Abstract
Nitrogen (N) is an essential element both affecting rhizosphere microorganisms within soil and supporting plant nutrition; however, little is known about how the rhizosphere microbial community composition of tiger nut in sandy soil responds to nitrogen addition. In this study, high-throughput sequencing technology is employed to analyze the shifts in composition and co-occurrence networks of rhizosphere microbial communities in tiger nut after nitrogen addition in sandy farmland. Results reveal that nitrogen addition significantly increases several soil parameters, including total organic matter (SOC, 32.2%), total nitrogen (TN, 46.2%), alkali-hydro nitrogen (AN, 92.7%), β-1,4-glucosidase (BG, 12.6%), L-leucine aminopeptidase (LAP, 8.62%), β-1,4-xylosidase(XYL, 25.6%), and β-1,4-N-acetylglucosaminidase (NAG, 32.3%). Meanwhile, bacterial α-diversity decreases with nitrogen addition, while fungi remain unaffected. Network analysis indicates a reduction in connections between microorganisms; however, increasing stability is observed in the interaction network after nitrogen addition. Importantly, nitrogen addition leads to the enhancement of rhizosphere soil multifunctionality, with fungal diversity identified as the primary driver of soil multifunctionality. The positive impact of microbial diversity on soil multifunctionality outweighs the relative negative effects. This study sheds light on the nuanced effects of nitrogen addition on rhizosphere microbial diversity and its consequent impact on soil multifunctionality, with Acidobacteria, Proteobacteria and Ascomycota having positive effects, providing a comprehensive understanding of the complex environmental–plant–soil–microbe interactions in sandy farmland ecosystems.
1. Introduction
In the agricultural production process, nitrogen fertilizer is a focal point for fertilization strategies [1]. The significance of nitrogen lies in its dual role: it is a more crucial element required for the growth of crops than phosphorus and potassium [2]. It must also be considered that nitrogen absorbed by crops remains in the soil and affects soil function [3]. Hui [4] has shown that nitrogen addition can augment soil organic matter (SOM), contributing to the enhancement of long-term soil productivity. Furthermore, findings in the literature indicate that nitrogen fertilizer enhances soil microbial activity, facilitating the activation of insoluble phosphorus in the soil, thereby increasing its availability and bolstering overall soil function [5]. However, divergent perspectives exist in the literature. Some studies suggest that nitrogen fertilizer input leads to a reduction in soil pH, resulting in soil acidification. This process accelerates the depletion of alkaline ions in the soil, potentially diminishing microbial activity, deteriorating soil quality, and consequently decreasing soil function [6]. The influence of nitrogen application on soil functionality, as well as the role of nitrogen in regulating various soil functions, remains a topic of debate and requires additional research. Understanding these dynamics is crucial for developing precise fertilization strategies to optimize crop productivity and minimize adverse effects on soil quality and functionality. Soil microorganisms play pivotal roles in the intricacies of soil biogeochemical cycles, contributing significantly to various soil functions, including nutrient cycling, abiotic stress tolerance, and the suppression of soil-borne pathogens [7]. Moreover, these microorganisms actively contribute to crop growth and development, thereby influencing food production for human consumption and promoting the overall service function and stability of agricultural ecosystems [8,9]. Rhizosphere microorganisms, a specialized subset, exert their influence in the proximity of the root system. They are attracted to the rhizosphere through root exudates and specialized metabolites [8,10]. The interactions and symbiotic relationships among diverse microbial communities in the rhizosphere play crucial roles in nutrient cycling within the root zone, enhancing plant disease resistance, and ecological adaptation [11].
Notably, the richness and diversity of rhizosphere soil microbial communities have been identified as potential indicators of root zone soil health, quality, and functionality. These microbial communities are integral players in the carbon and nitrogen cycling of the plant–soil relationship, underscoring their importance in maintaining the equilibrium of the soil ecosystem [9,12]. Given these critical roles, understanding the impact of fertilization on rhizosphere microbial diversity holds significant importance. It emerges as a critical factor in accurately assessing soil multifunctionality, mirroring the holistic influence of fertilization practices on the intricate relationships within the plant–soil–microbe continuum.
Tiger nut (Cyperus esculentus L.) is an annual herbaceous plant belonging to the sedge family (Cyperaceae) and the Cyperus genus. Renowned for its remarkable characteristics, tiger nut exhibits exceptional resilience, as it is drought, salt and alkali tolerant as well as proficient in thriving in poor soil conditions [13,14]. The cultivation of tiger nut is widespread across various continents, including Europe, America, Asia, and Africa, for application in medicine, food, and cosmetics [15,16]. Numerous studies underscore its robust resistance to adversity, and adaption to diverse environments with a high yield in sandy soil farmland [13,17,18]. Notably, Tian [13] emphasized the critical role of nitrogen fertilizer in fostering the growth and development of tiger nut in sandy soil conditions. Additionally, Vittorio [19] highlighted the pivotal contribution of rhizosphere microorganisms in influencing plant growth, development, and nutrient utilization. Despite the wealth of research on tiger nut, investigations specifically focusing on the rhizosphere microorganisms of tiger nut cultivated in sandy soil remain limited.
In this study, we investigated the impact of nitrogen addition on soil ecosystem versatility through its influence on the composition of rhizosphere microbial communities. Our objective was to uncover how rhizosphere soil microbial diversity and soil versatility respond to nitrogen addition. To address these questions, we (1) evaluated the influence of nitrogen addition on the composition and diversity of rhizosphere soil microbial communities in sandy soil; (2) identified alterations in the symbiotic networks and community assembly of rhizosphere microbes subsequent to nitrogen addition; and (3) demonstrated the beneficial influences of soil environmental conditions and microbial diversity on soil versatility. Through these hypotheses, we aimed to gain comprehensive insights into the intricate dynamics between nitrogen addition, rhizosphere microbial communities, and soil versatility, contributing to a deeper understanding of the complex interplay within this ecological system.
