Analysis of Microbial Diversity in Rhizosphere Soil of Panax notoginseng under Different Water and Microbial Fertilizer Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Sample Collection
2.2. Illumina HiSeq Sequencing
2.2.1. OTU Analysis
2.2.2. Alpha and Beta Diversity Analysis
3. Results
3.1. Bacteria
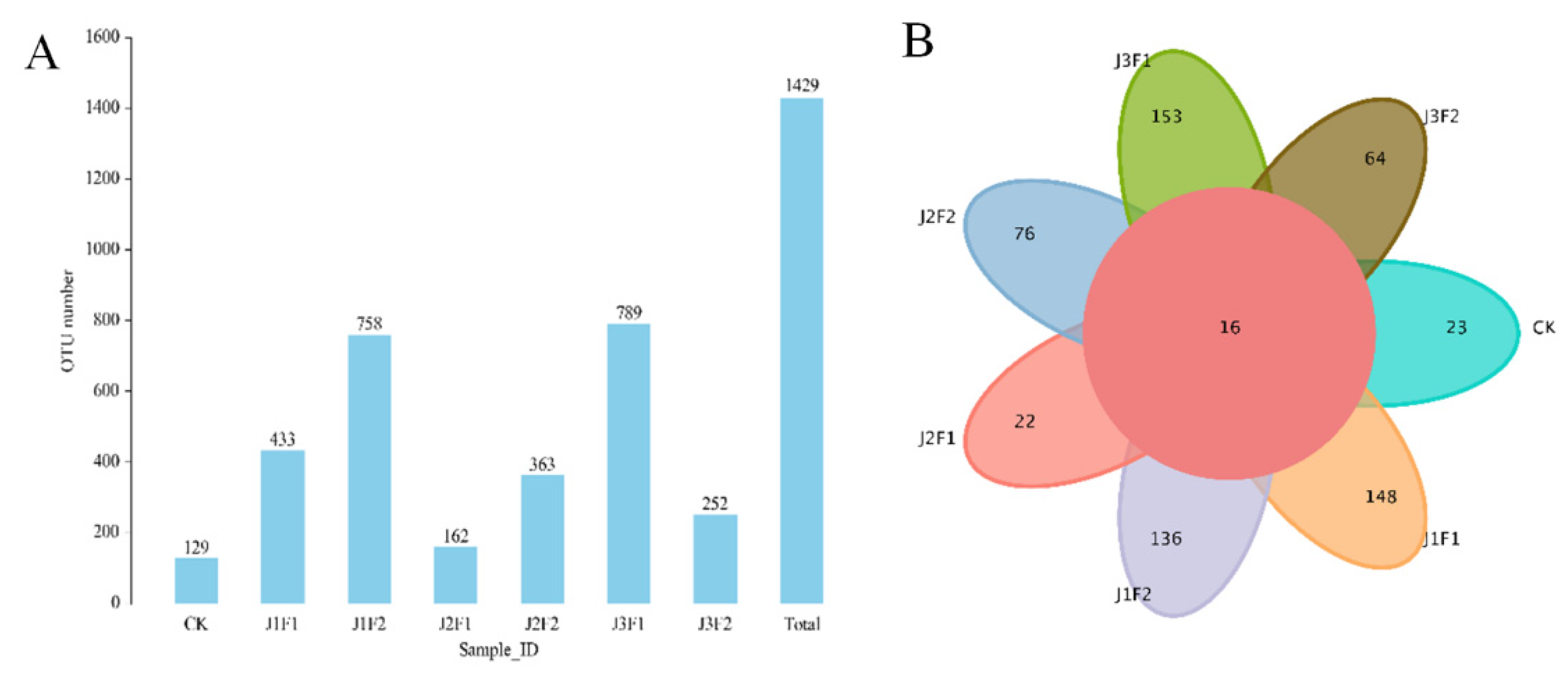
3.1.1. Bacterial OTU Analysis
3.1.2. Bacterial Alpha Diversity Analysis
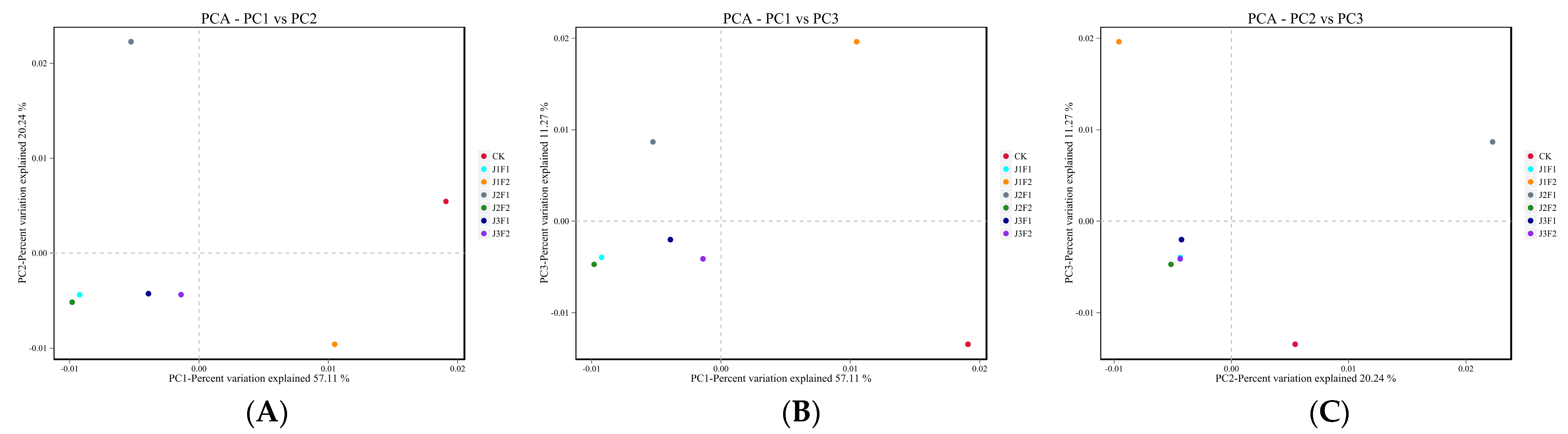
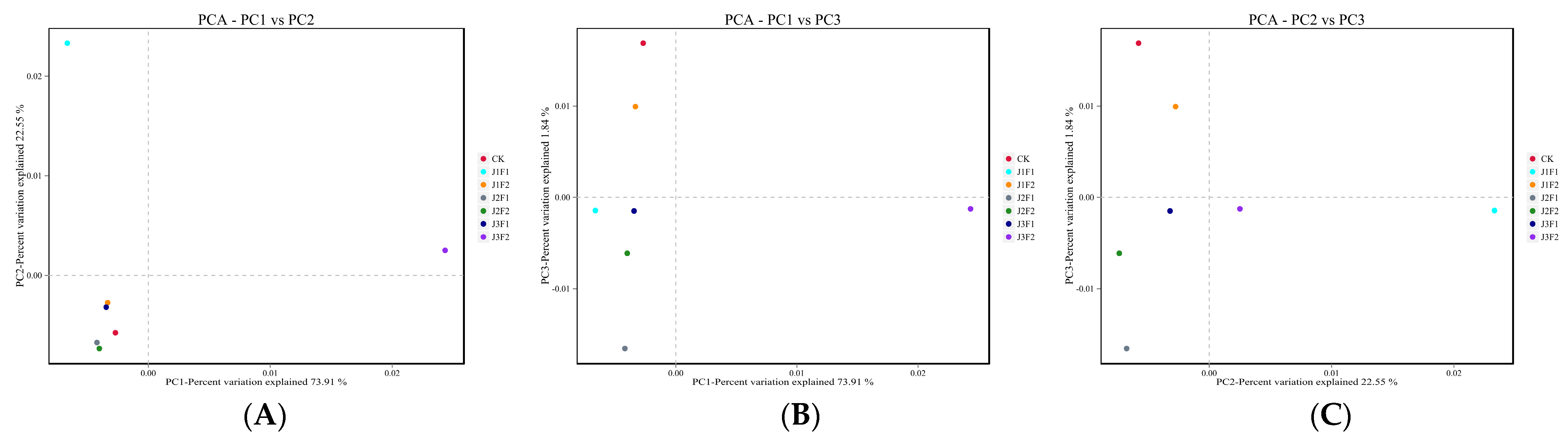
3.1.3. Bacterial Beta Analysis
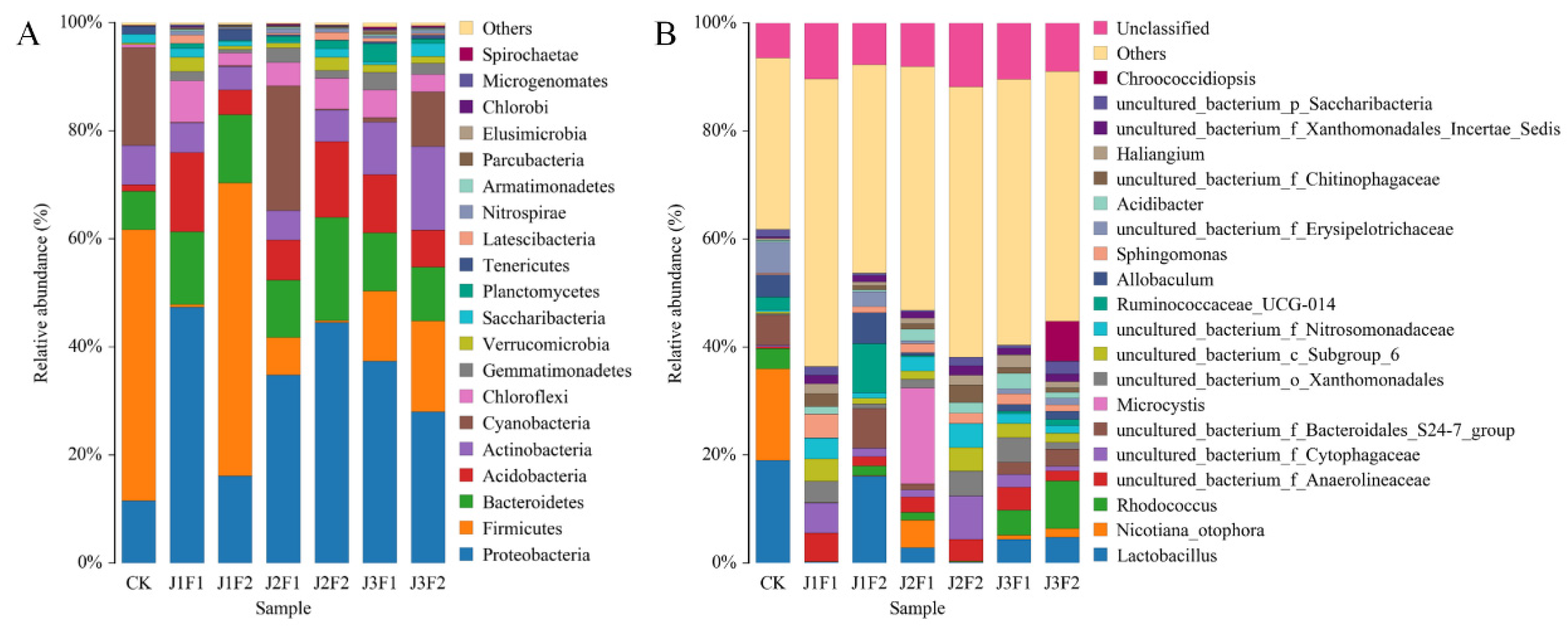
3.1.4. Bacterial Community Composition
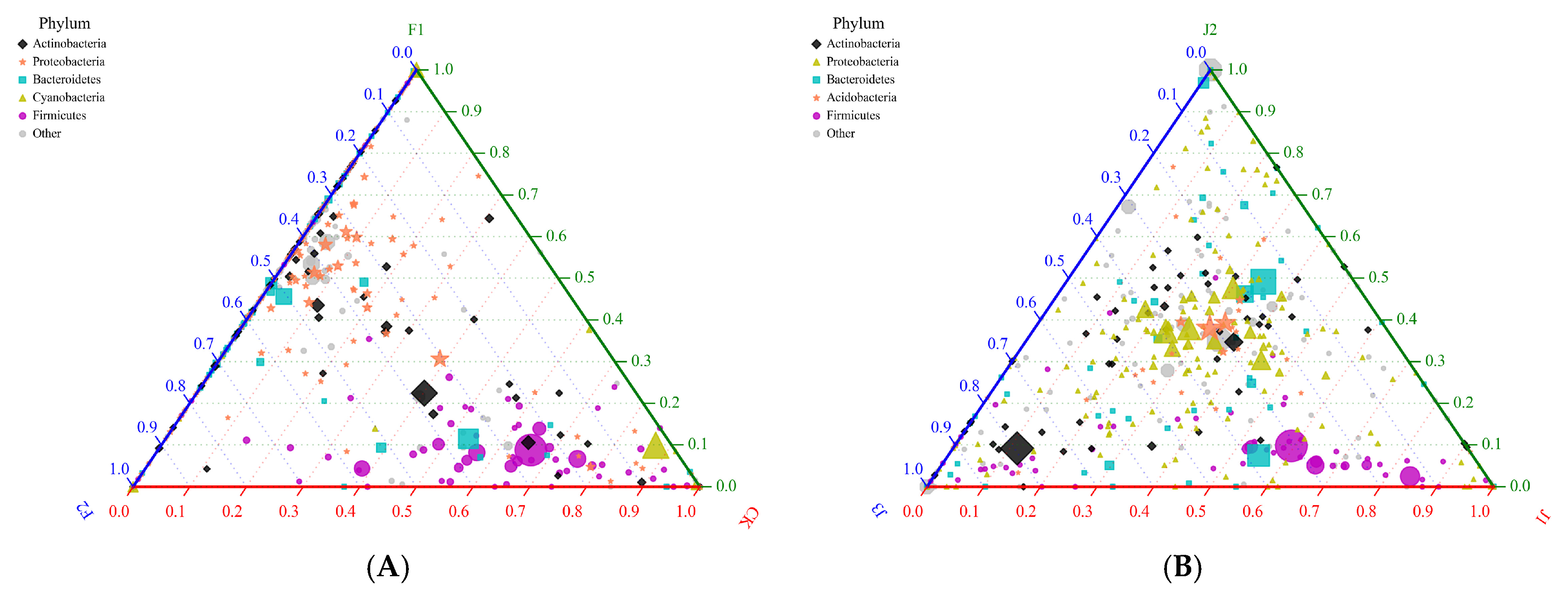
3.1.5. Bacterial Ternary Diagram
3.2. Fungi
3.2.1. Fungal OTU Analysis
3.2.2. Fungal Alpha Diversity Analysis
3.2.3. Fungal Beta Analysis
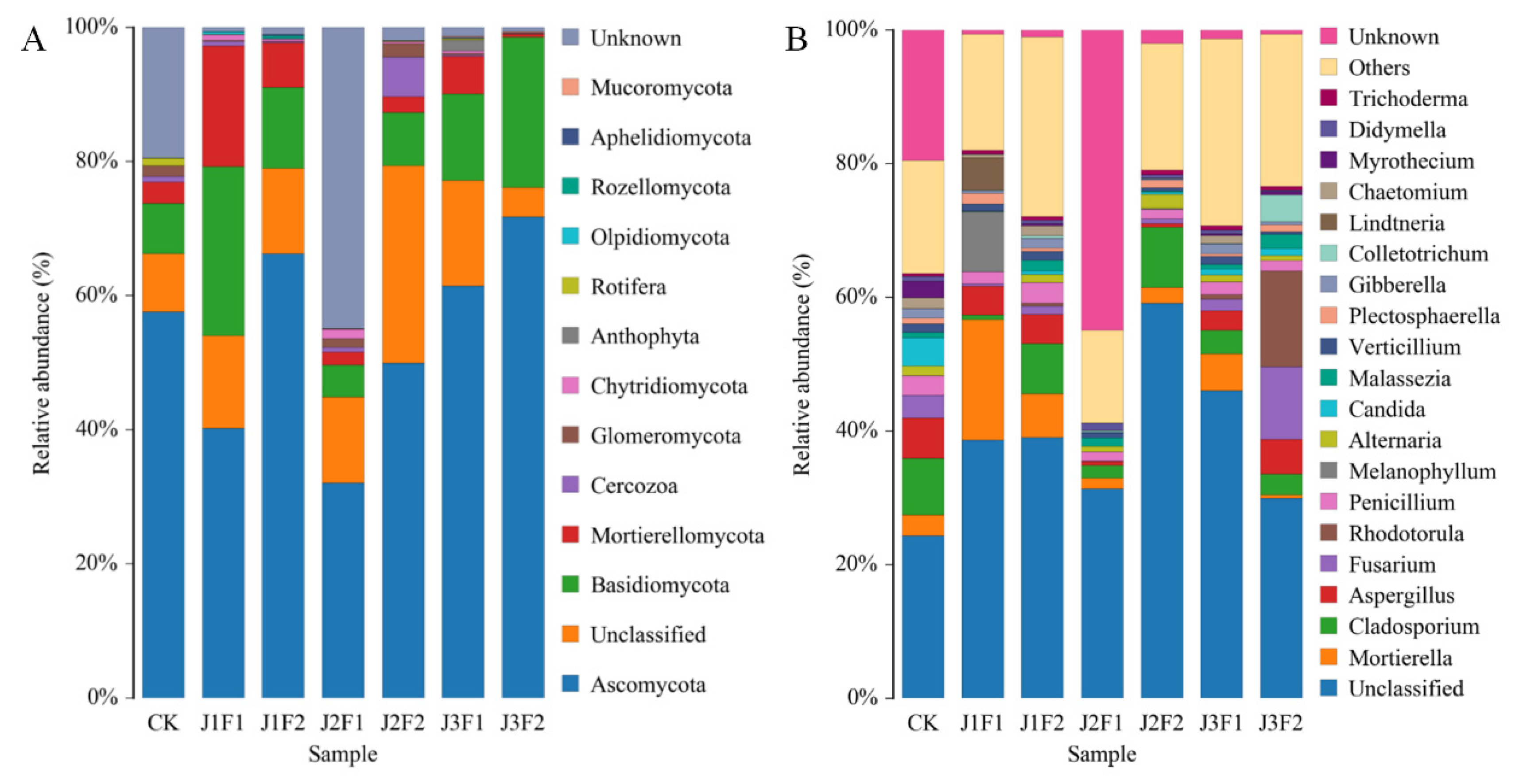
3.2.4. Fungal Community Composition
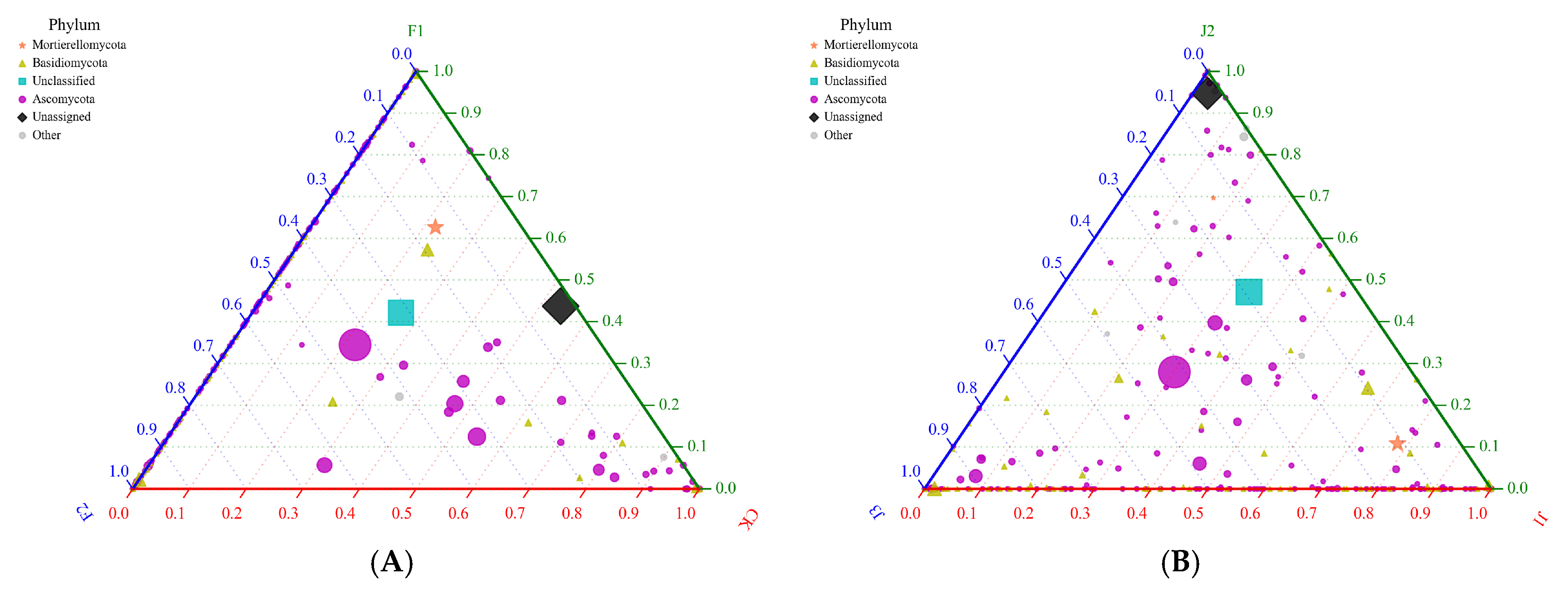
3.2.5. Fungal Ternary Diagram
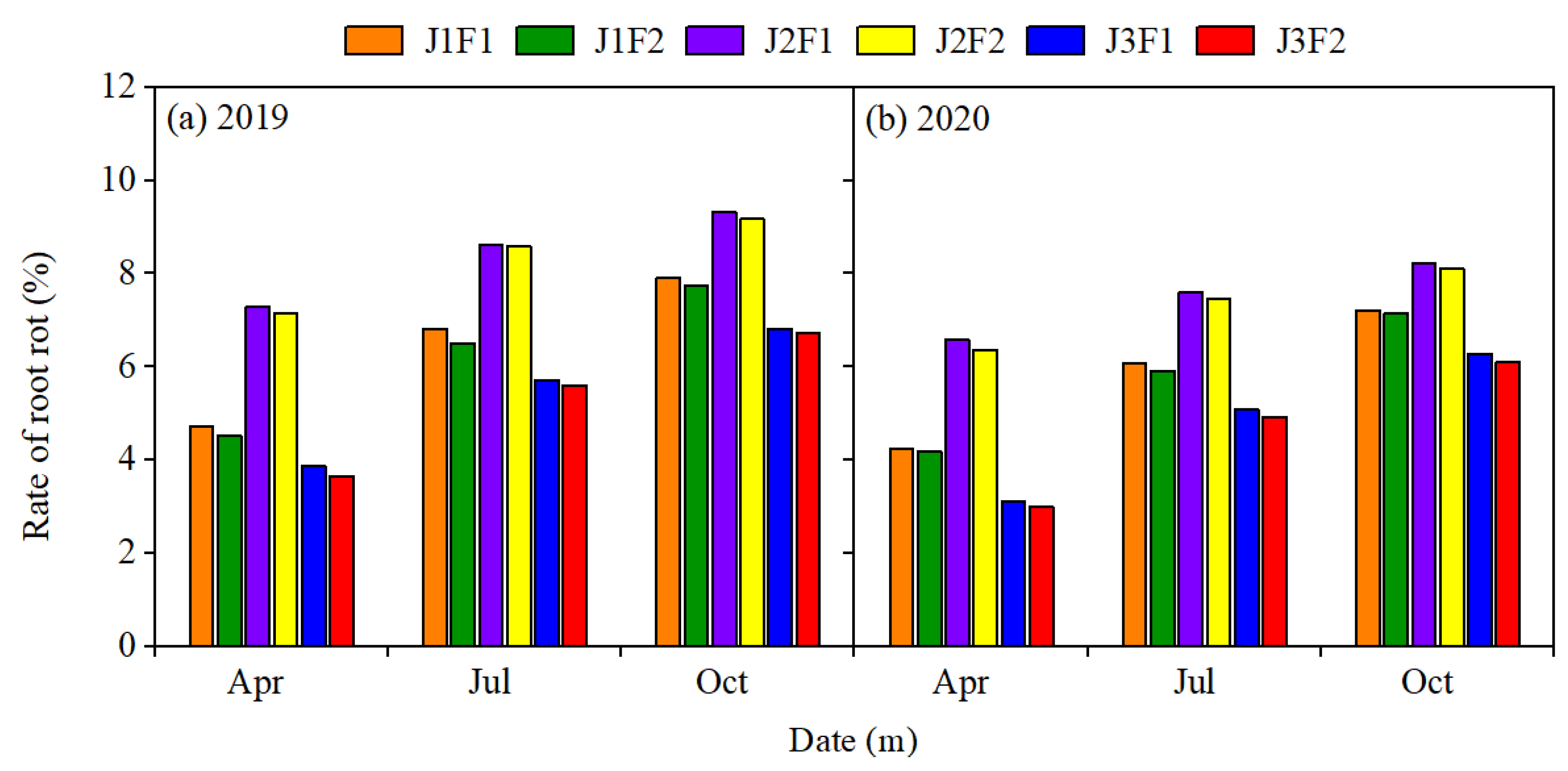
3.3. Panax Notoginseng Root Rot Incidence
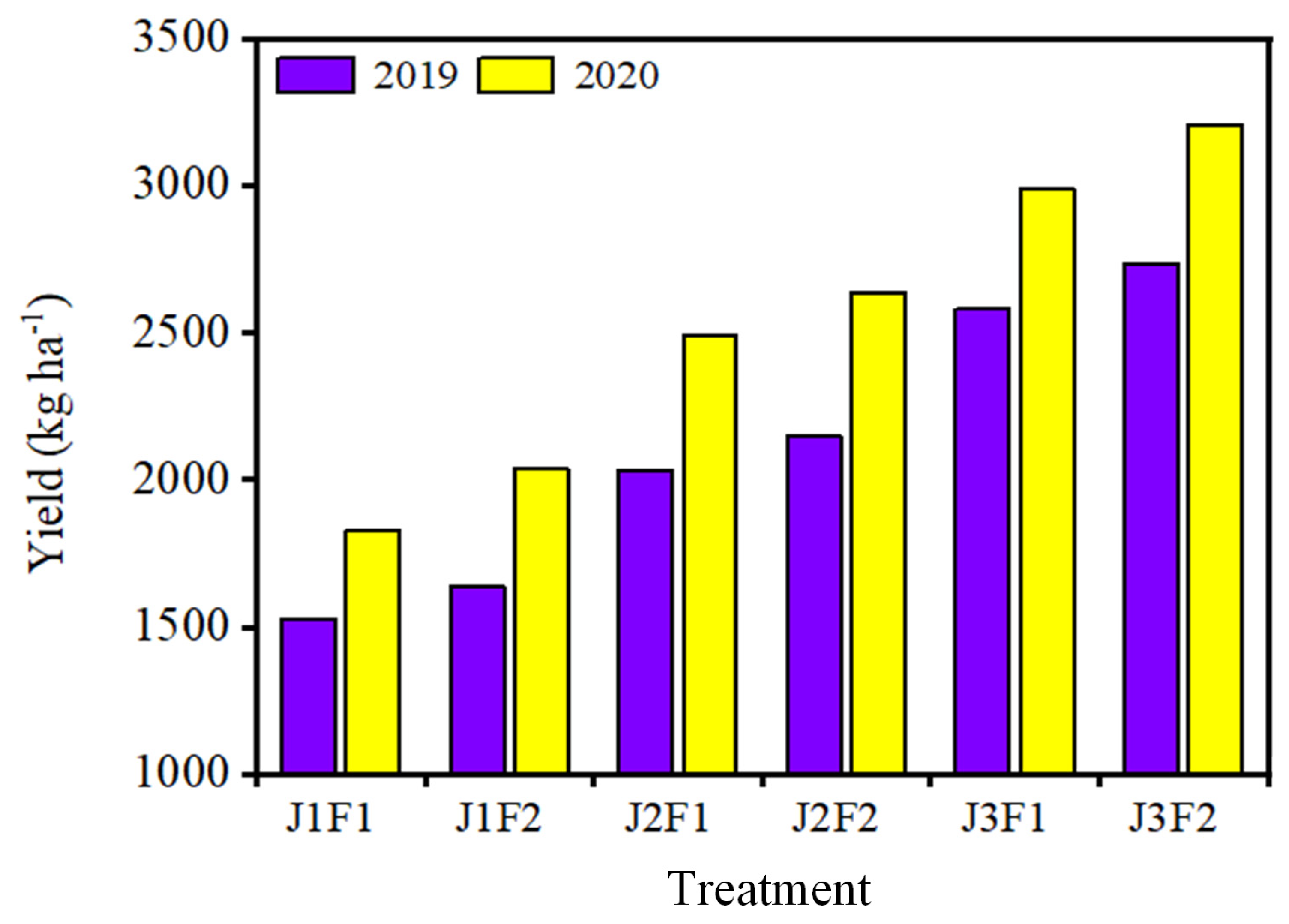
3.4. Panax Notoginseng Yield
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Canfora, L.; Salvati, L.; Benedetti, A.; Francaviglia, R. Is soil microbial diversity affected by soil and groundwater salinity? Evidences from a coastal system in central Italy. Environ. Monit. Assess. 2017, 189, 319. [Google Scholar] [CrossRef] [PubMed]
- Rillig, M.C.; Ryo, M.; Lehmann, A.; Aguilar-Trigueros, C.A.; Buchert, S.; Wulf, A.; Iwasaki, A.; Roy, J.; Yang, G. The role of multiple global change factors in driving soil functions and microbial biodiversity. Science 2019, 366, 886–890. [Google Scholar] [CrossRef] [PubMed]
