| CqAUX7, 8 | R, S | Transport of amino acids in root and stem | |
| Cq2, 3, 5 | R, S, L | Long-distance transport and distribution of amino acids in root, stem and leaf | |
| CqAUX6 | L, G | Transfer of amino acids in leaf and grain development | |
| ANT | CqANT1, 2 | R, S | Long-distance transport of amino acids in root and stem |
| ATLa | CqATLa6 | G | Development of grain and flower |
| CqATLa1, 2, 4, 7 | R | Amino acid uptake in root | |
| CqATLa12 | S | Transport of amino acids in the stem | |
| CqATLa3 | R, G | Amino acid uptake in root and grain development | |
| CqATLa5, 8 | R, S | Amino acid uptake in root and transport in stem | |
| CqATLa9, 10 | S, L | Amino acid transport in the stem; regulates amino acid distribution in the leaf | |
| CqATLa11 | R, S, L | Amino acid uptake in root; transport in the stem; regulate amino acid distribution in leaf | |
| ATLb | CqATLb2, 7, 8, 18, 20 | G | Distribution of amino acids in grain |
| CqATLb4, 9, 11 | L | Transfer of amino acids in the leaf | |
| CqATLb5 | S | Transport of amino acids in stem | |
| CqATLb15, 19, 28 | R | Amino acid uptake in root | |
| CqATLb13, 24 | R, G | Amino acid uptake in root and distribution in grain | |
| CqATLb10, 27 | R, S | Amino acid uptake in root and transport in stem | |
| CqATLb1, 21 | S, L | Transfer of amino acids from stem to leaf | |
| CqATLb25 | S, G | Transfer of amino acids from stem to leaf, and to grain | |
| CqATLb3, 6, 16, 17, 26 | R, S, L | Uptake of amino acids in root; long-distance transport from stem to leaf | |
| CqATLb22 | R, L, G | Amino acid uptake in root and transfer from leaf to grain | |
| CAT | CqCAT10, 14, 16 | G | Participate in grain development |
| CqCAT7, 12, 17, 18 | R | Uptake of amino acids in root | |
| CqCAT1, 3, 4, 5, 8, 13 | R, S | Amino acid uptake in root; transport in stem | |
| CqCAT15 | R, G | Amino acid uptake in root; distribution in grain | |
| CqCAT11 | L, G | Transfer and distribution of amino acids from leaf to grain | |
| CqCAT2, 6, 9 | R, S, L | Amino acid transport and distribution in leaf; maintain cellular ion balance | |
| ACT | CqBAT2 | R | Transport and distribution of amino acids in root |
| CqBAT4, 5, 8 | S | Transport of amino acids | |
| CqBAT1, 6 | R, G | Role in amino acid transport; grain development | |
| CqBAT3 | S, G | Amino acid transport in stem and grain development | |
| CqBAT9 | R, S, G | Long-distance transport of amino acids; grain development | |
| CqBAT7 | R, S, L | Long-distance transport and distribution of amino acids | |
| PHS | CqLAT3, 5, 7 | G | Distribution of amino acids in the grain |
| CqLAT2 | S | Transport of amino acids in the stem | |
| CqLAT1, 4, 6, 8 | R, S | Amino acid uptake in root; transport in stem | |
| TTP | CqTTP4 | R | Amino acid uptake in root |
| CqTTP3 | S | Transport of amino acids in stem | |
| CqTTP1, 2 | S, L | Transport and distribution of amino acids |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Huang, J.; Zhang, Y.; Yang, X.; Gou, T.; Ren, A.; Ding, P.; Wu, X.; Sun, M.; Gao, Z. Functional Framework of Amino Acid Transporters in Quinoa: Genome-Wide Survey, Homology, and Stress Response. Agronomy 2024, 14, 1648. https://doi.org/10.3390/agronomy14081648
Li L, Huang J, Zhang Y, Yang X, Gou T, Ren A, Ding P, Wu X, Sun M, Gao Z. Functional Framework of Amino Acid Transporters in Quinoa: Genome-Wide Survey, Homology, and Stress Response. Agronomy. 2024; 14(8):1648. https://doi.org/10.3390/agronomy14081648
Chicago/Turabian StyleLi, Linghong, Jianxun Huang, Yulai Zhang, Xinhui Yang, Tong Gou, Aixia Ren, Pengcheng Ding, Xiangyun Wu, Min Sun, and Zhiqiang Gao. 2024. "Functional Framework of Amino Acid Transporters in Quinoa: Genome-Wide Survey, Homology, and Stress Response" Agronomy 14, no. 8: 1648. https://doi.org/10.3390/agronomy14081648
APA StyleLi, L., Huang, J., Zhang, Y., Yang, X., Gou, T., Ren, A., Ding, P., Wu, X., Sun, M., & Gao, Z. (2024). Functional Framework of Amino Acid Transporters in Quinoa: Genome-Wide Survey, Homology, and Stress Response. Agronomy, 14(8), 1648. https://doi.org/10.3390/agronomy14081648