2. Materials and Methods
2.1. The Study Site
This study was conducted in Xing’an County, Kashi city, Xinjiang Province (38°38′ N, 77°06′ W), which is located on the southern edge of the Taklimakan Desert characterized as consisting of sandy soils (>95% sand) with low nutrient contents. This area experiences a temperate continental climate, a mean annual temperature of 11.4 °C, and a mean annual precipitation of 56.6 mm, with approximately 80% of the precipitation occurring between July and September. Selected physical and chemical properties of the 0–20 cm soil layer are shown in Table 1.

Table 1.
Selected physical and chemical properties of the uppermost 20 cm soil layer before fertilization in 2022.
2.2. Experimental Design
The experimental block design was established in 2018, with a total of 12 blocks, including two treatments and six block replicates. This study involved a design featuring two N fertilizers and six replicate treatments with N application rates of 0 and 20 g N m−2 (termed control and N addition). The experimental field was subjected to continuous cropping. Tiger nuts (Zhongyousha No. 1) were cultured, with each plot measuring 15 × 10 m, with 2 × 0.5 m guard rows set between plots. The plants were spaced 10 cm apart, with a row spacing of 30 cm. During this study, the weeds were regularly removed. Urea was used as the form of N and was applied through drip irrigation. The N0 plots received only water. Irrigation was conducted 15 times over the entire reproductive period, with each tube containing two rows spaced 60 cm apart, delivering an irrigation volume of 5250 m3 ha−1.
When the tiger nuts reached maturity, 15–20 well-grown crops were randomly selected from each fertilization plot using the five-point sampling method in 2022. The surface litter was cleared using a shovel, and the soil surrounding the root system was delicately loosened with a hoe. A vertical excavation was then made around the roots to a depth of approximately 20 cm until most of the roots became visible. Subsequently, the rhizosphere soil (located within 2 mm of the root surface) was meticulously collected using either a small, sterilized spoon or brush [20]. This soil was then placed into sterile sampling bags, which were labeled, sealed, and promptly stored in a sampling container at low temperature. A total of twelve rhizosphere soil were transported under cool conditions to the lab, where each soil sample was separated into three portions: one was left to air-dry at ambient temperature to analyze soil physicochemical characteristics, another was refrigerated at 4 °C for assessing the levels of ammonium and nitrate nitrogen as well as the activity of soil extracellular enzymes, and the last portion was kept in a deep freezer for future DNA extraction.
2.3. Soil Property Analysis and Function Assessment
Soil pH and electrical conductivity (EC) were determined using calibrated probes and a multiparameter meter (HQ40D, HACH, Loveland, USA) in deionized water at a ratio of 1:2.5 (w/v). Soil total organic C (SOC) and total N (TN) concentrations were determined by combustion (Vario MACRO cube, Elementar, Frankfurt, Germany) [13], and alkali-hydro nitrogen (AN) was determined by the alkali hydrolysis diffusion method [13]. Soil nitrate nitrogen (NO3−-N) and ammonium nitrogen (NH4+-N) were extracted with potassium chloride and subjected to continuous flow analyzer analysis (AA3, Seal Analytical, Norderstedt, Germany) [21,22].
Activities of five soil extracellular enzyme activities at 25 °C involved in C and N cycling of soil were measured. Briefly, we weighed 2.75 g (dry weight equivalent) of field soil sample to a small glass dish, added 91 mL of acetate buffer (50 mM, pH = 6.0), and then blended for 1 min. We combined 800 μL of the soil suspension with 200 μL of fluorometric substrates, which were enzyme-specific (Table S1), into a 96-well microplate. This setup included four replicates alongside a negative control (comprising buffer and substrate) and a blank (buffer mixed with suspension), all of which were then incubated in darkness at 25 °C. For determining the activity levels of soil enzymes, standard curves were established by blending 800 μL of the soil suspensions with 200 μL of calibration solutions (either 4-methylumbelliferone or 7-amino-4-methylcoumarin for LAP), spanning concentrations from 0 to 100 μM. The assays for soil extracellular enzyme activities (EEAs) were executed promptly to measure the activity levels of enzymes already secreted by soil microbes. Details on quality control are provided in Table S2. We recorded fluorescence intensities using a microplate reader (Biotek Synergy 2, Winooski, VT, USA) [21,22].
In this study, a comprehensive set of nine variables (SOC: soil total organic C, AN: alkali-hydro nitrogen, NO3−-N: soil nitrate nitrogen, NH4+-N: soil ammonium nitrogen, BG: β-1,4-glucosidase, XYL: cellobiohydrolase, CBH: β-1,4-xylosidase, NAG: β-1,4-N-acetylglucosaminidase, respectively, and LAP: L-leucine aminopeptidase) were carefully chosen as representative indicators of soil functions. These variables were specifically selected due to their efficacy in serving as proxies for essential processes in farmland ecosystems, encompassing soil carbon and nitrogen cycling, sequestration, and the productivity of soil fertility [23,24]. To quantify the intricate relationships between biodiversity and soil functions, three distinct approaches were employed: single-function analysis, averaging, and turnover assessment [25]. Soil multifunctionality was quantified through Z score transformation, a method employed in previous studies, and subsequently. The averages of these transformed variables were calculated by an approach established in [26,27]. Before proceeding with the analyses, the collected data underwent scrutiny for normal distribution using the Shapiro–Wilk test. In cases where data were found not to follow a normal distribution, appropriate normal transformations were applied to ensure the robustness of subsequent statistical analyses.