- Kuang, S.; Su, Y.; Wang, H.; Yu, W.; Lang, Q.; Matangi, R. Soil Microbial Community Structure and Diversity around the Aging Oil Sludge in Yellow River Delta as Determined by High-Throughput Sequencing. Archaea 2018, 2018, 7861805. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Zhang, G.; Yu, Z.; Ding, H.; Xu, Y.; Zhang, Z. Effect of Drought Stress and Developmental Stages on Microbial Community Structure and Diversity in Peanut Rhizosphere Soil. Int. J. Mol. Sci. 2019, 20, 2265. [Google Scholar] [CrossRef] [PubMed]
- Praeg, N.; Pauli, H.; Illmer, P. Microbial Diversity in Bulk and Rhizosphere Soil of Ranunculus glacialis Along a High-Alpine Altitudinal Gradient. Front. Microbiol. 2019, 10, 1429. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Gaston, K.J. Global patterns in biodiversity. Nature 2000, 405, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.C.; Jiang, T.; Liu, Y.X.; Bai, Y.C.; Reed, J.; Qu, B.; Goossens, A.; Nützmann, H.W.; Bai, Y.; Osbourn, A. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 2019, 364, eaau6389. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Rio, T.G.D.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef]
- Fu, H.; Yuan, G.; Jeppesen, E.; Ge, D.; Li, W.; Zou, D.; Huang, Z.; Wu, A.; Liu, Q. Local and regional drivers of turnover and nestedness components of species and functional beta diversity in lake macrophyte communities in China. Sci. Total Environ. 2019, 687, 206–217. [Google Scholar] [CrossRef]
- Zhang, Y.; Zheng, Y.; Xia, P.; Xun, L.; Liang, Z. Impact of continuous Panax notoginseng plantation on soil microbial and biochemical properties. Sci. Rep. 2019, 9, 13205. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Swenson, N.G.; Ji, N.; Mi, X.; Ren, H.; Guo, L.; Ma, K. Differential soil fungus accumulation and density dependence of trees in a subtropical forest. Science 2019, 366, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Willis, A.D. Rarefaction, Alpha Diversity, and Statistics. Front. Microbiol. 2019, 10, 2407. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; NISCComparative Sequencing Program Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; Green, E.D. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef]
