Incorporation of Photoperiod Insensitivity and High-Yield Genes into an Indigenous Rice Variety from Myanmar, Paw San Hmwe
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth
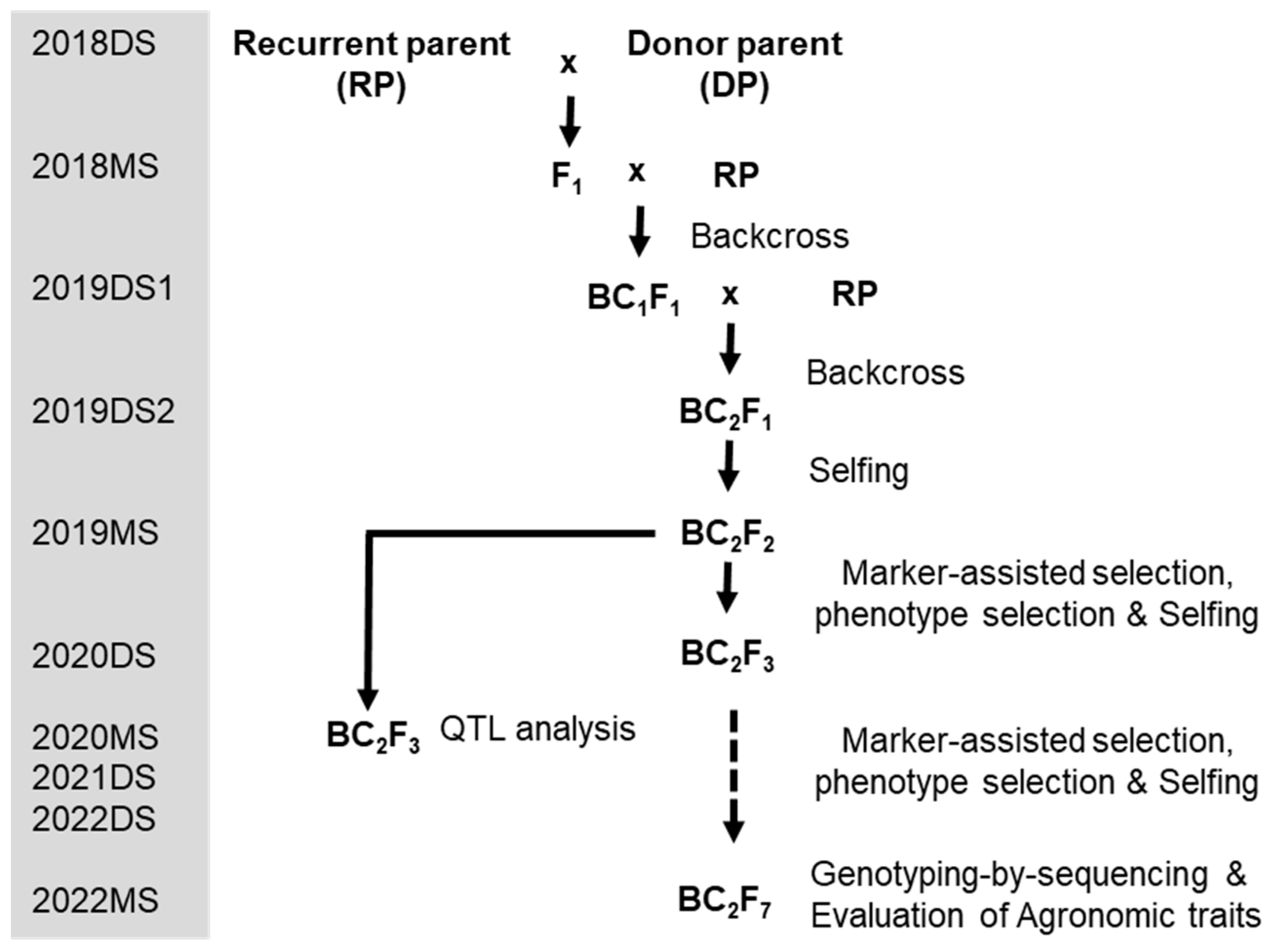
2.2. Development of Promising Lines
2.3. GBS
2.4. QTL Analysis of Days to Heading (DTH) in the BC2F3 Population
2.5. Foreground Selection of Gn1a and WFP Alleles
2.6. Recurrent Parent Genome Recovery (RPGR) Analysis
2.7. Field Trials and Evaluation of the Agronomic Traits and Yield Performance
2.8. Evaluation of the Physical Properties and Cooking and Eating Qualities
2.9. Data Analyses
3. Results
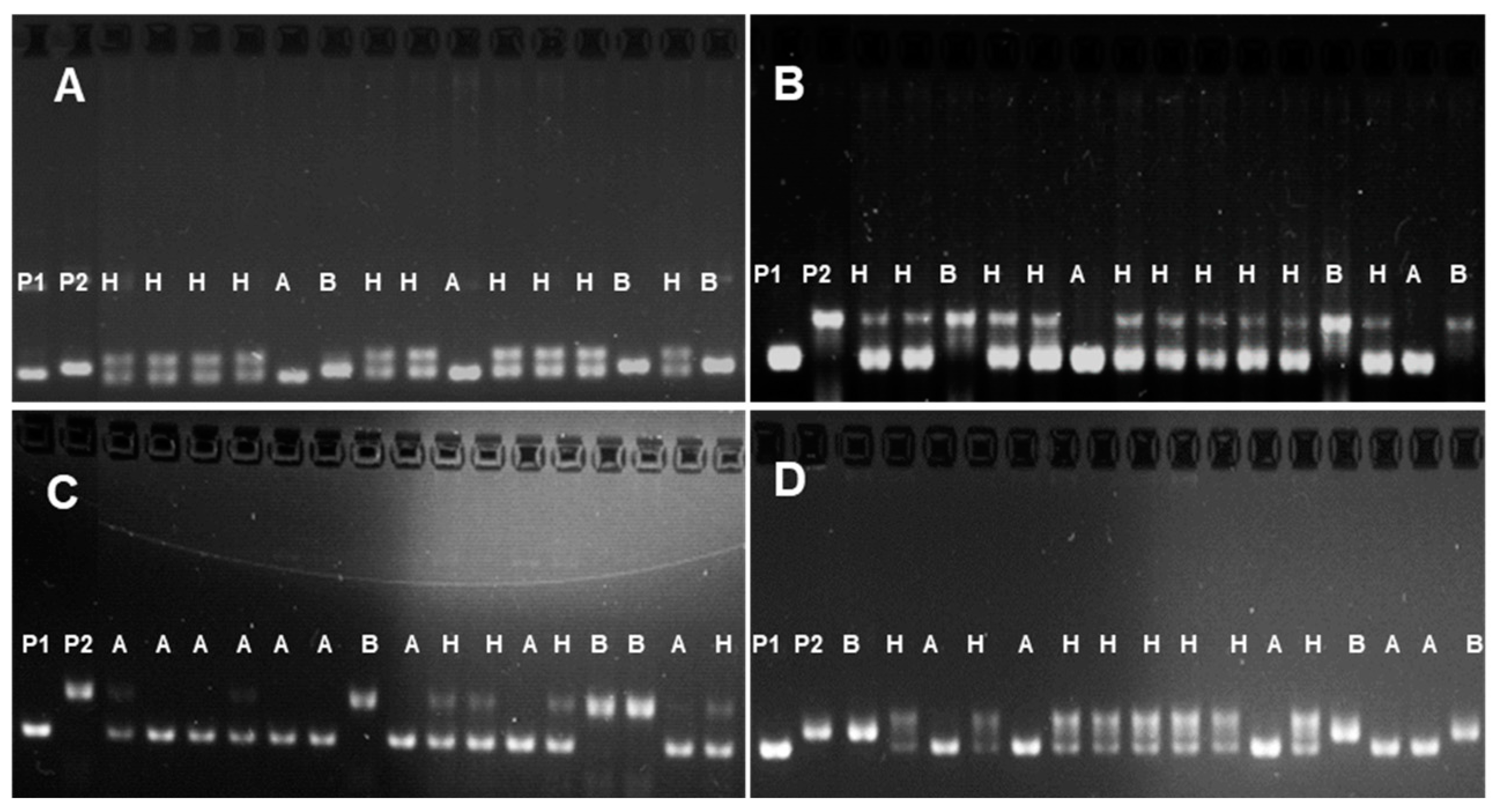
3.1. MAB for the Development of Promising Lines
3.2. RPGR Analysis of the Promising Lines
3.3. Introgression of Early Heading (Ehd) Genes into the Promising Lines
3.4. QTL Analysis of Days to Heading in the BC2F3 Population
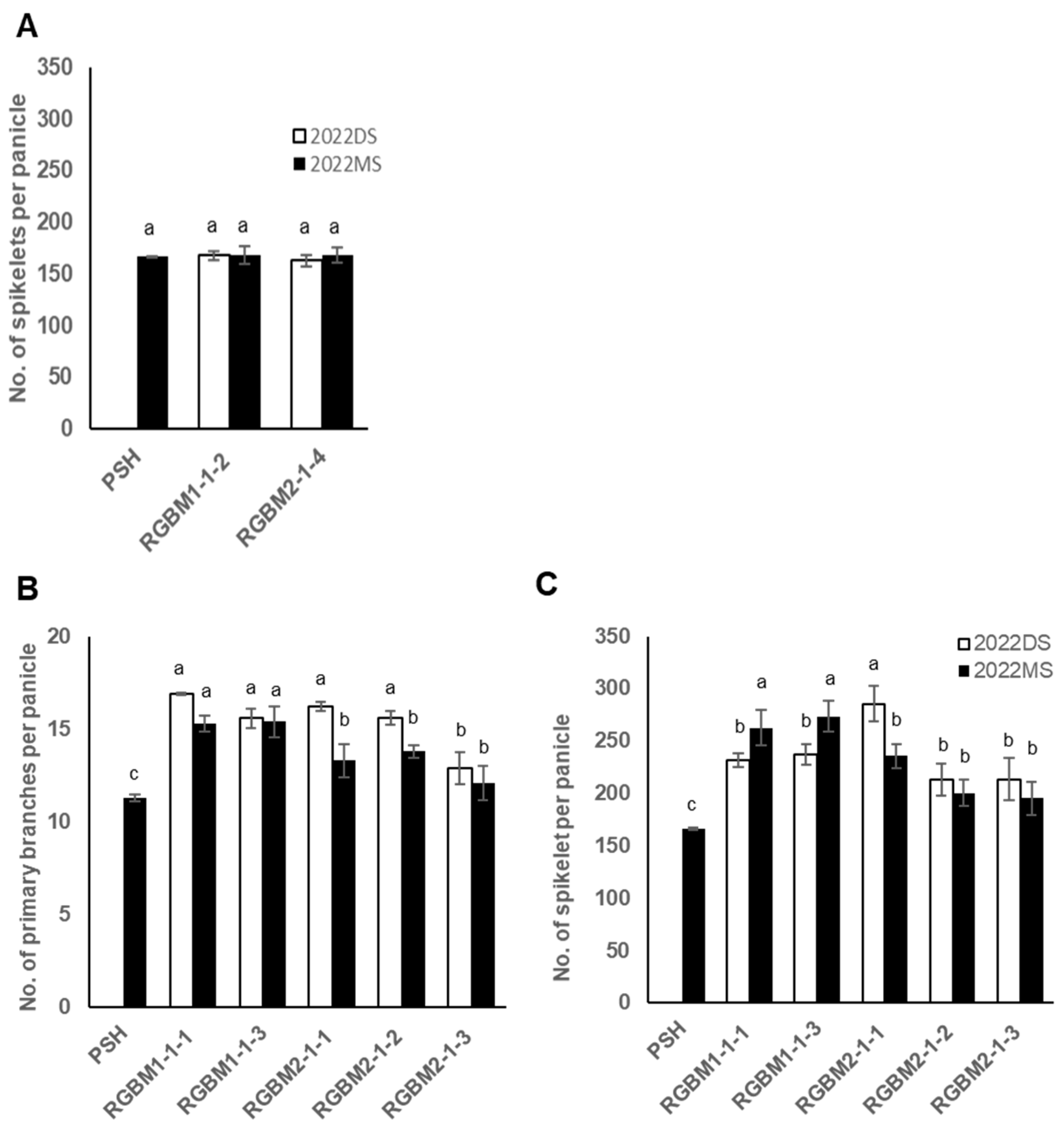
3.5. Effect of Gn1a Allele on the Spikelet Number
3.6. Effects of the Combination of Gn1a and WFP Alleles on the Spikelet Number and Primary Branching
3.7. Agronomic Traits and Yield Performance of the Promising Lines
3.8. Evaluation of the Physical Properties and Cooking and Eating Qualities of the Selected Promising Lines
3.9. Evaluation of the Aroma-Associated Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oo, K.S.; Kongjaimun, A.; Khanthong, S.; Yi, M.; Myint, T.T.; Korinsak, S.; Siangliw, J.L.; Myint, K.M.; Vanavichit, A.; Malumpong, C.; et al. Characterization of Myanmar Paw San Hmwe accessions using functional genetic markers. Rice Sci. 2015, 22, 53–64. [Google Scholar] [CrossRef]
- Singh, R.K.; Singh, U.S.; Khush, G.S.; Rohilla, R.; Singh, J.P.; Singh, G.; Shekhar, K.H. Small and medium grained aromatic rices of India. In Aromatic Rices; Singh, R.K., Singh, U.S., Khush, G.S., Eds.; Science Publishers: Enfield, CT, USA; Oxford & IBH Publishing: New Delhi, India, 2000; pp. 155–179. [Google Scholar]