2.4. DNA Extraction and High-Throughput Sequencing
The Fast DNA Spin kit (MP Biomedicals, Santa Ana, CA, USA) was used to extract microbial DNA from 0.5 g of fresh soil, and the concentration and purity of the extracted DNA were determined using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA). High-throughput sequencing was performed on satisfactory DNA samples. PCR amplification was conducted using the bacterial universal primers 515F (5′-GTGCCAGCMGCCGCGG-3′) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′) and the fungal universal primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′). PCR amplification was performed on bacterial V4–V5 regions and fungal ITS1 regions. PCR was performed using TransGen AP221–02 with TransStart Fastpfu DNA polymerase. The pooled samples were then sequenced on an Illumina HiSeq 3000 platform (Illumina, San Diego, CA, USA).
2.5. Sequence Preprocessing
Bacterial and fungal amplicon sequences were analyzed with EasyAmplicon v1.14 [28]. Raw amplicon reads were merged, primers and barcodes were removed, and then quality control was performed to obtain high-quality amplicon sequences in VSEARCH v2.15 [29]. These high-quality sequences were denoised into amplicon sequence variants (ASVs), and representative sequences were picked in USEARCH v11.0.667 [30]. The bacterial and fungal ASVs were aligned to the RDP v18 [31], UNITE v8.2 [32], and NCBI nucleotide [33] databases to remove chimera sequences with the UCHIME algorithm [34]. The taxonomies of bacterial and fungal representative sequences were determined with RDP v18 and UNITE v8.2 using the SINTAX algorithm [35]. Bacterial ASVs assigned to chloroplasts and mitochondria as well as archaea were removed from the dataset [36,37]. After taxonomic identification, we obtained 12,073 ASVs for the bacteria and 1072 ASVs for the fungi. All samples were rarefied to minimum sequencing depths of 8776 for bacteria and 14,642 for fungi for downstream analyses.
2.6. Statistical Analysis
The results of all analyses are presented as the means ± standard deviations (SD) based on six replications. Data comparison, including soil chemical attributes, α-diversity metrics, and soil microbial taxa relative abundances across treatments, was performed using a two-way ANOVA in IBM SPSS v22.0 (IBM Corp., Armonk, NY, USA). To gauge microbial community diversity, five diversity indices were utilized: observed ASV species, Shannon, ACE, Richness, and Chao1. The Kruskal–Wallis test, executed in R, facilitated the assessment of microbial community composition differences between the treatments, considering p-values < 0.05 as statistically significant.
The association between individual soil functionalities, overall soil multifunctionality, and rhizosphere microbial diversity was analyzed using an ordinary least squares (OLS) linear regression model. Bacterial and fungal α-diversity were represented by the total ASV counts for bacteria and fungi at each site, respectively. A stepwise Akaike information criterion (AIC) model selection approach was adopted to construct linear additive models for each soil function, identifying a minimal set of microbial taxa (8 bacterial and 2 fungal phyla) influential on soil functionalities. Relationship between the number of soil functions and the proportion of the microbial pool that impacted these functions positively or negatively was also estimated [25]. The trade-offs of positive and negative effects or whether one type was dominant was determined by merging these two relationships.
To pinpoint soil characteristics and microbial elements (microbial diversity and dominant phyla) driving soil multifunctionality, random forest (RF) modeling was employed, utilizing R’s “A3” and “rfPermute” packages. The “rdacca.hp” package facilitated a hierarchical partitioning analysis, clarifying the relative significance of environmental and microbial factors in influencing soil multifunctionality [38].
3. Results
3.1. Response of Rhizosphere Soil Nutrients and Soil Extracellular Enzyme Activities to N Addition
The addition of nitrogen (N) fertilizer to the rhizospheric soil markedly enhanced its nutrient profiles and the activities of extracellular enzymes, pivotal for soil health and plant growth. Specifically, the concentrations of AN, SOC, TN, NO3−-N, and NH4+-N in the soil witnessed substantial increases of 92.7%, 32.2%, 46.2%, 126.3%, and 162.8%, respectively, when compared to the control group (Figure 1), indicating significant nutrient enrichment due to N fertilization (p < 0.05). In terms of extracellular enzyme activities, which are crucial for the cycling of carbon (C) and nitrogen (N) in the soil, there was a noticeable uptick following the application of nitrogen fertilizer. The activities of BG, LAP, NAG, and XYL saw increases of 12.6%, 8.62%, 32.3%, and 25.6%, respectively, compared to the control treatment. These enzymes play critical roles in the decomposition of organic matter, thereby facilitating nutrient cycling and availability in the soil ecosystem. A lower soil pH, indicating increased acidity, can affect the availability of various nutrients and potentially harm microbial diversity and activity. Similarly, a decreased C:N ratio could indicate an imbalance in the soil’s organic matter composition, potentially affecting soil structure and fertility.
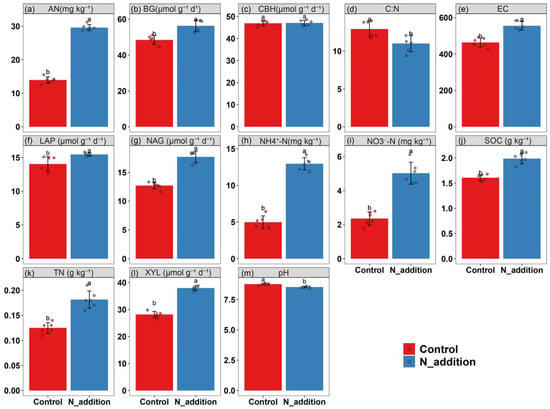
Figure 1.