- Dubois, P.C.A.; Trynka, G.; Franke, L.; Hunt, K.A.; Romanos, J.; Curtotti, A.; Zhernakova, A.; Heap, G.A.; Ádány, R.; Aromaa, A.; et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010, 42, 295–302. [Google Scholar] [CrossRef]
- Ye, Q.; Park, J.; Laurence, J.S.; Parthasarathy, R.; Misra, A.; Spencer, P. Ternary phase diagram of model dentin adhesive exposed to over-wet environments. J. Dent. Res. 2011, 90, 1434–1438. [Google Scholar] [CrossRef]
- Aguirre-von-Wobeser, E.; Rocha-Estrada, J.; Shapiro, L.R.; de la Torre, M. Enrichment of Verrucomicrobia, Actinobacteria and Burkholderiales drives selection of bacterial community from soil by maize roots in a traditional milpa agroecosystem. PLoS ONE 2018, 13, e0208852. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, M.; Gao, J.; Pu, T.; Bilal, M.; Wang, Y.; Zhang, X. Antifungal activity screening of soil actinobacteria isolated from Inner Mongolia, China. Biol. Control 2018, 127, 78–84. [Google Scholar] [CrossRef]
- Soo, R.M.; Hemp, J.; Hugenholtz, P. Evolution of photosynthesis and aerobic respiration in the cyanobacteria. Free Radic. Biol. Med. 2019, 140, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Pankratova, E.M. Functioning of cyanobacteria in soil ecosystems. Eurasian Soil Sci. 2006, 39, S118–S127. [Google Scholar] [CrossRef]
- Wolińska, A.; Kuźniar, A.; Zielenkiewicz, U.; Izak, D.; Szafranek-Nakonieczna, A.; Banach, A.; Błaszczyk, M. Bacteroidetes as a sensitive biological indicator of agricultural soil usage revealed by a culture-independent approach. Appl. Soil Ecol. 2017, 119, 128–137. [Google Scholar] [CrossRef]