- Thet, K.M. Myanmar Paw San rice varieties. In Proceedings of the Past, Present and Future of Myanmar Paw San Hmwe Conference, Naypyitaw, Myanmar, 14–15 June 2018; Myanmar Rice Federation (MRF): Yangon, Myanmar, 2018; pp. 134–143. [Google Scholar]
- Juliano, B.O.; Perez, C.M.; Kaosa-Ard, M. Grain quality characteristics of export rices in selected markets. Cereal Chem. 1990, 67, 192–197. [Google Scholar]
- Webb, B.D. Rice quality and grades. In Rice: Production and Utilization; Luh, B.S., Ed.; AVI Pub Co.: Westport, CT, USA, 1980; pp. 543–565. [Google Scholar]
- Cagampang, G.B.; Perez, C.M.; Juliano, B.O. A gel consistency test for the eating quality of rice. J. Sci. Food Agric. 1973, 24, 1589–1594. [Google Scholar] [CrossRef]
- Little, R.R.; Hilder, G.B.; Dawson, E.H. Differential effect of dilute alkali on 25 varieties of milled white rice. Cereal Chem. 1958, 35, 111–126. [Google Scholar]
- Vergara, B.S.; Chang, T.T. The Flowering Response of the Rice Plant to Photoperiod, a Review of the Literature, 4th ed.; International Rice Research Institute: Los Banos, Philippines, 1985. [Google Scholar]
- Nishida, H.; Okumoto, Y.; Nakagawa, H.; Ichitani, K.; Inoue, H.; Tanisaka, T. Analysis of tester lines for rice (Oryza sativa L.) heading-time genes using reciprocal photoperiodic transfer treatments. Ann. Bot. 2001, 88, 527–536. [Google Scholar] [CrossRef]
- Khush, G.S. Green revolution: The way forward. Nat. Rev. Genet. 2001, 2, 815–822. [Google Scholar] [CrossRef]
- Singh, V.P. The Basmati Rice of India. In Aromatic Rices; Singh, R.K., Singh, U.S., Khush, G.S., Eds.; Science Publishers: Enfield, CT, USA; Oxford & IBH Publishing: New Delhi, India, 2000; pp. 135–154. [Google Scholar]
- Sarkarung, S.; Somrith, B.; Chitrakorn, S. Aromatic Rices of Thailand. In Aromatic Rices; Singh, R.K., Singh, U.S., Khush, G.S., Eds.; Science Publishers: Enfield, CT, USA; Oxford & IBH Publishing: New Delhi, India, 2000; pp. 180–183. [Google Scholar]
- Khush, G.S. New plant type of rice for increasing the genetic yield potential. In Rice Breeding and Genetics; Nanda, J.S., Ed.; Science Publishers: Boca Raton, FL, USA, 2000; pp. 99–108. [Google Scholar]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef]