Effects of N fertilization on soil factors and soil extracellular enzyme activities (a–m). Note: AN: alkali-hydro nitrogen (a), BG: β-1,4-glucosidase (b), CBH: cellobiohydrolase (c), C:N: soil C:N (d), EC: soil electrical conductivity (e), LAP: L-leucine aminopeptidase (f), NAG: β-1,4-N-acetylglucosaminidase, (g) NH4+-N: soil ammonium nitrogen (h), NO3−-N: soil nitrate nitrogen (i), SOC: soil total organic carbon (j), TN: soil total nitrogen (k), XYL: β-1,4-xylosidase (l) and pH: soil pH (m). Lowercase letters denote significant differences based on the t-test (n = 6, p < 0.05).
3.2. Responses of Rhizosphere Microbial Community Composition and Diversity to N Addition
As shown in Figure S2, Proteobacteria, Actinobacteria, Firmicutes, Chloroflexota, and Bacteroidota were the dominant species at the phylum level in the rhizosphere bacterial community. Nitrogen addition increased the abundances of Actinobacteria and Proteobacteria but decreased the abundances of Firmicutes and Actinobacteria. Ascomycota and Basidiomycota were the most abundant fungal phyla in the rhizosphere. Nitrogen addition increased the abundances of Ascomycota and Chytridiomycota but decreased the abundance of Basidiomycota. Nitrogen application significantly reduced the α diversity of the rhizosphere bacterial microbial community (p < 0.05) but had no significant effect on the α diversity of the rhizosphere fungal community (Figure 2). The PCoA results showed that N fertilization significantly affected the composition of the rhizosphere microbial formation of tiger nut (p = 0.007 and p = 0.006) (Figure 3). In the analysis, PCoA explained 49.08% and 61.38% of the bacterial and fungal data variation, respectively, and both rhizosphere bacterial and fungal communities were significantly affected by NAG and C:N (Figure S2).
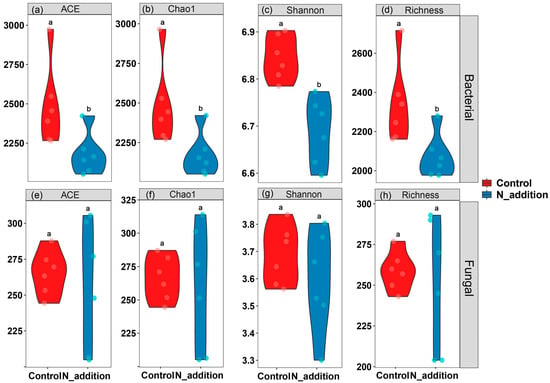
Figure 2.
Alpha diversity of rhizosphere bacterial (a–d) and fungal (e–h) microbial communities under N fertilization. Lowercase letters denote significant differences based on the t-test comparisons (n = 6, p < 0.05).
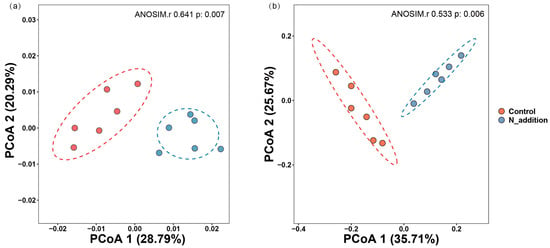
Figure 3.
Principal coordinate analysis (PCoA) based on Bray–Curtis distances to visualize the patterns of rhizosphere bacterial (a) and fungal (b) microbial communities under nitrogen (N) fertilization.
3.3. Influences of N Addition on the Complexity of the Microbial Co-Occurrence Network in Rhizosphere Soil
The networks of both the control and N addition groups exhibited scale-free patterns, with the degrees of distribution adhering to power law distributions and R2 values ranging from 0.82 to 0.86 (Figure S4). Notably, the research findings underscored significant disparities in the co-occurrence network topology between the control and N addition treatment groups. Although the number of nodes is smaller than the control group (Figure 4), with the increase in N, the proportion of bacteria in the composition of the microbial network increased by 3.34% (Figure 5a), and the positive correlation of microbial interactions in the network significantly increased (Figure 5b). The N addition treatment notably heightened the interactions within both the bacterial and bacterial–fungal networks (Figure 5c). Under N addition conditions, the network robustness, as indicated by natural connectivity, was markedly higher than that of the control group (Figure 5d), while the network complexity exhibited an inverse relationship (Figure 5e). Unlike the control group, which displayed small-world characteristics with a higher average clustering coefficient and shorter average path length, N addition treatment altered these parameters. These findings suggest that under nutrient-limited conditions, the increased interactions between fungi and bacteria facilitated more efficient nutrient acquisition from the soil (Figure 5f). In conclusion, N addition induced a shift in the network complexity of the microbial community.
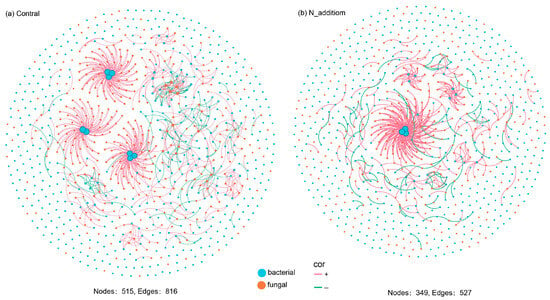
Figure 4.
Co-occurrence patterns of rhizosphere soil bacterial and fungal communities in (a) control and (b) N addition.
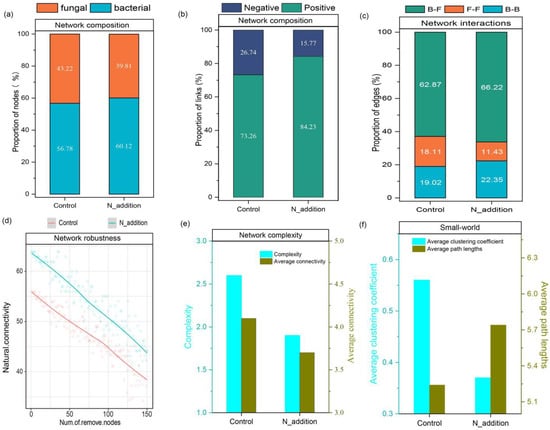
Figure 5.
Network composition and interactions were characterized using the proportions of nodes (a), links (b), and edges (c). The network robustness (d) was assessed by determining the proportions of species randomly removed from each network. Network complexity was captured through measures of complexity and average connectivity (e). The presence of small-world properties was indicated by a high average clustering coefficient and small average path lengths (f). In the context of edges, (b–f) represented connections between bacteria and fungi in the network.
3.4. Rhizosphere Microbial Diversity–Soil Multifunctionality Relationships
In our investigation, we explored the intricate relationship between microbial diversity, including both α and β diversity and various taxonomic groups, and the functioning of the soil, examining both individual soil functions (Figures S5 and S6) and overall soil multifunctionality (Figure 4). Notably, our results revealed a significant positive correlation between soil multifunctionality and bacterial β diversity (R2 = 0.84, p = 0.001) (Figure 6c). Additionally, significant negative correlations between soil multifunctionality and bacterial α diversity we observed (R2 = 0.59, p = 0.002) as well as fungal β diversity (R2 = 0.72, p = 0.001) (Figure 6a,d). Delving into specific bacterial taxa, Actinobacteria (R2 = 0.77, p = 0.001) and Proteobacteria exhibited noteworthy positive correlations with soil multifunctionality (R2 = 0.84, p < 0.001), while Actinobacteria (R2 = 0.73, p = 0.001) and Firmicutes (R2 = 0.71, p = 0.001) demonstrated significant negative correlations (Figure 6e–h). Turning to fungal taxa, Ascomycota (R2 = 0.87, p = 0.001) displayed a marked positive correlation with soil multifunctionality, whereas Basidiomycota exhibited a notable negative correlation (R2 = 0.75, p = 0.002) (see Figure 6i,j).
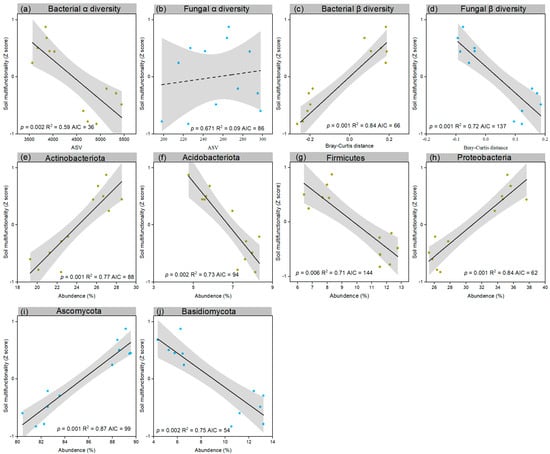
Figure 6.
Correlations between microbial diversity and soil multifunctionality (Z score). The associations between soil multifunctionality and various microbial diversity indices were investigated, including bacterial α diversity (a), fungal α diversity (b), bacterial β diversity (c), fungal β diversity (d), and specific microbial phyla, including Actinobacteria (e), Actinobacteria (f), Firmicutes (g), Proteobacteria (h), Ascomycota (i), and Basidiomycota (j), across the sampling sites (n = 12). Fitted linear ordinary least squares (OLS) models are represented by lines, with solid lines indicating statistically significant relationships (p < 0.05) and dashed lines denoting nonsignificant relationships (p > 0.05). The shaded areas depict the 95% confidence intervals of the fit. The Akaike information criterion (AIC) represents the goodness of fit for the linear regression analysis. Yellow represents bacteria, while blue represents fungi.
Through quantitative analysis, this study scrutinized the correlations between soil multifunctionality and the positive and negative impacts of microbial populations at the phylum level. The findings elucidated that as the accumulation of positively influencing microorganisms increased, the relationship tended toward saturation, with approximately 46.4% of phylum-level microorganisms exhibiting positive effects on soil multifunctionality, while approximately 29.7% had negative effects. When considering overall soil multifunctionality, all phylum-level microorganisms had positive effects, with approximately 87.5% exhibiting negative effects (Figure 7a,b). Furthermore, the research revealed a proportional increase in positive effects with the escalation of negative effects (Figure 7c), and the effect size of positive impacts slightly surpassed that of negative impacts (Figure 7d). In summary, the augmentation in soil multifunctionality post nitrogen addition was attributed to the heightened diversity of phylum-level rhizosphere microbial communities.
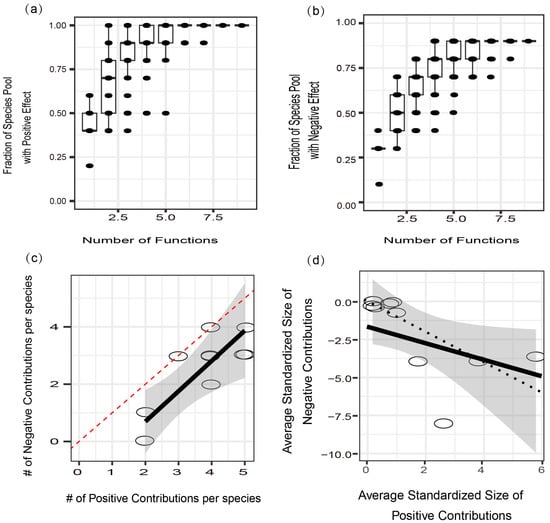
Figure 7.
Associations among soil functions, microbial species pool fraction, and the balance of positive and negative effects. The connections between the number of functions considered and the proportion of the microbial species pool were explored and categorized at the phylum level as exerting either positive (a) and negative (b) effects on soil functions. Additionally, the investigation delved into the relationship between the total numbers of positive and negative contributions made by microbial species to various soil functions (c). This study further examined the correlation between the average positive and negative effects of each microbial species (d). Each data point represents a microbial species at the phylum level. The shaded areas depict the 95% confidence interval of the fit, and the dashed line represents a 1:1 relationship.
3.5. Potential Drivers of Root Zone Soil Multifunctionality
The relative contributions of microbial diversity and environmental factors to root zone soil multifunctionality were assessed through random forest (RF) regression, with the model explaining an impressive 84% of the variance in soil multifunctionality. The RF model underscored the substantial influence of microbial diversity on rhizosphere soil multifunctionality in desertification farmland, with fungal β diversity, Firmicutes, Acidobacteria, and Ascomycota demonstrating significant effects (p < 0.05). These results underscored the pivotal role of microorganisms in steering soil multifunctionality in sandy farmland. Further insights from hierarchical partitioning analysis revealed that microbial diversity exerted the most significant individual impact on soil multifunctionality in the root zone, accounting for 32% of the variation, followed by soil properties (Figure 8b).
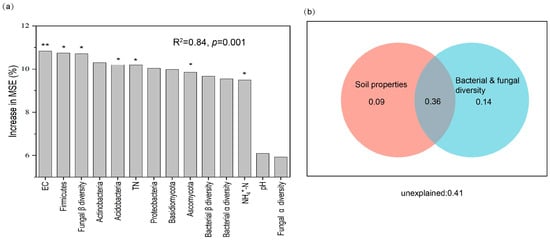
Figure 8.
Key Influences on soil multifunctionality and their operational mechanisms. Through the application of random forest analysis (a) and hierarchical partitioning (b), the average predictor importance concerning the environmental and rhizosphere microbial drivers of soil multifunctionality were unveiled, specifically focusing on dominant phyla representing more than 5% of the total microbial community. Significance levels were as follows: * p < 0.05, ** p < 0.01.
4. Discussion
Rhizosphere microorganisms constitute a crucial element of the rhizosphere microenvironment, actively engaging in soil nutrient transformation, elemental cycling, and the formation of soil structure. Additionally, they play a pivotal role in enhancing plant productivity by aiding in the acquisition of soil nutrients [39]. When plants absorb nutrients, they release root exudates that influence microorganism growth and development [40]. The findings of this study highlight that the relief of soil nitrogen limitation induced a shift in microbial life strategies, notably reflected in the increased abundances of Actinobacteria and Proteobacteria in the rhizosphere of tiger nut. This observation aligns with the nutrient enrichment hypothesis, wherein Actinobacteria, capable of obtaining water and nutrients through hyphal formation, demonstrate increased abundance, aiding in their survival under adverse conditions [41]. Moreover, Actinobacteria contribute to plant nutrition and enhance plant drought resistance. Conversely, Proteobacteria provide nutrients to plants through phosphorus solubilization, nitrogen fixation, and organic matter decomposition [42]. Notably, the abundance of oligotrophic fungi decreased, while that of copiotroph fungi increased after N addition.
Similarly, changes in the soil environment are primary drivers of variations in soil microbial diversity [40]. This study observed that under nitrogen fertilization treatment, bacterial α-diversity was significantly lower compared to the control group (Figure 2a–d). In contrast, fungal diversity remained unchanged (Figure 2e–h). This disparity may stem from nitrogen fertilization altering the soil’s environmental conditions, transitioning the nutrient limitation from a dual carbon-nitrogen constraint to solely carbon, thereby affecting the bacterial community composition. Conversely, fungi, being less impacted by external environmental changes and more by their intrinsic nutritional requirements, demonstrate reduced sensitivity to such environmental alterations [42,43,44]. These changes signified a shift in fungal growth strategies, conducive to the augmentation of fungal diversity. Similarly, PCoA reveals that nitrogen addition significantly influences both bacterial and fungal β diversity (Figure 3a,b), corroborating this hypothesis [9,43]. These findings underscore the importance of nutrient limitation as a key factor shaping microbial community composition [44]. These results collectively emphasize the intricate interplay between nutrient availability and the strategies employed by rhizosphere microorganisms, shedding light on their adaptive responses to changes in nutrient conditions.
