Abstract
Active accumulated temperature (AAT) serves as a pivotal metric for assessing soybean adaptation across diverse climatic conditions, particularly in the northeastern regions of China. This study embarked on a genome-wide association analysis (GWAS) to elucidate the genetic determinants influencing AAT and its impact on flowering time among soybean varieties. Leveraging a panel of 140 elite soybean varieties encompassing both Chinese and European early-maturity groups and employing high-density genotyping, significant associations were identified on chromosome 6. Notably, a key gene, Glyma.06g204500, emerged as a central component, exhibiting strong linkage to the well-established E1 locus, alongside three distinct haplotypes. This investigation underscores Glyma.06g204500’s potential role in mediating soybean’s response to temperature fluctuations, offering novel insights into the genetic mechanisms underpinning soybean adaptation to local environmental conditions.
1. Introduction
Soybean (Glycine max [L.] Merr.) is a short-day plant originating from China that is sensitive to photoperiod and temperature [1,2]. With global warming and the high demand for soybean, expanding north and adapting to high-latitude environments are the key points of soybean breeding projects [3]. To describe how soybean varieties can adapt to a local environment and be introduced to others areas, the maturity group (MG) system was founded in America and is utilized in most main soybean producing areas [4], whereas active accumulated temperature (AAT) is used in Northeast China [5]. Northeast China is the most crucial and biggest soybean-harvesting area in China [6], and is located from 40°25′ to 48°40′ north latitude. Breeders and farmers in this area take advantage of AAT to describe how the plants adapt to certain areas and categorize the area into different subareas to describe the maturity group [7].
Flowering is treated as a signal of starting the reproductive period of a plant, and therefore, the time to flowering is recognized as a crucial agronomic trait in adaptation [8]. Soybean is believed to originate from the south part of China, where the active accumulated temperature is high and the day length is short. Broad adaptability across latitudes is due to genetic variation in major gene loci and quantitative trait loci (QTLs) controlling flowering and maturity. For example, dominant alleles at loci E1, E2, E3, E4, E7, E8, and E10 confer late flowering, while dominant alleles at E6, E9, E11, and J promote early flowering [9,10]. E1 acts as a central regulator, repressing FLOWERING LOCUS T orthologs, which are crucial for initiating flowering. E2 encodes a GIGANTEA ortholog affecting the circadian clock and flowering time. E3 and E4 encode phytochrome A homologs, crucial for sensing red and far-red light, influencing flowering under long-day conditions. E6 and J are involved in promoting flowering under short-day conditions, conferring a ‘long-juvenile’ trait that is beneficial in tropical environments. Several FT-like genes play roles in the control of flowering time, with GmFT2a and GmFT5a being major functional members influencing flowering under different day lengths. The time to flowering is a complex network in which circadian clock components, photoreceptors, and various genes interact to regulate it. E1 is highlighted as a critical hub integrating signals from light and the circadian clock to control flowering time through the repression or activation of FT orthologs [10].
These genes and their interaction create a sophisticated genetic framework governing soybean’s flowering time in response to photoperiodic signals. But there are few studies that focus on the adaptation to temperature changes. Yang et al. identified three genes in 294 wild soybean accessions distributed at chromosomes 2, 4, and 9 that are associated with the pathways of photosynthesis light reaction and actin filament binding [11]. Zhu et al. located a homolog of the vernalization pathway gene GmFRL1 in soybean [12]. Although the interaction of the gene and environment was established, the genetic mechanism of how the gene reacts to environmental change is still not clear.
Figuring out how the active accumulated temperature influences the time to flowering is important to exchange the germplasm globally and for breeders to cultivate new varieties. To achieve this goal, this study collected 140 varieties from the elite early Chinese and European soybean varieties and sowed them in five locations in Europe. This short report predicted a hub gene related to AAT, which was beneficial for understanding how soybeans adapt to different temperature changes.