- McKee, L.S.; Martínez-Abad, A.; Ruthes, A.C.; Vilaplana, F.; Brumer, H. Focused Metabolism of beta-Glucans by the Soil Bacteroidetes Species Chitinophaga pinensis. Appl. Environ. Microbiol. 2019, 85, e02231-18. [Google Scholar] [CrossRef] [PubMed]
- Lladó, S.; Žifčáková, L.; Větrovský, T.; Eichlerová, I.; Baldrian, P. Functional screening of abundant bacteria from acidic forest soil indicates the metabolic potential of Acidobacteria subdivision 1 for polysaccharide decomposition. Biol. Fertil. Soils 2016, 52, 251–260. [Google Scholar] [CrossRef]
- de Chaves, M.G.; Silva, G.G.Z.; Rossetto, R.; Edwards, R.A.; Tsai, S.M.; Navarrete, A.A. Acidobacteria Subtreatments and Their Metabolic Potential for Carbon Degradation in Sugarcane Soil Amended With Vinasse and Nitrogen Fertilizers. Front. Microbiol. 2019, 10, 1680. [Google Scholar] [CrossRef]
- Zhang, Y.; Cong, J.; Lu, H.; Li, G.; Qu, Y.; Su, X.; Zhou, J.; Li, D. Community structure and elevational diversity patterns of soil Acidobacteria. J. Environ. Sci. 2014, 26, 1717–1724. [Google Scholar] [CrossRef]
- Schrempf, H. Actinobacteria within soils: Capacities for mutualism, symbiosis and pathogenesis. FEMS Microbiol. Lett. 2013, 342, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Wolińska, A.; Górniak, D.; Zielenkiewicz, U.; Kuźniar, A.; Izak, D.; Banach, A.; Błaszczyk, M. Actinobacteria Structure in Autogenic, Hydrogenic and Lithogenic Cultivated and Non-Cultivated Soils: A Culture-Independent Approach. Agronomy 2019, 9, 598. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, C.; Folly, Y.M.E.; Chang, J.; Wang, Y.; Zhou, L.; Zhang, H.; Liu, Y. Morphological and Transcriptomic Analysis of the Inhibitory Effects of Lactobacillus plantarum on Aspergillus flavus Growth and Aflatoxin Production. Toxins 2019, 11, 636. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.F.; Xing, P.; Liu, S.; Wu, Q.L. Enhanced Microbial Interactions and Deterministic Successions during Anoxic Decomposition of Microcystis Biomass in Lake Sediment. Front. Microbiol. 2019, 10, 2474. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Yang, Z. Studies on diversity of higher fungi in Yunnan, southwestern China: A review. Plant Divers. 2018, 40, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Challacombe, J.F.; Hesse, C.N.; Bramer, L.M.; McCue, L.A.; Lipton, M.; Purvine, S.; Nicora, C.; Gallegos-Graves, L.V.; Porras-Alfaro, A.; Kuske, C.R. Genomes and secretomes of Ascomycota fungi reveal diverse functions in plant biomass decomposition and pathogenesis. BMC Genom. 2019, 20, 976. [Google Scholar] [CrossRef] [PubMed]
- Barrera, V.A.; Martin, M.E.; Aulicino, M.; Martínez, S.; Chiessa, G.; Saparrat, M.C.; Gasoni, A.L. Carbon-substrate utilization profiles by Cladorrhinum (Ascomycota). Rev. Argent. Microbiol. 2019, 51, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Frey, S.D.; Knorr, M.; Parrent, J.L.; Simpson, R.T. Chronic nitrogen enrichment affects the structure and function of the soil microbial community in temperate hardwood and pine forests. For. Ecol. Manag. 2004, 196, 159–171. [Google Scholar] [CrossRef]
- Lienhard, P.; Terrat, S.; Prévost-Bouré, N.C.; Nowak, V.; Régnier, T.; Sayphoummie, S.; Panyasiri, K.; Tivet, F.; Mathieu, O.; Levêque, J.; et al. Pyrosequencing evidences the impact of cropping on soil bacterial and fungal diversity in Laos tropical grassland. Agron. Sustain. Dev. 2014, 34, 525–533. [Google Scholar] [CrossRef]
- Desalegn, G.; Turetschek, R.; Kaul, H.P.; Wienkoop, S. Microbial symbionts affect Pisum sativum proteome and metabolome under Didymella pinodes infection. J. Proteom. 2016, 143, 173–187. [Google Scholar] [CrossRef]
- Buhtz, A.; Witzel, K.; Strehmel, N.; Ziegler, J.; Abel, S.; Grosch, R. Perturbations in the Primary Metabolism of Tomato and Arabidopsis thaliana Plants Infected with the Soil-Borne Fungus Verticillium dahliae. PLoS ONE 2015, 10, e0138242. [Google Scholar] [CrossRef] [PubMed]
- Pasche, J.S.; Taylor, R.J.; Gudmestad, N.C. Colonization of Potato by Colletotrichum coccodes: Effect of Soil Infestation and Seed Tuber and Foliar Inoculation. Plant Dis. 2010, 94, 905–914. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, J.; Bi, S.; Chen, H.; Chen, T.; Yang, R.; Li, M.; Fu, Y.; Jia, A.Q. Anti-Biofilm and Antivirulence Activities of Metabolites from Plectosphaerella cucumerina against Pseudomonas aeruginosa. Front. Microbiol. 2017, 8, 769. [Google Scholar] [CrossRef] [PubMed]
- Elansary, H.O.; El-Ansary, D.O.; Al-Mana, F.A. 5-Aminolevulinic Acid and Soil Fertility Enhance the Resistance of Rosemary to Alternaria dauci and Rhizoctonia solani and Modulate Plant Biochemistry. Plants 2019, 8, 585. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, N.; Vitale, S.; Turrà, D.; Reverberi, M.; Fanelli, C.; Vinale, F.; Marra, R.; Ruocco, M.; Pascale, A.; d’Errico, G.; et al. Root Exudates of Stressed Plants Stimulate and Attract Trichoderma Soil Fungi. Mol. Plant Microbe Interact. 2018, 31, 982–994. [Google Scholar] [CrossRef] [PubMed]
- Barilli, E.; Cobos, M.J.; Rubiales, D. Clarification on Host Range of Didymella pinodes the Causal Agent of Pea Ascochyta Blight. Front. Plant Sci. 2016, 7, 592. [Google Scholar] [CrossRef] [PubMed]
- Zaki, O.; Weekers, F.; Thonart, P.; Tesch, E.; Kuenemann, P.; Jacques, P. Limiting factors of mycopesticide development. Biol. Control 2020, 144, 104220. [Google Scholar] [CrossRef]
- Awad, N.E.; Kassem, H.A.; Hamed, M.A.; El-Feky, A.M.; Elnaggar, M.A.; Mahmoud, K.; Ali, M.A. Isolation and characterization of the bioactive metabolites from the soil derived fungus Trichoderma viride. Mycology 2018, 9, 70–80. [Google Scholar] [CrossRef]
- Yadav, D.R.; Kim, S.W.; Adhikari, M.; Um, Y.H.; Kim, H.S.; Kim, C.; Lee, H.B.; Lee, Y.S. Three New Records of Mortierella Species Isolated from Crop Field Soil in Korea. Mycobiology 2015, 43, 203–209. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Pawłowska, J.; Letcher, P.M.; Kirk, P.M.; Humber, R.A.; Schüßler, A.; Wrzosek, M.; Muszewska, A.; Okrasińska, A.; Istel, Ł.; et al. Notes for genera: Basal clades of Fungi (including Aphelidiomycota, Basidiobolomycota, Blastocladiomycota, Calcarisporiellomycota, Caulochytriomycota, Chytridiomycota, Entomophthoromycota, Glomeromycota, Kickxellomycota, Monoblepharomycota, Mortierellomycota, Mucoromycota, Neocallimastigomycota, Olpidiomycota, Rozellomycota and Zoopagomycota). Fungal Divers. 2018, 92, 43–129. [Google Scholar]
- Hoysted, G.A.; Kowal, J.; Jacob, A.; Rimington, W.R.; Duckett, J.G.; Pressel, S.; Orchard, S.; Ryan, M.H.; Field, K.J.; Bidartondo, M.I. A mycorrhizal revolution. Curr. Opin. Plant Biol. 2018, 44, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zuccaro, A.; Lahrmann, U.; Langen, G. Broad compatibility in fungal root symbioses. Curr. Opin. Plant Biol. 2014, 20, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Egidi, E.; Delgado-Baquerizo, M.; Plett, J.M.; Wang, J.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K. A few Ascomycota taxa dominate soil fungal communities worldwide. Nat. Commun. 2019, 10, 2369. [Google Scholar] [CrossRef] [PubMed]

| Treatments | CK | J1F1 | J1F2 | J2F1 | J2F2 | J3F1 | J3F2 |
|---|---|---|---|---|---|---|---|
| Irrigation/m3 | 0.25 | 0.125 | 0.125 | 0.375 | 0.375 | Alternation | Alternation |
| Microbial Fertilizer/kg | 0 | 1.426 | 1.971 | 1.426 | 1.971 | 1.426 | 1.971 |
| Treatments | OTU | ACE | Chao1 | Simpson | Shannon | Coverage |
|---|---|---|---|---|---|---|
| CK | 481 | 497.3891 | 514.8333 | 0.0635 | 4.1825 | 0.9994 |
| J1F1 | 1279 | 1327.4364 | 1335.5962 | 0.0051 | 6.1185 | 0.9971 |
| J1F2 | 1029 | 1057.8396 | 1099.0227 | 0.0252 | 5.2412 | 0.9983 |
| J2F1 | 1364 | 1416.8629 | 1428.6271 | 0.0388 | 5.3527 | 0.9968 |
| J2F2 | 1222 | 1270.5589 | 1274.0099 | 0.0052 | 6.049 | 0.9975 |
| J3F1 | 1225 | 1240.5259 | 1244.6154 | 0.0074 | 6.0033 | 0.9987 |
| J3F2 | 1129 | 1141.9841 | 1153.0698 | 0.0166 | 5.701 | 0.9989 |
| Treatments | OTU | ACE | Chao1 | Simpson | Shannon | Coverage |
|---|---|---|---|---|---|---|
| CK | 129 | 219.5933 | 175.4286 | 0.0451 | 3.9208 | 0.9994 |
| J1F1 | 433 | 483.4292 | 473.625 | 0.0304 | 4.5707 | 0.9996 |
| J1F2 | 758 | 759.3125 | 761.5 | 0.0072 | 5.7662 | 0.9999 |
| J2F1 | 162 | 194.7184 | 189.0833 | 0.1898 | 3.1832 | 0.9996 |
| J2F2 | 363 | 431.0311 | 395.5 | 0.0197 | 4.8948 | 0.9993 |
| J3F1 | 789 | 793.5483 | 798.2308 | 0.0067 | 5.7597 | 0.9997 |
| J3F2 | 252 | 333.4158 | 343.0 | 0.0398 | 4.187 | 0.9999 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, L.; Kong, L.; Yang, Q.; Nian, H.; Liang, J. Analysis of Microbial Diversity in Rhizosphere Soil of Panax notoginseng under Different Water and Microbial Fertilizer Conditions. Agronomy 2024, 14, 922. https://doi.org/10.3390/agronomy14050922
Yao L, Kong L, Yang Q, Nian H, Liang J. Analysis of Microbial Diversity in Rhizosphere Soil of Panax notoginseng under Different Water and Microbial Fertilizer Conditions. Agronomy. 2024; 14(5):922. https://doi.org/10.3390/agronomy14050922
Chicago/Turabian StyleYao, Leilei, Lei Kong, Qiliang Yang, Hongjuan Nian, and Jiaping Liang. 2024. "Analysis of Microbial Diversity in Rhizosphere Soil of Panax notoginseng under Different Water and Microbial Fertilizer Conditions" Agronomy 14, no. 5: 922. https://doi.org/10.3390/agronomy14050922
APA StyleYao, L., Kong, L., Yang, Q., Nian, H., & Liang, J. (2024). Analysis of Microbial Diversity in Rhizosphere Soil of Panax notoginseng under Different Water and Microbial Fertilizer Conditions. Agronomy, 14(5), 922. https://doi.org/10.3390/agronomy14050922