Nitrogen addition notably decreases the network connectivity and complexity of microorganisms, as evidenced by the lower average degree and longer average path length (Figure 4 and Figure 5e,f). Intriguingly, despite the overall reduction in network complexity, the fungal–bacterial interactions and network stability increased (Figure 5a–c). A comparison with random networks underscores that the empirical network structure is nonrandom co-occurrence, indicating a regulatory influence on the formation of fungal–bacterial interactions. In the control treatment, the rhizosphere microbial interaction network exhibits a shorter average path length and a larger average clustering coefficient, implying higher energy flow and nutrient cycling efficiency in this network [45,46]. However, with increased soil nutrient availability post nitrogen addition, microorganisms seem to alter their reliance on mutual interactions for nutrient acquisition, leading to an increase in the average path length. These observations align with the “hunger game” hypothesis, suggesting that microorganisms, in nutrient-limited environments, engage in “cooperative clustering” to alleviate resource limitations to sustain their life activities [12,38]. As nutrient limitation is relieved, this cooperative relationship diminishes. This strategy supports hypothesis two, indicating that nitrogen addition reduces rhizosphere microbial interactions and decreases network complexity. Our study’s results highlight that positive correlations among microorganisms increases, while network complexity decreased, suggesting a shift from “antagonism” to “symbiosis” among microorganisms. Additionally, findings from Chen [8] support the idea that, with the alleviation of nutrient limitation, positive correlations in the fungal–bacterial interaction network increase while complexity decreases. This study underscored the significance of nutrient demand as a driving factor in regulating microbial interactions.
This article explored the regulatory impact of nitrogen addition on soil multifunctionality mediated by rhizosphere microorganisms in sandy soil, focusing on tiger nut cultivation. The results revealed a negative correlation between the α diversity of bacterial communities and soil multifunctionality following nitrogen addition. This finding contrasts with the generally observed positive correlation between biodiversity and ecosystem multifunctionality, although some studies support such a negative relationship [9,47]. This could have several potential explanations. First, possible explanations include the microbial diversity–soil multifunctionality relationship on ecosystem heterogeneity. For example, soil multifunctionality correlates positively with the composition and abundance of fungi in forest ecosystems [48]. Another contributing factor could be a shift in microbial nutrient acquisition strategies from co-nutrient nutrition to oligotrophic nutrition. Such a shift is exemplified in degraded alpine grasslands, where the dominant populations of bacteria and fungi change with increasing degrees of degradation. Additionally, as microbial abundance decreases, specific microorganisms with unique functions may become more influential. In karst areas, for instance, an increase in the abundance of Chloroflexi was associated with elevated soil available nitrogen content [47]. Moreover, in nutrient-scarce and imbalanced decertified farmland, the dormant state of many microorganisms may contribute to the observed negative correlation between microbial diversity and soil multifunctionality. These complexities highlight the context-dependent nature of the relationship between microbial diversity and soil functioning in different ecosystems.
Through random forest analysis, this study reveals that bacterial and fungal communities at the phylum level exerted significant driving effects on soil multifunctionality (Figure 8a). Interestingly, the observed changes in rhizosphere microbial communities do not uniformly align with alterations in soil multifunctionality across diverse ecosystems [9,27,49], suggesting that the regulation of soil multifunctionality by microbial communities might be intricately influenced by factors such as niche differentiation, the soil environment, and survival strategies. Notably, within the fungal taxa examined in this study, the Ascomycota group emerged as having a positive impact on the regulation of soil multifunctionality. This finding implies a potential key role for Ascomycota in plant residue decomposition, aligning with prior research conducted by Li [48] In their study, the abundance of saprophytic fungi, including Ascomycota, increased with forest age during forest succession. This increase in saprophytic fungal abundance was associated with the restoration of soil multifunctionality, supporting the notion that these fungi contribute significantly to overall soil functionality [50]. These findings collectively underscore the nuanced relationship between microbial communities and soil multifunctionality, suggesting that the impact of microbial regulation on multifunctionality is context-dependent and influenced by various ecological factors.
N addition is a common agricultural practice aimed at enhancing plant growth, but it also has profound effects on soil ecosystems, particularly on microbial communities that play crucial roles in soil health and ecosystem functions. The addition of nitrogen has been observed to significantly influence soil microbial dynamics, affecting both fungi and bacteria, albeit in different ways [51]. Fungi have shown a predominant response to soil enzymatic activities, specifically enzymes such as N-acetylglucosaminidase (NAG), cellobiohydrolase (CBH), and β-glucosidase (BG) (Figure S3). These enzymes are involved in the decomposition of organic matter, nutrient cycling, and the overall maintenance of soil health, indicating fungi’s critical role in soil biochemical processes [9]. On the other hand, the response of bacteria to nitrogen addition is more closely associated with changes in soil physicochemical properties, such as pH, electrical conductivity (EC), and total nitrogen (TN) (Figure S3). These properties are fundamental to soil health, influencing water retention, nutrient availability, and the overall structure and function of microbial communities [43]. The findings suggest that while fungi are more directly involved in the biochemical processing of soil, bacteria are sensitive indicators of the soil’s physical and chemical environment. Among the soil physicochemical properties, EC and TN stand out as key non-biological drivers that influence both the diversity of soil microbes and the multifunctionality of sandy soil (Figure 8a). This underscores the substantial role of these properties in shaping microbial diversity and, by extension, the multifunctional capacity of soils to support various ecosystem services such as nutrient cycling, water filtration, and plant growth support [8,43]. The impact of EC changes on microbial activity and soil functions is particularly significant. High soil salinity, indicated by elevated EC values, can disrupt microbial life by causing osmotic stress, altering nutrient availability, and even inhibiting the growth of less tolerant microbial species [27]. This can lead to shifts in microbial community composition and function, affecting soil’s ability to process organic matter and cycle nutrients effectively. Supporting these observations, research demonstrated that alterations in soil EC values, resulting from prolonged fertilization practices, can lead to significant changes in soil microbial community composition and carbon metabolism processes. It emphasizes the need for a nuanced understanding of these interactions to manage soil health and fertility sustainably, ensuring that agricultural practices do not inadvertently compromise soil’s ecological functions and its ability to support life.