2. Materials and Methods
2.1. Elite Soybean Cultivars
A set of 140 early-maturity soybean varieties was collected for the present study (see descriptive information in Supplementary Table S1, and Yao et al. 2023) [13]. The total set consisted of 63 Chinese (CN) and 77 European (EU) elite cultivars. European cultivars were provided from soybean breeding companies in Austria, France, Germany, Hungary, Italy, Poland, Serbia, Romania, Switzerland, and Ukraine, spanning a wide range of geographic adaptation and maturity groups from 0000 to II. Chinese soybean cultivars were provided from breeding institutions located in Northeast China, comprising soybean maturity groups 000 to III (i.e., Northeastern Spring (NEsp) or Northern Spring (Nsp) cultivars according to the Chinese soybean ecotype classification).
2.2. Experimental Design and Data Collection
The experiment was carried out from the latitude and longitude of 45.33678, 19.840971 to 51.691992, 17.070175 at five locations in Tulln (Austria) short for AT, Kühnham (Germany) short for DE, Nyon (Switzerland) short for CH, DLón (Poland) short for PL, and Novi Sad (Serbia) short for SRB in 2019 (Supplementary Table S2). The experiment was arranged in a complete randomized block design with two replications. The fertilization and weed and pest control were controlled normally as local conditions.
2.3. DNA Extraction and SNP Genotyping
Young leaf tissue was collected from each accession, and total genomic DNA was isolated using the CTAB method. After that, all samples’ DNA was genotyped by ZDX 1, which was the same as Yao’s method [13]. The ZDX 1 [13] was designed by COMPASS BIOTECH-NOLOGY (Beijing Compass Biotechnology Co., Ltd., Beijing, China) and the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences.
2.4. Statistical Analyses
The date to flowering was scored at the first flower on any node of the plants, and the AAT was calculated from the sowing date to the time to flowering [5,14] as the following:
where n is the number of days to flowering, Ti is the average temperature for the ith day, and B is the base temperature for soybean growth, which is usually 10 °C. A descriptive statistical analysis involving the parameters of the mean, range, kurtosis, skewness, standard deviation (SD), and coefficient of variation (CV%) for AAT was carried out using the R software (version 4.3.1).
To dismiss the environmental influence to the AAT, best linear unbiased prediction (BLUP) values of AAT for each variety across the six locations were calculated and used for GWAS analysis.
The GWAS analysis was performed with the R package rMVP [15] in the R software version 4.3.1 (R core team 2023). Haplotypes were analysed with geneHapR [16].
3. Results
The range (maximum and minimum value), mean, SD, CV, kurtosis, and skewness of the AAT for the GWAS panel of 140 accessions in the five different environments are presented in Table 1. The AAT had the largest range in Poland, with a minimum AAT of 797.00 °C and a maximum of 1796.00 °C; the coefficient of variance value was 21.76%. The narrowest AAT range was found in Serbia, with a minimum value of 1056.00 °C and a maximum value of 1395.00 °C; the coefficient of variance value was 5.07%.

Table 1.
AAT distribution in six locations.
BLUP values for the accumulation temperature of the time to flowering were calculated and showed normal distribution, with an average active accumulated temperature of 1089.69 °C (Supplementary Figure S1). To identify the gene related to the accumulation temperature of flowering, 140 varieties from Europe and China were genotyped by the ZDX1 array; 157,353 SNPs distributed over the 20 chromosomes strongly passed the quality control (Supplementary Figure S2). The phenotype and genotype data were utilized for a genome-wide association study with the FarmCPU model. Quantile–quantile plotting indicated the significant SNPs associated with AAT. When the threshold was set as −log10(P) ≥ 4.0, 17 significant SNPs were detected across chromosome 6, and these SNPs were categorized into two LD blocks (Figure 1A,B). Most of the significant SNPs were categorized into the first LD block and only five SNPs were in the second LD block.
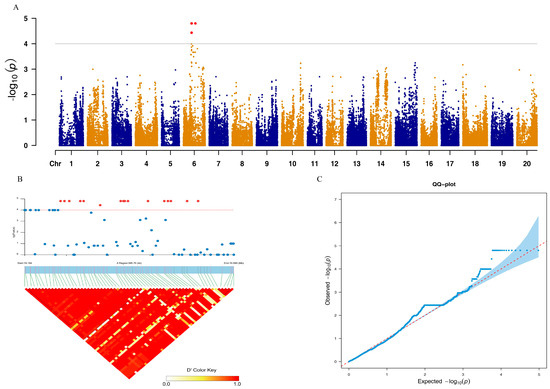
Figure 1.
GWAS results of the AAT. (A) Manhattan plot for the 20 chromosomes; threshold was set as −log10(P) ≥ 4.0, and significant SNPs are filled in red. (B) LD block analysis for the significant SNPs with an interval range of 75 kb. (C) Quantile–quantile plot (Q-Q plot) for GWAS analysis.
Considering the LD decay of the population in this collection, 75 kb intervals upstream and downstream of the significant SNPs were treated as the key area for the candidate genes’ selection; 16 genes were found in this area, seven of which could be annotated by Soybase (Table 2). The most significant SNP was located on gene Glyma.06g204500, with an annotation of a Haloacid dehalogenase-like hydrolase (HAD) superfamily protein. Thus, it was considered as the hub gene for the response to temperature differences.

Table 2.
Candidate gene annotation based on Soybase.
The haplotype analysis revealed that there are three haplotypes in the 140 soybean collections. There was no significant difference between H001 and H002 (Figure 2), which have the same locus on SNP_06_19278170, but when the locus changed from C to T, a significant difference could be observed according to the accumulation temperature for time to flowering. Therefore, the gene Glyma.06g204500 can be the hub gene for adjusting the accumulation temperature for the time to flowering.
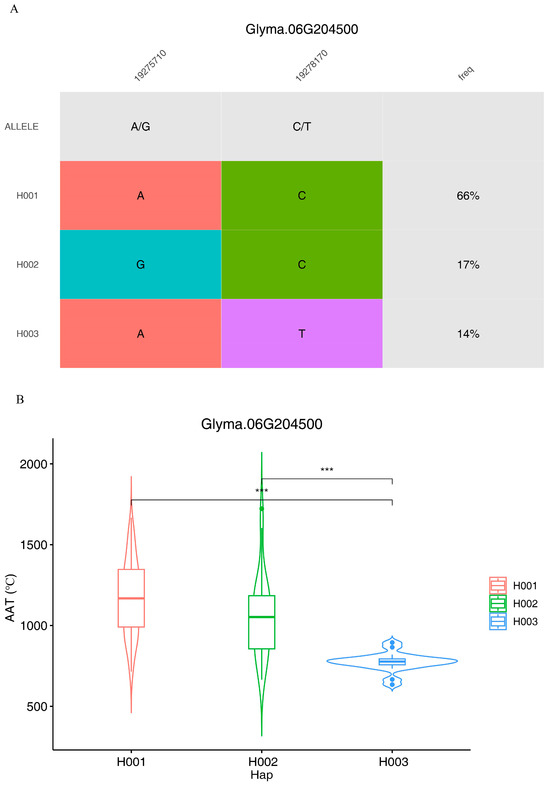
Figure 2.
Haplotype analysis for the gene Glyma.06g204500. (A) Haplotype variation; (B) the AAT distribution with the variant haplotypes. *** p < 0.001.
4. Discussion
Flowering is one of the most important traits to describe how soybean varieties adapt to the ecological environment [17]. Multiple quantitative trait loci (QTLs) and genes have been identified that regulate the photoperiod pathways and influence the time to flowering [18,19,20,21], but only a few that reveal how the time to flowering is affected by temperature have been reported, especially the active accumulated temperature.
Soybeans thrive in a temperature range of 17–32 °C, with variations depending on latitude, cultivars, and cultivation methods. The biological minimum temperature for soybean growth is considered to be around 17–18 °C. Temperatures below 15 °C can slow down plant growth and inhibit leaf and shoot formation, and temperatures below 10 °C can disrupt the flowering process. Low-temperature stress from germination to emergence, which is a sensitive period of soybean growth, can affect protein and cell membrane metabolism adversely. This kind of damage can cause the elongation of soybean stems to encounter obstacles, thereby affecting the flowering period [22,23].
Our study employed 140 elite Chinese and European early-maturity soybeans to mine the key gene related to the active accumulated temperature. The genotype panel was well structured and the diversity of the collection ranged widely [13]. An area was identified upstream of the E1 locus at chromosome 6, which contained 16 genes. Gene Ontology annotation and haplotype analysis indicated that Glyma.06g204500 was a hub gene related to the AAT. Three haplotypes can be categorized in the collection and linked with the E1 locus. The accessions carrying H001, H002, and H003 haplotypes corresponded with e1-as, E1, and e1-nl, respectively (genotyping results for the E1 locus are not shown). Thus, we assume that the gene Glyma.06g204500 is the hub gene in responding to AAT changes. Notably, an intriguing observation was made regarding entry 17, ERICA, categorized under maturity group 000, which achieved the flowering stage at the minimal active accumulated temperature (AAT) of 714 °C in Switzerland. This variety exhibited stability in AAT across Austria, Germany, and Poland. Furthermore, ERICA possesses the e1-nl allele and the H003 haplotype of the identified hub gene, underscoring the genetic foundation of its temperature adaptation and flowering time. When the sowing area was relocated southward to Serbia, the accumulated average temperature (AAT) required for ERICA increased to 1158 °C. This elevation signifies that the flowering process in soybeans is governed by a multifaceted physiological mechanism that cannot be accurately modeled by simplistic approaches. The intricate genetic basis underlying this process necessitates further investigation. The observed variance in flowering time across different geographical locations underscores the complexity of the regulatory network controlling this trait in soybeans. Consequently, it is imperative to conduct comprehensive studies focusing on the genetic mechanisms that influence flowering, to enhance our understanding and facilitate the breeding of soybeans adapted to diverse environmental conditions.
E1 is a specialized transcription factor integrating the photoperiod pathways in legume plants [24,25]. E1 is known to encode a protein related to the B3 domain and is strongly induced by the photoperiod factor (day length) [25]. Recently, it was reported that E1 can respond to high temperatures under short-day conditions. The flowering process can be accelerated from 25 degrees Celsius to 30 degrees Celsius, but will be delayed when the temperature reaches 35 degrees Celsius [26]. Our results indicated that the gene Glyma.06g204500 could be another factor influencing E1 by responding to temperature changes. However, the genetic mechanism of how E1 is influenced by Glyma.06g204500 is still not clear, and further studies should be carried out to reveal how E1 takes part in controlling the thermomorphogensis process to accelerate or delay the flowering time of soybean.
With global warming, extreme temperatures occur in high-latitude areas frequently during the soybean growing season. Understanding how soybean reacts to temperature changes is beneficial for global food safety and the protein supply chain.
5. Conclusions
The exploration of genetic influencers on the active accumulated temperature (AAT) and its subsequent effect on the flowering time in soybeans has yielded significant insights. Through comprehensive genome-wide association analysis, this study pinpointed crucial genetic markers on chromosome 6, emphasizing the role of Glyma.06g204500 in soybean’s adaptative response to varying temperatures. The association between this gene and the E1 locus, along with the identification of three variant haplotypes, advances our understanding of the intricate genetic architecture governing soybean’s phenological responses. This research not only reinforces the complexity of the flowering process in soybeans but also highlights potential genetic targets for enhancing soybean adaptability through breeding. As the global climate continues to evolve, the findings of this study will be instrumental in guiding soybean breeding strategies aimed at ensuring food security and sustaining agricultural productivity in changing environmental conditions.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy14040833/s1, Table S1: List of 140 varieties in this study; Table S2: Environment code for the experiment carried out in Europe in 2019; Figure S1: Distribution of AAT of time to flowering; Figure S2: Dentist of SNPs passed quality control.
Author Contributions
Conceptualization, X.Y.; methodology, X.Y; software, X.Y.; validation, X.Y. and D.Z.; formal analysis, X.Y.; investigation, X.Y; resources, X.Y.; data curation, X.Y; writing—original draft preparation, X.Y; writing—review and editing, X.Y; visualization, X.Y.; supervision, D.Z; project administration, D.Z.; funding acquisition, D.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the National Key R&D Program of China (2021YFD1201100); Heilongjiang Province Unveiled Project (2021ZXJ05B011); the German Federal States of Baden-Württemberg (Ministry for Rural Areas and Consumer Protection Baden-Württemberg) and Bavaria (Bavarian State Ministry of Food, Agriculture and Forestry); the Swiss Confederation (Agroscope); and Saatgut Austria (Seed Association of Austria). Open access funding of this publication is provided by the University of Natural Resources and Life Sciences, Vienna (BOKU), Vienna, Austria.
Data Availability Statement
Data is contained within the article and Supplementary Material.
Acknowledgments
The authors acknowledge financial support from the National Key R&D Program of China (2021YFD1201100). Furthermore, we gratefully acknowledge the support provided by the Friedrich Haberlandt Scholarship committee, administrative support by Donau Soja, and financial support from the German Federal States of Baden-Württemberg (Ministry for Rural Areas and Consumer Protection Baden-Württemberg) and Bavaria (Bavarian State Ministry of Food, Agriculture and Forestry), the Swiss Confederation (Agroscope), and Saatgut Austria (Seed Association of Austria). The authors are also grateful to soybean breeding companies and institutions for providing the seeds of the soybean cultivars used in the present study. We also acknowledge the contributions of Timea Jakab of Gabonakutató Nonprof. Közhasznú Kft., Hungary. Helpful advice provided by Matthias Krön (Vienna, Austria) and Johann Vollmann is also gratefully acknowledged. And we also acknowledge all the effort from our cooperate partners from China (Chinese academy of agriculture science) and Vuk DORDEVIC (IFVCNS, Serbia), Jerzy NAWRACALA (Poznan, Ploand), Christine RIEDEL (LFL Bayern, Germany), Claude-Alain BETRIX (AGROSCOPE, Switzerland) and Martin Pachner (BOKU, Austria).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Vollmann, J.; Škrabišová, M. Going north: Adaptation of soybean to long-day environments. J. Exp. Bot. 2023, 74, 2933–2936. [Google Scholar] [CrossRef] [PubMed]
- Döttinger, C.A.; Hahn, V.; Leiser, W.L.; Würschum, T. Do We Need to Breed for Regional Adaptation in Soybean?—Evaluation of Genotype-by-Location Interaction and Trait Stability of Soybean in Germany. Plants 2023, 12, 756. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, S.; Kong, F.; Lin, X.; Lu, S. Altered regulation of flowering expands growth ranges and maximizes yields in major crops. Front. Plant Sci. 2023, 14, 1094411. [Google Scholar] [CrossRef] [PubMed]
- Mourtzinis, S.; Conley, S.P. Delineating Soybean Maturity Groups across the United States. Agron. J. 2017, 109, 1397–1403. [Google Scholar] [CrossRef]
- Wen, H.; Wu, T.; Jia, H.; Song, W.; Xu, C.; Han, T.; Sun, S.; Wu, C. Analysis of Relationship between Soybean Relative Maturity Group, Crop Heat Units and ≥10 °C Active Accumulated Temperature. Agronomy 2022, 12, 1444. [Google Scholar] [CrossRef]
- Di, Y.; You, N.; Dong, J.; Liao, X.; Song, K.; Fu, P. Recent soybean subsidy policy did not revitalize but stabilize the soybean planting areas in Northeast China. Eur. J. Agron. 2023, 147, 126841. [Google Scholar] [CrossRef]
- Li, R.; Guo, J.; Song, Y. Optimizing parameters of a non-linear accumulated temperature model and method to calculate linear accumulated temperature for spring maize in Northeast China. Theor. Appl. Climatol. 2020, 141, 1629–1644. [Google Scholar] [CrossRef]
- Li, H.; Du, H.; He, M.; Wang, J.; Wang, F.; Yuan, W.; Huang, Z.; Cheng, Q.; Gou, C.; Chen, Z.; et al. Natural variation of FKF1 controls flowering and adaptation during soybean domestication and improvement. New Phytol. 2023, 238, 1671–1684. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Zhai, H.; Wu, H.; Xu, K.; Watanabe, S.; Harada, K. The Synchronized Efforts to Decipher the Molecular Basis for Soybean Maturity Loci E1, E2, and E3 That Regulate Flowering and Maturity. Front. Plant Sci. 2021, 12, 632754. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Liu, B.; Weller, J.L.; Abe, J.; Kong, F. Molecular mechanisms for the photoperiodic regulation of flowering in soybean. J. Integr. Plant Biol. 2020, 63, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; LI, W.; Fan, C.; Liu, M.; Liu, J.; Liang, W.; Wang, L.; Di, S.; Fang, C.; Li, H.; et al. Genome-wide association study uncovers major genetic loci associated with flowering time in response to active accumulated temperature in wild soybean population. BMC Genom. 2022, 23, 749. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Leiser, W.L.; Hahn, V.; Würschum, T. The genetic architecture of soybean photothermal adaptation to high latitudes. J. Exp. Bot. 2023, 74, 2987–3002. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Xu, J.; Liu, Z.-X.; Pachner, M.; Molin, E.M.; Rittler, L.; Hahn, V.; Leiser, W.; Gu, Y.; Lu, Y.; et al. Genetic diversity in early maturity Chinese and European elite soybeans: A comparative analysis. Euphytica 2023, 219, 17. [Google Scholar] [CrossRef]
- Fehr, W.R.; Caviness, C.E.; Burmood, D.T.; Pennington, J.S. Stage of Development Descriptions for Soybeans, Glycine max (L.) Merrill 1. Crop Sci. 1971, 11, 929–931. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A Memory-efficient, Visualization-enhanced, and Parallel-accelerated Tool for Genome-wide Association Study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Jia, G.; Diao, X. geneHapR: An R package for gene haplotypic statistics and visualization. BMC Bioinform. 2023, 24, 199. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Lu, S.; Cai, Y.; Xu, X.; Zhang, L.; Chen, F.; Jiang, B.; Zhang, H.; Sun, S.; Zhai, H.; et al. Molecular breeding for improvement of photothermal adaptability in soybean. Mol. Breeding 2023, 43, 60. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, G.; Miller, M.J.; Steketee, C.J.; Jackson, S.A.; de Tunes, L.V.M.; Li, Z. Genetic control and allele variation among soybean maturity groups 000 through IX. Plant Genome 2021, 14, e20146. [Google Scholar] [CrossRef] [PubMed]
- Perfil’ev, R.; Shcherban, A.; Potapov, D.; Maksimenko, K.; Kiryukhin, S.; Gurinovich, S.; Panarina, V.; Polyudina, R.; Salina, E. Impact of Allelic Variation in Maturity Genes E1–E4 on Soybean Adaptation to Central and West Siberian Regions of Russia. Agriculture 2023, 13, 1251. [Google Scholar] [CrossRef]
- Wang, L.; Li, H.; He, M.; Dong, L.; Huang, Z.; Chen, L.; Nan, H.; Kong, F.; Liu, B.; Zhao, X. GIGANTEA orthologs, E2 members, redundantly determine photoperiodic flowering and yield in soybean. J. Integr. Plant Biol. 2023, 65, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Fang, C.; Liu, B.; Yang, H.; Kong, F. Origin, variation, and selection of natural alleles controlling flowering and adaptation in wild and cultivated soybean. Mol. Breeding 2023, 43, 36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xu, M.; Dwiyanti, M.S.; Watanabe, S.; Yamada, T.; Hase, Y.; Kanazawa, A.; Sayama, T.; Ishimoto, M.; Liu, B.; et al. A Soybean Deletion Mutant That Moderates the Repression of Flowering by Cool Temperatures. Front. Plant Sci. 2020, 11, 429. [Google Scholar] [CrossRef] [PubMed]
- Staniak, M.; Szpunar-Krok, E.; Kocira, A. Responses of Soybean to Selected Abiotic Stresses—Photoperiod, Temperature and Water. Agriculture 2023, 13, 146. [Google Scholar] [CrossRef]
- Zhai, H.; Wan, Z.; Jiao, S.; Zhou, J.; Xu, K.; Nan, H.; Liu, Y.; Xiong, S.; Fan, R.; Zhu, J.; et al. GmMDE genes bridge the maturity gene E1 and florigens in photoperiodic regulation of flowering in soybean. Plant Physiol. 2022, 189, 1021–1036. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Zhai, H.; Liu, B.; Kong, F.; Yuan, X.; Wu, H.; Cober, E.R.; Harada, K. Molecular identification of genes controlling flowering time, maturity, and photoperiod response in soybean. Plant Syst. Evol. 2012, 298, 1217–1227. [Google Scholar] [CrossRef]
- Tang, Y.; Lu, S.; Fang, C.; Liu, H.; Dong, L.; Li, H.; Su, T.; Li, S.; Wang, L.; Cheng, Q.; et al. Diverse flowering responses subjecting to ambient high temperature in soybean under short-day conditions. Plant Biotechnol. J. 2023, 21, 782–791. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).