- Song, X.J.; Huang, W.; Shi, M.; Zhu, M.Z.; Lin, H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef]
- Weng, J.; Gu, S.; Wan, X.; Gao, H.; Guo, T.; Su, N.; Lei, C.; Zhang, X.; Cheng, Z.; Guo, X.; et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008, 18, 1199–1209. [Google Scholar] [CrossRef]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef]
- Huang, X.; Qian, Q.; Liu, Z.; Sun, H.; He, S.; Luo, D.; Xia, G.; Chu, C.; Li, J.; Fu, X. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef]
- Kim, S.-R.; Ramos, J.M.; Hizon, R.J.M.; Ashikari, M.; Virk, P.S.; Torres, E.A.; Nissila, E.; Jena, K.K. Introgression of a functional epigenetic OsSPL14WFP allele into elite indica rice genomes greatly improved panicle traits and grain yield. Sci. Rep. 2018, 8, 3833. [Google Scholar] [CrossRef]
- Yamada, S.; Kurokawa, Y.; Nagai, K.; Angeles-Shim, R.B.; Yasui, H.; Furuya, N.; Yoshimura, A.; Doi, K.; Ashikari, M.; Sunohara, H. Evaluation of backcrossed pyramiding lines of the yield-related gene and the bacterial leaf blight resistant genes. J. Intl. Cooper Agric. Dev. 2020, 18, 18–28. [Google Scholar]
- Feng, X.; Wang, C.; Nan, J.; Zhang, X.; Wang, R.; Jiang, G.; Yuan, Q.; Lin, S. Updating the elite rice variety Kongyu 131 by improving the Gn1a locus. Rice 2017, 10, 35. [Google Scholar] [CrossRef]
- Reyes, V.P.; Angeles-Shim, R.B.; Mendioro, M.S.; Manuel, M.C.C.; Lapis, R.S.; Shim, J.; Sunohara, H.; Nishiuchi, S.; Kikuta, M.; Makihara, D.; et al. Marker-assisted introgression and stacking of major QTLs controlling grain number (Gn1a) and number of primary branching (WFP) to NERICA cultivars. Plants 2021, 10, 844. [Google Scholar] [CrossRef]
- Hasan, M.M.; Rafii, M.Y.; Ismail, M.R.; Mahmood, M.; Rahim, H.A.; Alam, M.A.; Ashkani, S.; Malek, M.A.; Latif, M.A. Marker-assisted backcrossing: A useful method for rice improvement. Biotechnol. Biotechnol. Equip. 2015, 29, 237–254. [Google Scholar] [CrossRef]
- Chen, S.; Lin, X.H.; Xu, C.G.; Zhang, Q. Improvement of bacterial blight resistance of ‘Minghui 63’, an elite restorer line of hybrid rice, by molecular marker-assisted selection. Crop Sci. 2000, 40, 239–244. [Google Scholar] [CrossRef]
- Septiningsih, E.M.; Pamplona, A.M.; Sanchez, D.L.; Neeraja, C.N.; Vergara, G.V.; Heuer, S.; Ismail, A.M.; Mackill, D.J. Development of submergence-tolerant rice cultivars: The Sub1 locus and beyond. Ann. Bot. 2009, 103, 151–160. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple Genotyping-by-Sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Arbelaez, J.D.; Moreno, L.T.; Singh, N.; Tung, C.-W.; Maron, L.G.; Ospina, Y.; Martinez, C.P.; Grenier, C.; Lorieux, M.; McCouch, S. Development and GBS-genotyping of introgression lines (ILs) using two wild species of rice. O. meridionalis and O. rufipogon, in a common recurrent parent, O. sativa cv. Curinga. Mol. Breed. 2015, 35, 81. [Google Scholar] [CrossRef]
- Begum, H.; Spindel, J.E.; Lalusin, A.; Borromeo, T.; Gregorio, G.; Hernandez, J.; Virk, P.; Collard, B.; McCouch, S.R. Genome-wide association mapping for yield and other agronomic traits in an elite breeding population of tropical rice (Oryza sativa). PLoS ONE 2015, 10, e0119873. [Google Scholar] [CrossRef]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Poland, J.A.; Brown, P.J.; Sorrells, M.E.; Jannink, J.L. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 2012, 7, e32253. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Fragoso, C.A.; Heffelfinger, C.; Zhao, H.; Dellaporta, S.L. Imputing genotypes in biallelic populations from low-coverage sequence data. Genetics 2016, 202, 487–495. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Broman, K.W.; Wu, H.; Sen, S.; Churchill, G.A. R/qtl: QTL mapping in experimental crosses. Bioinformatics 2003, 19, 889–890. [Google Scholar] [CrossRef] [PubMed]
- Benchimol, L.; Souza, C.L.D., Jr.; Souza, A.P.D. Microsatellite-assisted backcross selection in maize. Genet. Mol. Biol. 2005, 28, 789–797. [Google Scholar] [CrossRef]
- Okadome, H.; Toyoshima, H.; Ohtsubo, K. Multiple measurements of physical properties of individual cooked rice grains with a single apparatus. Cereal Chem. 1999, 76, 855–860. [Google Scholar] [CrossRef]
- Okadome, H.; Toyoshima, H.; Ohtsubo, K. Many-sided evaluation of physical properties of cooked rice grains with a single apparatus. J. Jpn. Soc. Food Sci. 1995, 43, 1004–1011. [Google Scholar] [CrossRef]
- Takeyama, S.; Toyama, R.; Fujiwara, Y.; Arakawa, Y. Physical properties of steamed cakes made of miscellaneous cereals. Res. Bull. Iwate Ind. Res. Inst. 1998, 5, 28. [Google Scholar]
- Matsue, Y. On a sensory test of cooked rice in which 10 cultivars were evaluated by 13 panel members. J. Home Econ. 1992, 43, 1027–1032. [Google Scholar]
- Zhou, S.; Zhu, S.; Cui, S.; Hou, H.; Wu, H.; Hao, B.; Cai, L.; Xu, Z.; Liu, L.; Jiang, L.; et al. Transcriptional and post-transcriptional regulation of heading date in rice. New Phytol. 2021, 230, 943–956. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, L.M.T.; Fitzgerald, T.L.; Henry, R.J.; Jin, Q.; Waters, D.L.E. The gene for fragrance in rice. Plant Biotechnol. J. 2005, 3, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Vanavichit, A.; Tragoonrung, S.; Toojinda, T.; Wanchana, S.; Kamolsukyunyong, W. Transgenic rice plants with reduced expression of Os2AP and elevated levels of 2-acetyl-1-pyrroline. U.S. Patent 7,319,181, 15 January 2008. [Google Scholar]
- Myint, K.M.; Courtois, B.; Risterucci, A.M.; Frouin, J.; Soe, K.; Thet, K.M.; Vanavichit, A.; Glaszmann, J.C. Specific patterns of genetic diversity among aromatic rice varieties in Myanmar. Rice 2012, 5, 20. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Furuta, T.; Saw, O.M.; Win, K.T.; Hlaing, M.M.; Yasui, H.; Ashikari, M.; Thein, M.S.; Yoshimura, A.; Yamagata, Y. Development of genomic and genetic resources facilitating molecular genetic studies on untapped Myanmar rice germplasms. Breed. Sci. 2024, in press.
- Dormatey, R.; Sun, C.; Ali, K.; Coulter, J.A.; Bi, Z.; Bai, J. Gene Pyramiding for Sustainable Crop Improvement against Biotic and Abiotic Stresses. Agronomy 2020, 10, 1255. [Google Scholar] [CrossRef]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Chu, J.; Zhao, Y.; Beier, S.; Schulthess, A.W.; Stein, N.; Philipp, N.; Röder, M.S.; Reif, J.C. Suitability of single-nucleotide polymorphism arrays versus genotyping-by-sequencing for genebank genomics in wheat. Front. Plant Sci. 2020, 11, 42. [Google Scholar] [CrossRef]
- Kurokawa, Y.; Noda, T.; Yamagata, Y.; Angeles-Shim, R.; Sunohara, H.; Uehara, K.; Furuta, T.; Nagai, K.; Jena, K.K.; Yasui, H.; et al. Construction of a versatile SNP array for pyramiding useful genes of rice. Plant Sci. 2016, 242, 131–139. [Google Scholar] [CrossRef]
- Neeraja, C.N.; Maghirang-Rodriguez, R.; Pamplona, A.; Heuer, S.; Collard, B.C.; Septiningsih, E.M.; Vergara, G.; Sanchez, D.; Xu, K.; Ismail, A.M.; et al. A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theor. Appl. Genet. 2007, 115, 767–776. [Google Scholar] [CrossRef]
- Yi, M.; Nwe, K.T.; Vanavichit, A.; Chai-arree, W.; Toojinda, T. Marker assisted backcross breeding to improve cooking quality traits in Myanmar rice cultivar Manawthukha. Field Crop Res. 2009, 113, 178–186. [Google Scholar] [CrossRef]
- Sundaram, R.M.; Vishnupriya, M.R.; Biradar, S.K.; Laha, G.S.; Reddy, G.A.; Rani, N.S.; Sarma, N.P.; Sonti, R.V. Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica 2008, 160, 411–422. [Google Scholar] [CrossRef]



| Marker | Forward Primer Sequence | Reverse Primer Sequence | Target Gene | Chr. | Position (Mb) * |
|---|---|---|---|---|---|
| RM5423 | ATCCCACTTGCAGACGTAGG | ACAGCAGCAAGGTGCCTC | Gn1a | 1 | 2.17 |
| RM8098 | GATACCGTTACGCCTATTCAGTAGTGGG | CTTGATGAACTTGATTTCGTTGCAGTACA | Gn1a | 1 | 7.25 |
| RM531 | GAAACATCCCATGTTCCCAC | TCGGTTTTTCAGACTCGGTC | WFP | 8 | 22.47 |
| RM264 | GTTGCGTCCTACTGCTACTTC | GATCCGTGTCGATGATTAGC | WFP | 8 | 27.93 |
| Line Name | Designation | Total No. of SNPs | Recurrent Parent Genome Recovery Range of BC2F7 Population (%) | Recurrent Parent Genome Recovery of Selected BC2F7 Lines (%) |
|---|---|---|---|---|
| RGBM1-1-1 | Gn1a + WFP + Ehd-ST12-1 | 912 | 83.7–84.0 | 83.9 |
| RGBM1-1-3 | Gn1a + WFP + Ehd-ST12-2 | 922 | 84.4–87.5 | 87.2 |
| RGBM2-1-1 | Gn1a + WFP + Ehd-ST12-3 | 916 | 79.8–80.5 | 79.8 |
| RGBM2-1-2 | Gn1a + WFP + Ehd-ST12-4 | 915 | 84.4–88.6 | 88.6 |
| RGBM2-1-3 | Gn1a + WFP + Ehd-ST12-5 | 921 | 88.8–89.8 | 89.0 |
| RGBM1-1-2 | Gn1a + Ehd-ST12-1 | 919 | 87.6–88.4 | 87.7 |
| RGBM2-1-4 | Gn1a + Ehd-ST12-2 | 922 | 73.7–76.5 | 76.5 |
| Line Name | DTH | CL (cm) | TN | PL (cm) | PB | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| DS | MS | DS | MS | DS | MS | DS | MS | DS | MS | |
| PSH | - | 123 ± 0.0 | - | 134.3 ± 1.9 | - | 9.5 ± 0.5 | - | 25.7 ± 0.7 | - | 11.3 ± 0.3 |
| ST12 | 85 ± 0 | 78 ± 0.0 | 60.6 ± 3.5 | 65.3 ± 3.5 | 10.1 ± 0.5 | 9.20 ± 1.5 | 23.0 ± 1.9 | 23.4 ± 0.5 | 17.7 ± 0.5 | 15.4 ± 1.9 |
| RGBM1-1-1 | 112 ± 0 * | 94 ± 0.0 * | 103.4 ± 2.3 * | 106.8 ± 3.8 * | 8.4 ± 0.3 | 11.0 ± 1.6 | 26.8 ± 0.6 * | 27.2 ± 1.9 * | 16.9 ± 0.1 * | 15.3 ± 1.2 * |
| RGBM1-1-3 | 104 ± 0 * | 87 ± 0.0 * | 108.3 ± 2.5 * | 111.5 ± 1.6 * | 8.5 ± 0.8 | 10.5 ± 0.7 | 27.9 ± 0.6 * | 27.6 ± 1.5 * | 15.6 ± 1.1 * | 15.4 ± 1.6 * |
| RGBM2-1-1 | 100 ± 0 * | 83 ± 0.0 * | 98.4 ± 5.5 * | 101.3 ± 4.0 * | 10.6 ± 1.8 | 9.8 ± 0.6 | 26.1 ± 1.5 | 23.3 ± 1.0 * | 16.2 ± 0.5 * | 13.3 ± 1.8 * |
| RGBM2-1-2 | 102 ± 0 * | 89 ± 0.0 * | 96.4 ± 1.3 * | 95.9 ± 1.9 * | 11.6 ± 1.1 | 11.4 ± 1.9 | 23.8 ± 1.5 * | 22.4 ± 0.3 * | 15.6 ± 0.8 * | 13.8 ± 0.7 * |
| RGBM2-1-3 | 100 ± 0 * | 85 ± 0.0 * | 94.6 ± 4.2 * | 101.8 ± 3.9 * | 11.8 ± 1.7 | 12.4 ± 1.7 * | 25.5 ± 0.7 | 23.2 ± 0.2 * | 12.9 ± 1.7 * | 12.1 ± 1.9 * |
| RGBM1-1-2 | 105 ± 0 * | 90 ± 0.0 * | 102.8 ± 2.4 * | 105.5 ± 1.9 * | 12.1 ± 0.9 * | 11.8 ± 1.6 | 27.7 ± 0.2 * | 25.1 ± 0.4 | 9.1 ± 0.2 * | 8.9 ± 0.19 * |
| RGBM2-1-4 | 105 ± 0 * | 94 ± 0.0 * | 89.8 ± 1.1 * | 88.9 ± 5.6 * | 11.5 ± 1.0 | 12.4 ± 1.5 * | 25.9 ± 0.7 | 24.0 ± 1.1 * | 9.2 ± 0.2 * | 8.6 ± 0.37 * |
| Line Name | NS | PF | TGW (g) | GY (kg/ha) | ||||
|---|---|---|---|---|---|---|---|---|
| DS | MS | DS | MS | DS | MS | DS | MS | |
| PSH | - | 166.6 ± 1.9 | - | 81.8 ± 1.3 | - | 30.2 ± 0.3 | - | 5882.4 ± 142.0 |
| ST12 | 235.5 ± 26.2 | 195.6 ± 26.2 | 69.6 ± 5.6 | 81.7 ± 4.0 | 21.2 ± 0.2 | 21.6 ± 0.8 | 6301.6 ± 791.2 | 5233.4 ± 438.5 |
| RGBM1-1-1 | 231.8 ± 13.8 * | 262.6 ± 34.3 * | 65.1 ± 5.6 * | 72.8 ± 5.7 * | 28.7 ± 0.7 * | 28.4 ± 0.5 * | 6851.0 ± 960.1 * | 7029.1 ± 959.1 * |
| RGBM1-1-3 | 237.1 ± 20.2 * | 273.6 ± 29.4 * | 66.9 ± 3.9 * | 71.9 ± 4.7 * | 29.6 ± 0.2 | 29.4 ± 0.9 | 6086.9 ± 721.8 | 5829.6 ± 459.0 |
| RGBM2-1-1 | 285.4 ± 34.1 * | 235.6 ± 23.4 * | 66.9 ± 1.8 * | 72.2 ± 4.1 * | 28.4 ± 0.1 * | 29.6 ± 0.2 | 5493.4 ± 217.5 | 7240.1 ± 699.9 * |
| RGBM2-1-2 | 213.1 ± 30.2 * | 200.6 ± 25.9 * | 74.4 ± 5.5 | 84.6 ± 0.6 | 29.3 ± 0.5 * | 29.4 ± 0.2 * | 5774.8 ± 491.8 | 7349.8 ± 814.7 * |
| RGBM2-1-3 | 213.7 ± 26.1 * | 195.4 ± 31.6 * | 77.5 ± 2.9 | 83.3 ± 3.9 | 29.6 ± 0.5 | 30.2 ± 0.3 * | 5961.7 ± 807.8 | 7096.7 ± 160.3 * |
| RGBM1-1-2 | 167.9 ± 8.9 | 168.4 ± 16.8 | 79.1 ± 2.6 | 91.7 ± 0.9 * | 30.7 ± 0.3 | 31.2 ± 0.5 * | 6853.0 ± 643.0 * | 7408.8 ± 180.0 * |
| RGBM2-1-4 | 162.8 ± 11.4 | 168.0 ± 14.3 | 83.4 ± 7.7 | 91.6 ± 3.2 * | 25.8 ± 0.2 * | 26.9 ± 0.2 * | 6126.0 ± 1327.6 | 6296.0 ± 723.8 |
| Line Name | GL (mm) | GW (mm) | L/W Ratio | Appearance of Polished Rice | AC (%) | GC (mm) | GT (°C) | Elongation Ratio | Hardness (H1) | Stickiness (S1) | H1/S1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PSH | 7.5 | 3.0 | 2.5 | ~80% opaque | 21.3 | 31 | 75–79 | 2.3 | 969.4 | 51.7 | 18.9 |
| RGBM2-1-1 | 8.1 | 3.0 | 2.7 | ~80% opaque | 21.9 | 34 | 75–79 | 2.1 | 727.3 | 42.8 | 17.3 |
| RGBM2-1-3 | 8.3 | 3.0 | 2.7 | ~80% opaque | 22.7 | 30 | 75–79 | 2.3 | 1192.4 | 54.6 | 22.2 |
| RGBM1-1-2 | 8.3 | 3.5 | 2.4 | ~30% opaque | 16.0 | 53 | 75–79 | 1.7 | 753.8 | 88.8 | 8.4 |
| RGBM2-1-4 | 7.9 | 3.5 | 2.3 | ~90% opaque | 22.0 | 30 | 75–79 | 2.0 | 1377.4 | 85.8 | 16.0 |
| Line Name | Overall Eating Quality | Appearance | Taste | Aroma | Stickiness | Hardness |
|---|---|---|---|---|---|---|
| PSH | 0 | 0 | 0 | 0 | 0 | 0 |
| RGBM2-1-1 | 0.66 * | 0.53 * | 0.39 | 0.25 | 0.24 | −0.21 |
| RGBM2-1-3 | 0.72 * | 0.74 * | 0.76 * | 0.46 * | 0.38 * | −0.51 * |
| RGBM1-1-2 | 0.85 * | 1.14 * | 0.87 * | 0.42 * | 0.89 * | −0.96 * |
| RGBM2-1-4 | 0.76 * | 0.66 * | 0.51 * | 0.35 | 0.41 * | −0.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Win, K.T.; Hlaing, M.M.; Hlaing, A.L.L.; Maung, Z.T.Z.; Oo, K.N.; Nwe, T.; Moe, S.; Lin, T.; Saw, O.M.; Aung, T.; et al. Incorporation of Photoperiod Insensitivity and High-Yield Genes into an Indigenous Rice Variety from Myanmar, Paw San Hmwe. Agronomy 2024, 14, 632. https://doi.org/10.3390/agronomy14030632
Win KT, Hlaing MM, Hlaing ALL, Maung ZTZ, Oo KN, Nwe T, Moe S, Lin T, Saw OM, Aung T, et al. Incorporation of Photoperiod Insensitivity and High-Yield Genes into an Indigenous Rice Variety from Myanmar, Paw San Hmwe. Agronomy. 2024; 14(3):632. https://doi.org/10.3390/agronomy14030632
Chicago/Turabian StyleWin, Khin Thanda, Moe Moe Hlaing, Aye Lae Lae Hlaing, Zin Thu Zar Maung, Khaing Nwe Oo, Thinzar Nwe, Sandar Moe, Thein Lin, Ohm Mar Saw, Thado Aung, and et al. 2024. "Incorporation of Photoperiod Insensitivity and High-Yield Genes into an Indigenous Rice Variety from Myanmar, Paw San Hmwe" Agronomy 14, no. 3: 632. https://doi.org/10.3390/agronomy14030632
APA StyleWin, K. T., Hlaing, M. M., Hlaing, A. L. L., Maung, Z. T. Z., Oo, K. N., Nwe, T., Moe, S., Lin, T., Saw, O. M., Aung, T., Swe, M. S., Lar, S. M., Sin, E. S., Yamagata, Y., Angeles, E. R., Matsue, Y., Yasui, H., Thein, M. S., Win, N. K., ... Yoshimura, A. (2024). Incorporation of Photoperiod Insensitivity and High-Yield Genes into an Indigenous Rice Variety from Myanmar, Paw San Hmwe. Agronomy, 14(3), 632. https://doi.org/10.3390/agronomy14030632