Moreover, our study reveals that fungi play a more crucial role than bacteria in sandy soil, supporting hypothesis three. This outcome aligns with observations in various ecosystems, such as forests [48], farmland [9], and grasslands [12], where fungi, especially saprophytic fungi, have been recognized for their significant contribution to lignocellulose degradation. The pronounced environmental resilience and resistance of fungi compared to bacteria might explain their consistent roles across different ecosystems [43]. This study, therefore, adds to the growing body of evidence emphasizing the substantial impact of soil physicochemical characteristics and the pivotal role of fungi in influencing soil microbial communities and functions.
5. Conclusions
This study utilizes amplicon sequencing to investigate the impact of nitrogen fertilizer addition on rhizosphere microbial diversity and its consequent effects on soil multifunctionality. Our findings reveal that nitrogen fertilizer application precipitates a significant alteration in the multifunctionality of tiger nut rhizosphere soil by influencing the microbial community composition and interaction network. Network analysis elucidates that nitrogen addition reduces interactions between fungi and bacteria while concurrently enhancing the stability of the interaction network and helps tiger nut nutrient absorption after the reduction in microbial nutrient competition. These observations suggest that nitrogen addition has the potential to alleviate microbial competition, contributing to the maintenance of network stability. Additionally, analyses utilizing random forest (RF) and variance partitioning analysis (VPA) identify Firmicutes, Acidobacteria, and Ascomycota as pivotal contributors that significantly influenced soil multifunctionality and found that species detected at all phyla classification levels had a 100% positive contribution to soil versatility. These outcomes underscore the critical roles of both microbial diversity and specific taxa in shaping soil multifunctionality in sandy land. In conclusion, this research demonstrates that nitrogen addition profoundly influences soil multifunctionality by shaping microbial diversity and composition. These changes can have cascading effects on soil health, plant growth, and ecosystem sustainability. In conclusion, this study highlights the intricate relationships between nitrogen addition, microbial community dynamics, and soil physicochemical properties.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy14020368/s1, Table S1: The abbreviations of types, substrates, and functions of soil extracellular enzymes. Table S2. The QA/QC information for all measurements and analyzers used. Figure S1. Two different fertilization treatments and sampling methods in study area. Figure S2. Rhizosphere microbial community composition of bacteria (a) and fungal (b) at the phylum level under N addition. Figure S3. Redundancy analysis of environmental factors and rhizosphere bacterial (a) and fungal (b) microbial communities. The asterisks represent the significance (p < 0.05) factor via forward selection. Figure S4. Network topological feature of degree distribution pattern of rhizosphere microbial. Figure S5. Relationships between rhizosphere soil bacterial (a) and fungal (b) α diversity and individual soil function (n = 12). Solid and dashed lines denote statistically significant (two-sided p < 0.05) and nonsignificant (two-sided p > 0.05) relationships, respectively. The shaded areas show the 95% confidence interval of the fit. AN, TC, C: N, BG, CBH, XYL, NAG, LAP, NO3−-N and MBP were log transformed. Figure S6. Relationships between rhizosphere soil bacterial (a) and fungal (b) β diversity and individual soil function (n = 12). Solid and dashed lines denote statistically significant (two-sided p < 0.05) and nonsignificant (two-sided p > 0.05) relationships, respectively. The shaded areas show the 95% confidence interval of the fit. AN, TC, C: N, BG, CBH, XYL, NAG, LAP, NO3−-N and MBP were log transformed.
Author Contributions
J.W. was responsible for writing and editing, guiding the research and providing funding; L.L. was responsible for editing, providing some of the data and funding for the project; X.Z. came up with the idea, organized and analyzed the data, and completed the manuscript; J.C., J.L. and Z.C. contributed to editing. All authors have read and agreed to the published version of the manuscript.
Funding
The Central Guidance on Local Science and Technology Development Shihezi University innovation development project (CXFZ202307), the Tianshan Talent Program of Xinjiang province, Fund of Xinjiang Production and Construction Corps Province (grant no. 2060404), and the NSFC of China (grant no. 41763013).
Data Availability Statement
The raw sequencing data were deposited in the NCBI Sequence Read Archive under BioProject ID PRJNA1070446 (bacteria) and PRJNA1070540 (fungal).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Snapp, S.; Sapkota, T.B.; Chamberlin, J.; Cox, C.M.; Gameda, S.; Jat, M.L.; Marenya, P.; Mottaleb, K.A.; Negra, C.; Senthilkumar, K. Spatially differentiated nitrogen supply is key in a global food–fertilizer price crisis. Nat. Sustain. 2023, 6, 1268–1278. [Google Scholar] [CrossRef]
- Shahzad, A.N.; Qureshi, M.K.; Wakeel, A.; Misselbrook, T. Crop production in Pakistan and low nitrogen use efficiencies. Nat. Sustain. 2019, 2, 1106–1114. [Google Scholar] [CrossRef]
- Akhtar, K.; Wang, W.; Ren, G.; Khan, A.; Feng, Y.; Yang, G.; Wang, H. Integrated use of straw mulch with nitrogen fertilizer improves soil functionality and soybean production. Environ. Int. 2019, 132, 105092. [Google Scholar] [CrossRef] [PubMed]
- Hui, K.; Xi, B.; Tan, W.; Song, Q. Long-term application of nitrogen fertilizer alters the properties of dissolved soil organic matter and increases the accumulation of polycyclic aromatic hydrocarbons. Environ. Res. 2022, 215, 114267. [Google Scholar] [CrossRef] [PubMed]
- Ulm, F.; Gouveia, C.; Dias, T.; Cruz, C. N fertilization in a Mediterranean ecosystem alters N and P turnover in soil, roots and the ectomycorrhizal community. