Transcriptome Sequencing and Metabolome Analysis Reveals the Regulatory and Molecular Mechanisms of the Grain Filling Rate in Foxtail Millet (Setaria italica L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Investigation of Grain Development and GFR
2.3. RNA Sequencing and Transcriptomic Profiling Analysis
2.4. Metabolome Profiling Analysis
2.5. Conjoint Analysis of Transcriptomes and Metabolomes
2.6. Quantitative qRT-PCR
2.7. Statistical Analysis
3. Results
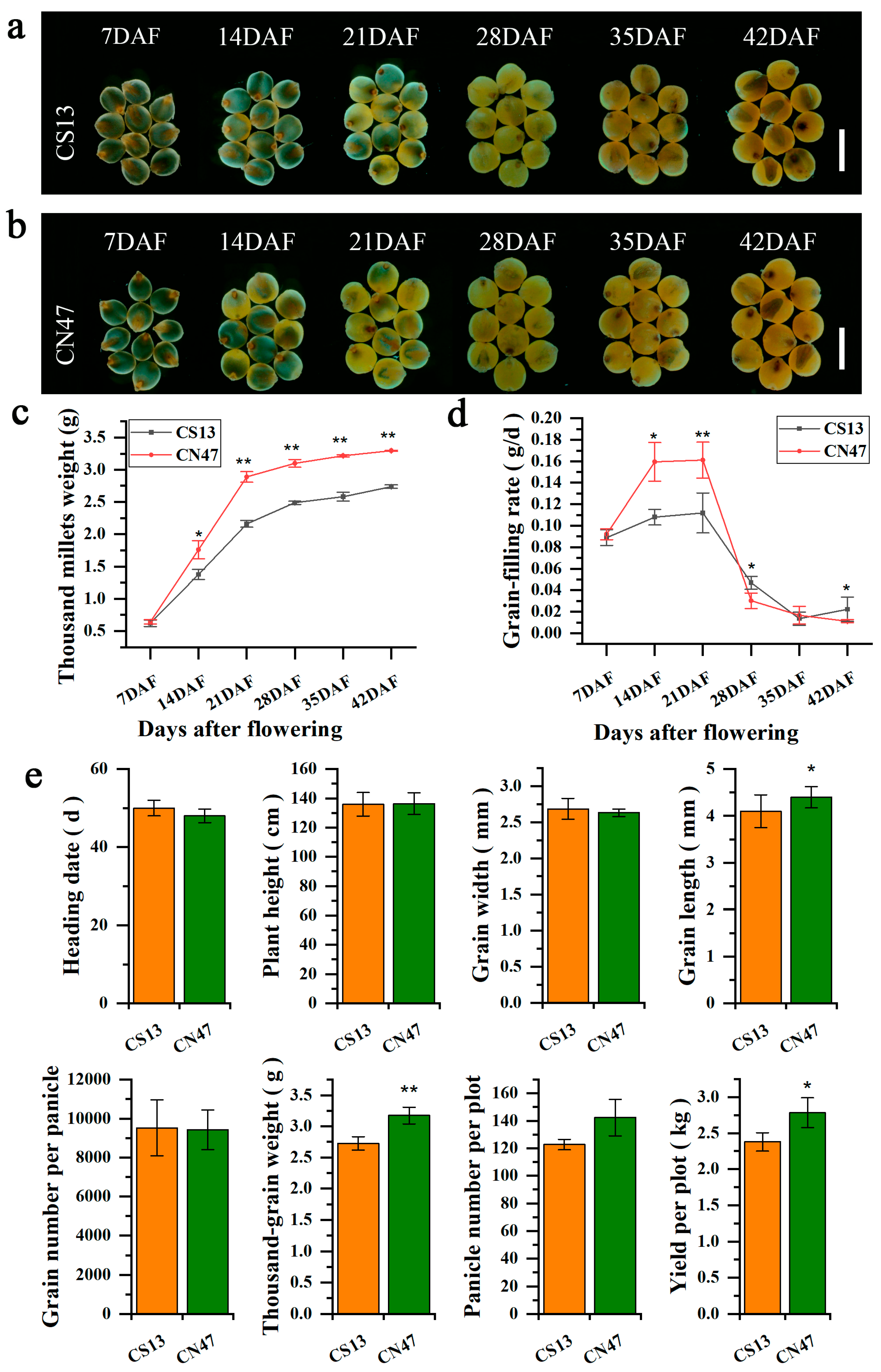
3.1. Characterization of Grain Filling Rate-Related Phenotypes in Foxtail Millet
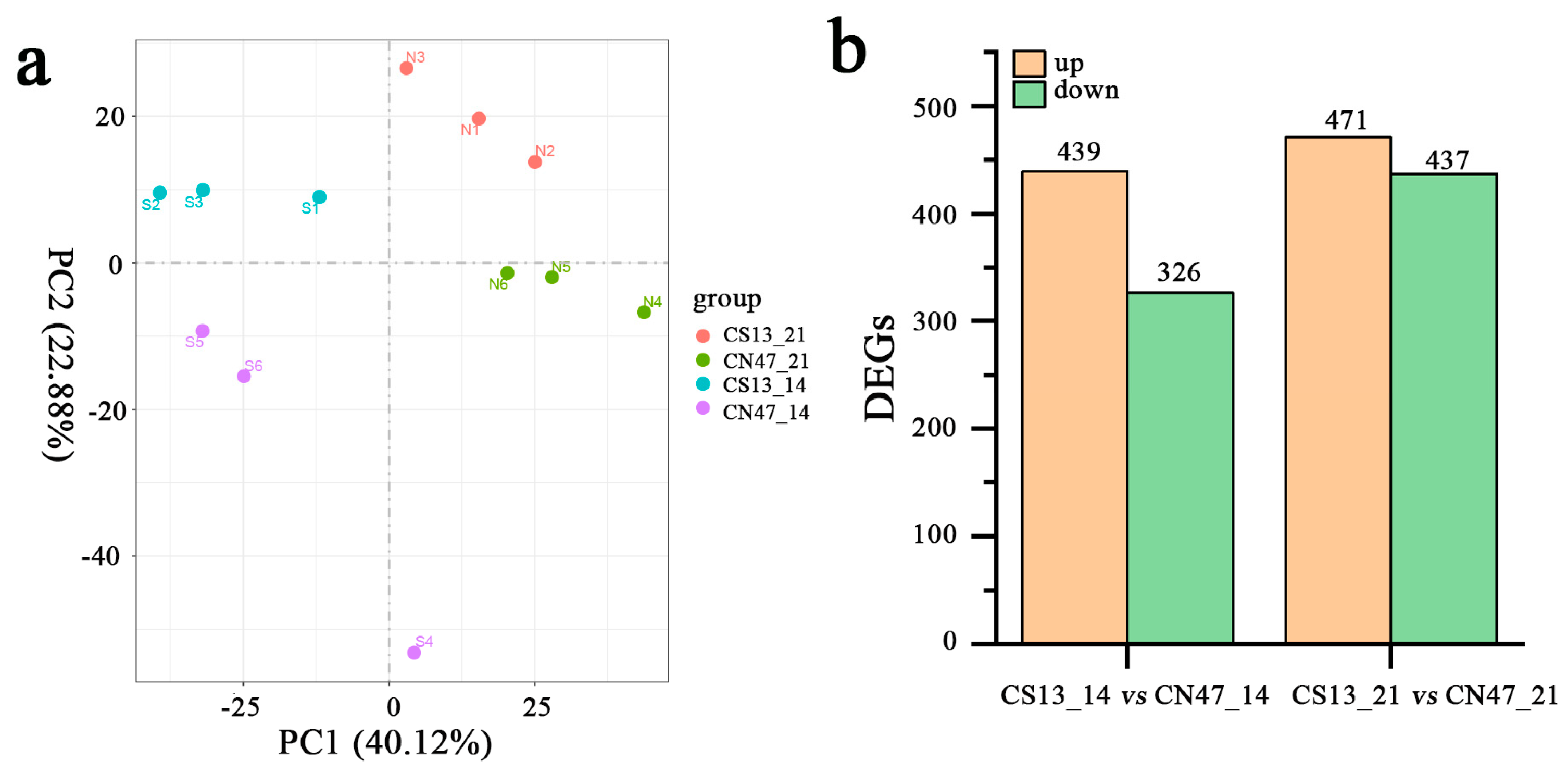
3.2. Transcriptomic Differences in Grains during Grain Filling Stages between Foxtail Millet Cultivars
3.3. Metabolite Differences in Grains during Grain Filling Stages between Foxtail Millet Cultivars
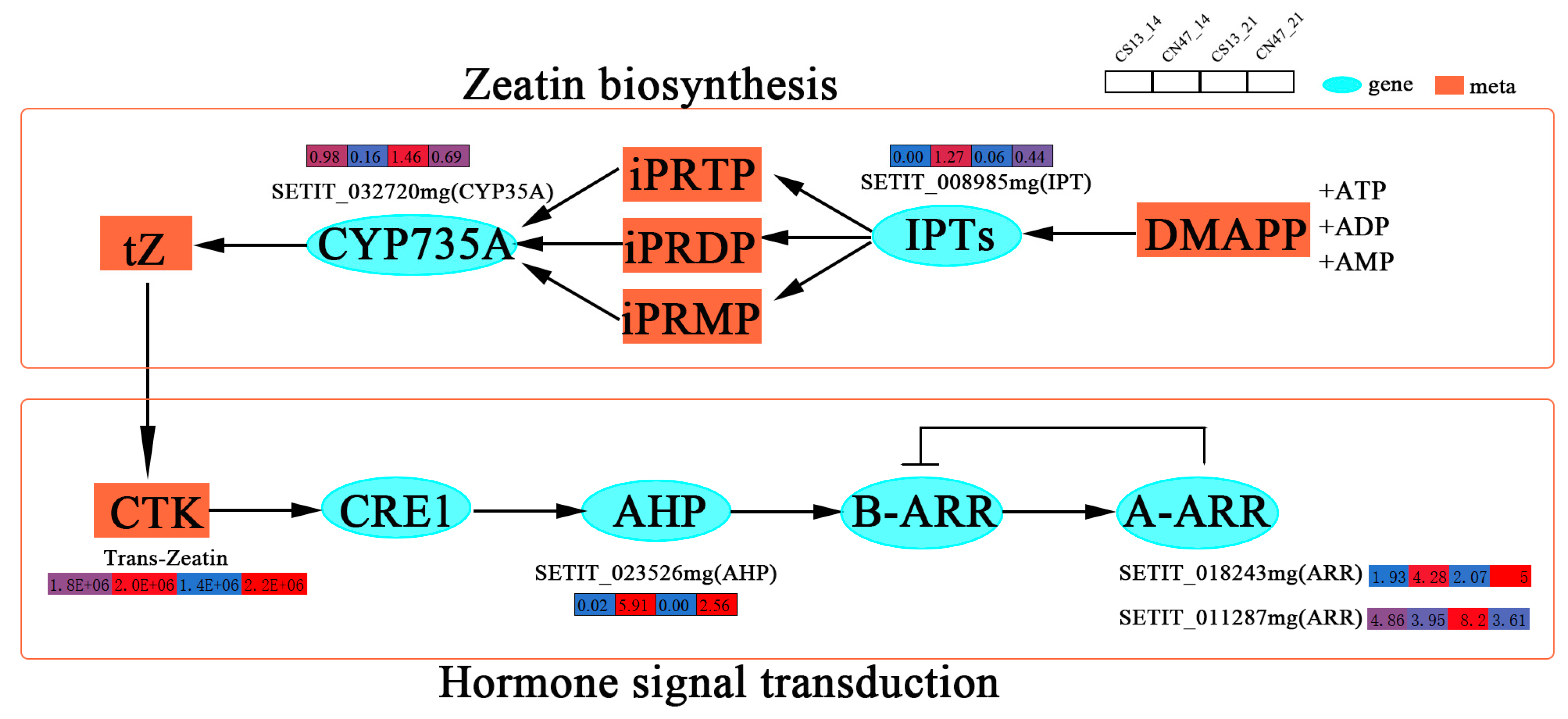
3.4. Integrated Transcriptome and Metabolome Analysis
3.5. IAA and CTK Were Key Regulators of the Grain Filling Rate
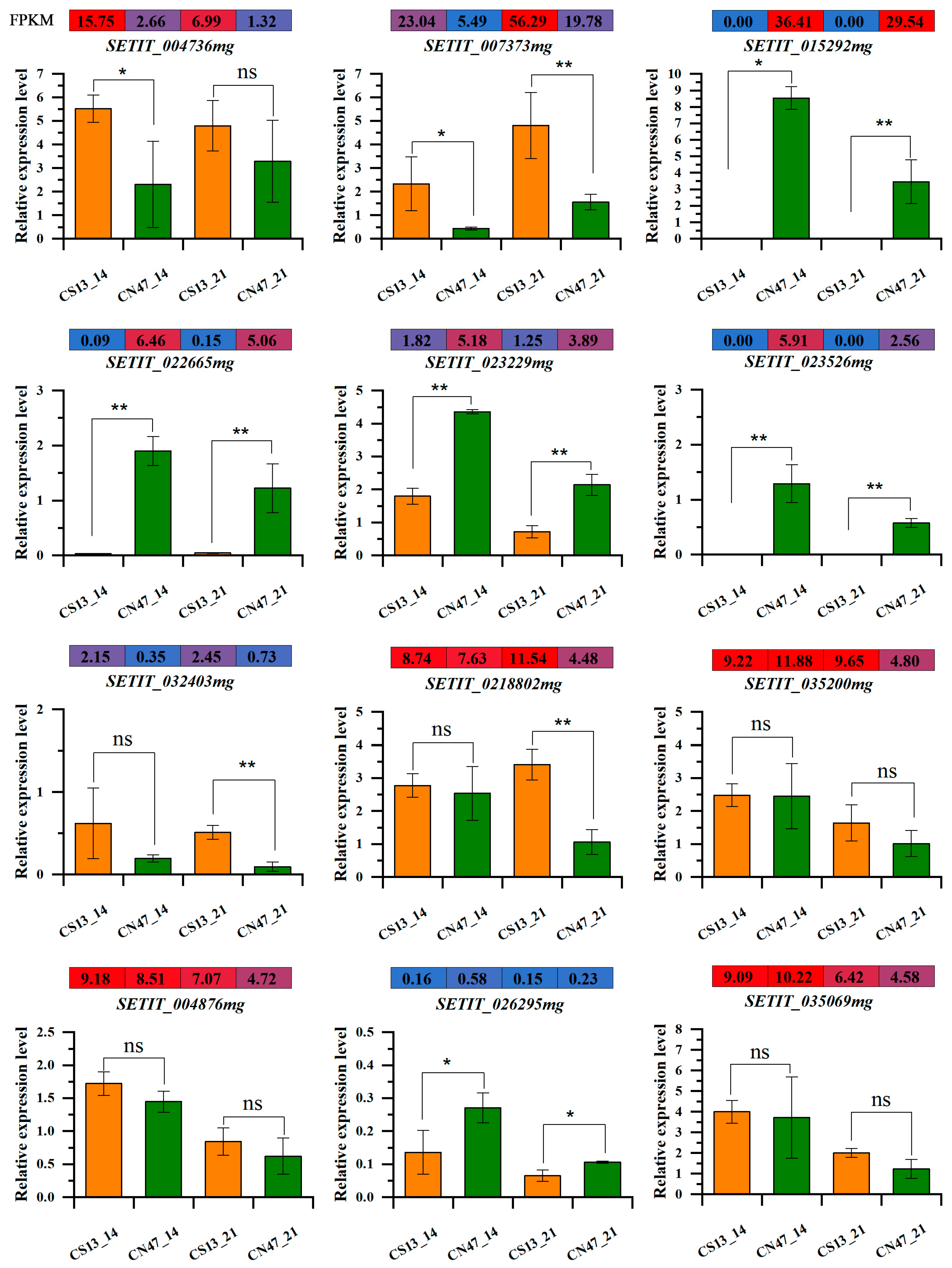
3.6. qRT-PCR Verification of RNA-Seq Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Lata, C.; Gupta, S.; Prasad, M. Foxtail millet: A model crop for genetic and genomic studies in bioenergy grasses. Crit. Rev. Biotechnol. 2013, 33, 328–343. [Google Scholar] [CrossRef] [PubMed]
- Memariani, Z.; Abbas, S.; Hassan, S.; Ahmadi, A.; Chabra, A. Naringin and naringenin as anticancer agents and adjuvants in cancer combination therapy: Efficacy and molecular mechanisms of action, a comprehensive narrative review. Pharmacol. Res. 2021, 171, 105264. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Purewal, S.; Sandhu, K.; Kaur, M.; Salar, R. Millets: A cereal grain with potent antioxidants and health benefits. J. Food Meas. Charact. 2019, 13, 793–806. [Google Scholar] [CrossRef]
- Sachdev, N.; Goomer, S.; Singh, L. Foxtail millet: A potential crop to meet future demand scenario for alternative sustainable protein. J. Sci. Food Agric. 2021, 101, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Sushree, S.; Rana, S.; Suranjika, S.; Muthamilarasan, M.; Parida, A.; Prasad, M. Genetic determinants of micronutrient traits in graminaceous crops to combat hidden hunger. Theor. Appl. Genet. 2021, 134, 3147–3165. [Google Scholar] [CrossRef] [PubMed]
- Amadou, I.; Amza, T.; Shi, Y.; Le, G. Chemical analysis and antioxidant properties of foxtail millet bran extracts. Songklanakarin J. Sci. Technol. 2011, 33, 509–515. [Google Scholar]
- Liang, S.; Yang, G.; Ma, Y. Chemical characteristics and fatty acid profile of foxtail millet bran oil. J. Am. Oil Chem. Soc. 2010, 87, 63–67. [Google Scholar] [CrossRef]
- Sharma, N.; Niranjan, K. Foxtail millet: Properties, processing, health benefits, and uses. Food Rev. Int. 2018, 34, 329–363. [Google Scholar] [CrossRef]
- Wang, D.; Su, M.; Hao, J.; Li, Z.; Dong, S.; Yuan, X.; Li, X.; Gao, L.; Chu, X.; Yang, G.; et al. Dynamic transcriptome landscape of foxtail millet grain development. Seed Biol. 2023, 2, 19. [Google Scholar] [CrossRef]
- Xing, Y.; Zhang, Q. Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 2010, 61, 421–442. [Google Scholar] [CrossRef]
- Li, X.; Pan, Z. A study on the grain filling characteristic of different weight wheat. Rev. China Agric. Sci. Technol. 2005, 7, 26–30. [Google Scholar] [CrossRef]
- Shouichi, Y.; Hara, T. Effects of air temperature and light on grain filling of an indica and a japonica rice (Oryza sativa L.) under controlled environmental conditions. J. Soil. Sci. Plant Nutr. 1977, 23, 15. [Google Scholar] [CrossRef]
- Wiegand, C.; Cuellar, J. Duration of grain filling and kernel weight of wheat as affected by temperature. Crop Sci. 1981, 21, 95–101. [Google Scholar] [CrossRef]
- Ma, J.; Ming, D.; Ma, W.; Xu, F. Effects of different nitrogen application periods on rice starch accumulation and starch synthesis Studies on the activity changes of related enzymes. Sci. Agric. Sin. 2005, 38, 290–296. [Google Scholar]
- Wang, Z.; Xu, Y.; Chen, T.; Zhang, H.; Yang, J.; Zhang, J. Abscisic acid and the key enzymes and genes in sucrose-to-starch conversion in rice spikelets in response to soil drying during grain filling. Planta 2015, 241, 1091–1107. [Google Scholar] [CrossRef]
- Sanford, D. Variation in kernel growth characters among soft red winter wheats. Crop Sci. 1985, 25, 626–630. [Google Scholar] [CrossRef]
- Mashiringwani, N.; Mashingaidze, K.; Kangai, J.; Olsen, K. Genetic basis of grain filling rate in wheat (Triticum aestivum L. emend. Thell.). Euphytica 1994, 76, 33–44. [Google Scholar] [CrossRef]
- Jones, D.; Peterson, M.; Geng, S. Association between grain filling rate and duration and yield components in rice. Crop Sci. 1979, 19, 641–644. [Google Scholar] [CrossRef]
- Zhao, Y.; Peng, T.; Sun, H.; Teotia, S.; Wen, H.; Du, Y.; Zhang, J.; Li, J.; Tang, G.; Xue, H.; et al. miR1432-OsACOT (acyl-CoA thioesterase) module determines grain yield via enhancing grain filling rate in rice. Plant Biotechnol. J. 2019, 17, 712–723. [Google Scholar] [CrossRef]
- Zhu, D.; Wang, H.; Liu, D.; Gao, D.; Lu, G.; Wang, J.; Gao, Z.; Lu, C. Characteristics of grain filling and dehydration in wheat. Sci. Agric. Sin. 2019, 52, 4251–4261. [Google Scholar] [CrossRef]
- Wei, X.; Jiao, G.; Lin, H.; Sheng, Z.; Shao, G.; Xie, L.; Tang, S.; Xu, Q.; Hu, P. GRAIN INCOMPLETE FILLING 2 regulates grain filling and starch synthesis during rice caryopsis development. J. Integr. Plant Biol. 2017, 59, 134–153. [Google Scholar] [CrossRef]
- Ishibashi, Y.; Okamura, K.; Miyazaki, M.; Phan, T.; Yuasa, T.; Iwaya-Inoue, M. Expression of rice sucrose transporter gene OsSUT1 in sink and source organs shaded during grain filling may affect grain yield and quality. Environ. Exp. Bot. 2014, 97, 49–54. [Google Scholar] [CrossRef]
- Bai, A.; Lu, X.; Li, D.; Liu, J.; Liu, C. NF-YB1-regulated expression of sucrose transporters in aleurone facilitates sugar loading to rice endosperm. Cell Res. 2016, 26, 384–388. [Google Scholar] [CrossRef]
- Sosso, D.; Luo, D.; Li, Q.; Sasse, J.; Yang, J.; Gendrot, G.; Suzuki, M.; Koch, K.; McCarty, D.; Chourey, P.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, J.; Zhang, Z.; Wu, Y. Transactivation of Sus1 and Sus2 by Opaque2 is an essential supplement to sucrose synthase-mediated endosperm filling in maize. Plant Biotechnol. J. 2020, 18, 1897–1907. [Google Scholar] [CrossRef]
- Wang, E.; Wang, J.; Zhu, X.; Hao, W.; Wang, L.; Li, Q.; Zhang, L.; He, W.; Lu, B.; Lin, H.; et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 2008, 40, 1370–1374. [Google Scholar] [CrossRef]
- Kuanar, S.; Molla, K.; Chattopadhyay, K.; Sarkar, R.; Mohapatra, P. Introgression of Sub1 (SUB1) QTL in mega rice cultivars increases ethylene production to the detriment of grain-filling under stagnant flooding. Sci. Rep. 2019, 9, 18567. [Google Scholar] [CrossRef]
- Zhu, G.; Ye, N.; Yang, J.; Peng, X.; Zhang, J. Regulation of expression of starch synthesis genes by ethylene and ABA in relation to the development of rice inferior and superior spikelets. J. Exp. Bot. 2011, 62, 3907–3916. [Google Scholar] [CrossRef]
- Zhang, W.; Cao, Z.; Zhou, Q.; Chen, J.; Xu, G.; Gu, J.; Liu, L.; Wang, Z.; Yang, J.; Zhang, H. Grain filling characteristics and their relations with endogenous hormones in large- and small-grain mutants of rice. PLoS ONE 2016, 11, e0165321. [Google Scholar] [CrossRef] [PubMed]
- Panda, B.; Sekhar, S.; Dash, S.; Behera, L.; Shaw, B. Biochemical and molecular characterisation of exogenous cytokinin application on grain filling in rice. BMC Plant Biol. 2018, 18, 89. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhang, M.; Zhou, Y.; Wang, Y.; Shen, J.; Chen, H.; Zhang, L.; Lü, B.; Liang, G.; Liang, J. The Rice G Protein gamma Subunit DEP1/qPE9-1 Positively Regulates Grain-Filling Process by Increasing Auxin and Cytokinin Content in Rice Grains. Rice 2019, 12, 91. [Google Scholar] [CrossRef]
- Liu, E.; Zeng, S.; Zhu, S.; Liu, Y.; Wu, G.; Zhao, K.; Liu, X.; Liu, Q.; Dong, Z.; Dang, X.; et al. Favorable Alleles of GRAIN-FILLING RATE1 increase the grain-filling rate and yield of rice. Plant Physiol. 2019, 181, 1207–1222. [Google Scholar] [CrossRef]
- Lin, Y.; Jiang, X.; Yang, T.; Yang, X.; Wang, Z.; Wu, F.; Liu, S.; Li, C.; Deng, M.; Ma, J.; et al. Identifcation and validation of stable quantitative trait loci for grain filling rate in common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2020, 133, 2377–2385. [Google Scholar] [CrossRef]
- Bennetzen, J.; Schmutz, J.; Wang, H.; Percifield, R.; Hawkins, J.; Pontaroli, A.; Estep, M.; Feng, L.; Vaughn, J.; Grimwood, J.; et al. Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 2012, 30, 555–561. [Google Scholar] [CrossRef]
- Jia, G.; Huang, X.; Zhi, H.; Zhao, Y.; Zhao, Q.; Li, W.; Chai, Y.; Yang, L.; Liu, K.; Lu, H.; et al. A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat. Genet. 2013, 45, 957–961. [Google Scholar] [CrossRef]
- Wang, T.; Song, H.; Li, P.; Wei, Y.; Hu, N.; Chen, Z.; Wang, W.; Liu, J.; Zhang, B.; Peng, R. Transcriptome Analysis Provides Insights into Grain Filling in Foxtail Millet (Setaria italica L.). Int. J. Mol. Sci. 2020, 21, 5031. [Google Scholar] [CrossRef]
- Wang, T.; Wang, S.; Tong, Y.; Wei, Y.; Li, P.; Hu, N.; Liu, Y.; Zhao, Z.; Zhao, Y.; Chen, H.; et al. Spatiotemporal dynamics of the foxtail millet transcriptome during grain filling. Physiol. Plant 2024, 176, e14157. [Google Scholar] [CrossRef]
- Wang, T.; Xing, L.; Song, H.; Wei, Y.; Li, P.; Lu, Q.; Hu, N.; Liu, Y.; Zhao, Y.; Liu, J.; et al. Large-scale metabolome analysis reveals dynamic changes of metabolites during foxtail millet grain filling. Food Res. Int. 2023, 165, 112516. [Google Scholar] [CrossRef]
- Wang, T.; Lu, Q.; Song, H.; Hu, N.; Wei, Y.; Li, P.; Liu, Y.; Zhao, Z.; Liu, J.; Zhang, B.; et al. DNA methylation and RNA-sequencing analysis show epigenetic function during grain filling in foxtail millet (Setaria italica L.). Front. Plant Sci. 2021, 12, 741415. [Google Scholar] [CrossRef]
- Song, H.; Wang, T.; Li, L.; Xing, L.; Xie, H.; Feng, B.; Liu, J. Comparative transcriptome analysis provides insights into grain filling commonalities and differences between foxtail millet [Setaria italica (L.) P. Beauv.] varieties with different panicle types. Peer J. 2022, 10, e12968. [Google Scholar] [CrossRef]
- Bolger, A.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, 106. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.; Blake, J.; Botstein, D.; Butler, H.; Cherry, J.; Davis, A.; Dolinski, K.; Dwight, S.; Eppig, J.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, 277–280. [Google Scholar] [CrossRef]
- Want, E.; Masson, P.; Michopoulos, F.; Wilson, I.; Theodoridis, G.; Plumb, R.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J. Global metabolic profiling of animal andhuman tissues via UPLC-MS. Nat. Protoc. 2012, 8, 17–32. [Google Scholar] [CrossRef]
- Wen, B.; Mei, Z.; Zeng, C.; Liu, S. metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinform. 2017, 18, 183. [Google Scholar] [CrossRef]
- Zhang, H.; Tan, G.; Yang, L.; Yang, J.; Zhang, J.; Zhao, B. Hormones in the grains and roots in relation to post-anthesis development of inferior and superior spikelets in japonica/indica hybrid rice. Plant Physiol. Biochem. 2009, 47, 195–204. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 2012, 5, 334–338. [Google Scholar] [CrossRef]
- Ma, X.; Dai, S.; Qin, N.; Zhu, C.; Qin, J.; Li, J. Genome-wide identification and expression analysis of the SAUR gene family in foxtail millet (Setaria italica L.). BMC Plant Biol. 2023, 23, 31. [Google Scholar] [CrossRef]
- Mok, M. Cytokinins and plant development—An overview. In Cytokinins, 1st ed.; Mok, M.C., Ed.; CRC Press: Boca Raton, FL, USA, 2019; pp. 15–66. [Google Scholar] [CrossRef]
- Ishimaru, T.; Matsuda, T.; Ohsugi, R.; Yamagishi, T. Morphological development of rice caryopses located at the different positions in a panicle from early to middle stage of grain filling. Funct. Plant Biol. 2003, 30, 1139–1149. [Google Scholar] [CrossRef]
- Liu, D.; Zhao, H.; Xiao, Y.; Zhang, G.; Cao, S.; Yin, W.; Qian, Y.; Yin, Y.; Zhang, J.; Chen, S.; et al. A cryptic inhibitor of cytokinin phosphorelay controls rice grain size. Mol. Plant 2022, 15, 293–307. [Google Scholar] [CrossRef]
- Zhao, Y. Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu. Rev. Plant Biol. 2018, 69, 417–435. [Google Scholar] [CrossRef]
- Slafer, G.; Foulkes, M.; Reynolds, M.; Murchie, E.; Carmo-Silva, E.; Flavell, R.; Gwyn, J.; Sawkins, M.; Griffiths, S. A ‘wiring diagram’ for sink strength traits impacting wheat yield potential. J. Exp. Bot. 2023, 74, 40–71. [Google Scholar] [CrossRef]
- Wang, G.; Kang, M.; Moreno, O. Genetic analyses of grain-filling rate and duration in maize. Field Crop Res. 1999, 61, 211–222. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, Z.; Cui, Z.; Hu, Y.; Wang, B.; Tang, J. Genetic analysis of grain filling rate using conditional QTL mapping in maize. PLoS ONE 2013, 8, e56344. [Google Scholar] [CrossRef]
- Hao, X.; Wang, G.; Wang, X.; Yang, H.; Cheng, Q.; Qin, Y. Breeding and cultivation techniques of millet variety Changsheng 13 suitable for mechanized production. China Seed Ind. 2019, 10, 74–76. [Google Scholar] [CrossRef]
- Fan, H.; Zhao, P.; Zhang, A.; Li, Y.; Wang, L.; Wang, R.; Guo, E. Breeding of high-quality herbicide-resistant millet variety Changnong 47 and its high-yield cultivation techniques. J. Hebei Agric. Sci. 2022, 26, 1–5. [Google Scholar]
- Garg, R.; Jain, M. RNA-Seq for Transcriptome Analysis in Non-model Plants. Methods Mol. Biol. 2013, 1069, 43–58. [Google Scholar] [CrossRef]
- Li, X.; Gao, J.; Song, J.; Guo, K.; Hou, S.; Wang, X.; He, Q.; Zhang, Y.; Yang, Y.; Tang, J.; et al. Multi-omics analyses of 398 foxtail millet accessions reveal genomic regions associated with domestication, metabolite traits, and anti-inflammatory effects. Mol. Plant 2022, 15, 1367–1383. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, J.; Qie, Q.; Yang, Y.; Hou, S.; Wang, X.; Li, X.; Han, Y. Comparative Analysis of Flavonoid Metabolites in Foxtail Millet (Setaria italica) with Different Eating Quality. Life 2021, 11, 578. [Google Scholar] [CrossRef]
- Schubert, H.; Blumenthal, R.; Cheng, X. Many paths to methyltransfer: A chronicle of convergence. Trends Biochem. Sci. 2003, 28, 329–335. [Google Scholar] [CrossRef]
- Rea, P. Plant ATP-Binding cassette transporters. Annu. Rev. Plant Biol. 2007, 58, 347–375. [Google Scholar] [CrossRef]
- Walter, S.; Kahla, A.; Arunachalam, C.; Perochon, A.; Khan, M.; Scofield, S.; Doohan, F. A wheat ABC transporter contributes to both grain formation and mycotoxin tolerance. J. Exp. Bot. 2015, 66, 2583–2593. [Google Scholar] [CrossRef]
- Bhati, K.; Alok, A.; Kumar, A.; Kaur, J.; Tiwari, S.; Pandey, A. Silencing of ABCC13 transporter in wheat reveals its involvement in grain development, phytic acid accumulation and lateral root formation. J. Exp. Bot. 2016, 67, 4379–4389. [Google Scholar] [CrossRef]
- Ma, B.; Cao, X.; Li, X.; Bian, Z.; Zhang, Q.; Fang, Z.; Liu, J.; Li, Q.; Liu, Q.; Zhang, L.; et al. Two ABCI family transporters, OsABCI15 and OsABCI16, are involved in grain-filling in rice. J. Genet. Genom. 2023; in press. [Google Scholar] [CrossRef]
- Gao, F.; Wang, K.; Liu, Y.; Chen, Y.; Chen, P.; Shi, Z.; Luo, J.; Jiang, D.; Fan, F.; Zhu, Y.; et al. Blocking miR396 increases rice yield by shaping inflorescence architecture. Nat. Plants 2015, 2, 15196. [Google Scholar] [CrossRef]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef]
- Li, H.; Lu, Q.; Deng, J.; Huang, J.; Cai, F.; Liang, C.; Chen, Q.; Wang, Y.; Zhu, L.; Zhang, X.; et al. Transcriptome analysis reveals key seed-development genes in common buckwheat (Fagopyrum esculentum). Int. J. Mol. Sci. 2019, 20, 4303. [Google Scholar] [CrossRef]
- Xu, X.; E, Z.; Zhang, D.; Yun, Q.; Zhou, Y.; Niu, B.; Chen, C. OsYUC11-mediated auxin biosynthesis is essential for endosperm development of rice. Plant Physiol. 2021, 185, 934–950. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, M.; Liang, J. RGB1 Regulates Grain Development and Starch Accumulation Through Its Effect on OsYUC11-Mediated Auxin Biosynthesis in Rice Endosperm Cells. Front. Plant Sci. 2021, 12, 585174. [Google Scholar] [CrossRef] [PubMed]
- Forestan, C.; Meda, S.; Varotto, S. ZmPIN1-mediated auxin transport is related to cellular differentiation during maize embryogenesis and endosperm development. Plant Physiol. 2010, 152, 1373–1390. [Google Scholar] [CrossRef] [PubMed]
- Adamowski, M.; Friml, J. PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Bailly, A.; Zwiewka, M.; Sovero, V.; Di, D.; Ge, P.; Oehri, J.; Aryal, B.; Hao, P.; Linnert, M.; et al. TWISTED DWARF1 mediates the action of auxin transport inhibitors on actin cytoskeleton dynamics. Plant Cell 2016, 28, 930–948. [Google Scholar] [CrossRef] [PubMed]
- Marchant, A.; Kargul, J.; May, S.; Delbarre, A.; Perrot-Rechenmann, C.; Bennett, M. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. Embo J. 2014, 18, 2066–2073. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.; Lee, Z.; Cho, H. ATP-binding cassette B4, an auxin-efflux transporter, stably associates with the plasma membrane and shows distinctive intracellular trafficking from that of PIN-FORMED proteins. Plant Physiol. 2012, 159, 642–654. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Zhang, F.; Zhu, Y.; Lan, D.; Yan, P.; Wang, Y.; Hu, Z.; Zhang, X.; Hu, J.; Niu, F.; et al. Auxin signaling module OsSK41- OsIAA10-OsARF regulates grain yield traits in rice. J. Integr. Plant Biol. 2023, 65, 1753–1766. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Huang, Y.; Qi, P.; Lian, G.; Hu, X.; Han, N.; Wang, J.; Zhu, M.; Qian, Q.; Bian, H. Functional analysis of auxin receptor OsTIR1/OsAFB family members in rice grain yield, tillering, plant height, root system, germination, and auxinic herbicide resistance. New Phytol. 2021, 229, 2676–2692. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, X.; Cheng, Y.; Du, X.; Teotia, S.; Miao, C.; Sun, H.; Fan, G.; Tang, G.; Xue, H.; et al. The miR167-OsARF12 module regulates grain filling and grain size downstream of miR159. Nat. Commun. 2023, 4, 100604. [Google Scholar] [CrossRef]
- Huang, X.; Lu, Z.; Zhai, L.; Li, N.; Yan, H. The Small Auxin-Up RNA SAUR10 Is Involved in the Promotion of Seedling Growth in Rice. Plants 2023, 12, 3880. [Google Scholar] [CrossRef]
- Yu, C.; Sun, C.; Shen, C.; Wang, S.; Liu, F.; Liu, Y.; Chen, Y.; Li, C.; Qian, Q.; Aryal, B.; et al. The auxin transporter, OsAUX1, is involved in primary root and root hair elongation and in Cd stress responses in rice (Oryza sativa L). Plant J. 2015, 83, 818–830. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Zhang, C.; Jiang, Y.; Huang, C.; Liu, Q.; Xiong, L.; Yang, W.; Chen, F. Nondestructive 3D image analysis pipeline to extract rice grain traits using X-ray computed tomography. Plant Phenom. 2020, 2020, 3414926. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Y.; Zhao, P.; Zhao, Y.; Liu, M.; Guo, E.; Wang, G.; Zhang, A. Transcriptome Sequencing and Metabolome Analysis Reveals the Regulatory and Molecular Mechanisms of the Grain Filling Rate in Foxtail Millet (Setaria italica L.). Agronomy 2024, 14, 1114. https://doi.org/10.3390/agronomy14061114
Han Y, Zhao P, Zhao Y, Liu M, Guo E, Wang G, Zhang A. Transcriptome Sequencing and Metabolome Analysis Reveals the Regulatory and Molecular Mechanisms of the Grain Filling Rate in Foxtail Millet (Setaria italica L.). Agronomy. 2024; 14(6):1114. https://doi.org/10.3390/agronomy14061114
Chicago/Turabian StyleHan, Yuetao, Peiyue Zhao, Yuan Zhao, Min Liu, Erhu Guo, Guoliang Wang, and Aiying Zhang. 2024. "Transcriptome Sequencing and Metabolome Analysis Reveals the Regulatory and Molecular Mechanisms of the Grain Filling Rate in Foxtail Millet (Setaria italica L.)" Agronomy 14, no. 6: 1114. https://doi.org/10.3390/agronomy14061114
APA StyleHan, Y., Zhao, P., Zhao, Y., Liu, M., Guo, E., Wang, G., & Zhang, A. (2024). Transcriptome Sequencing and Metabolome Analysis Reveals the Regulatory and Molecular Mechanisms of the Grain Filling Rate in Foxtail Millet (Setaria italica L.). Agronomy, 14(6), 1114. https://doi.org/10.3390/agronomy14061114





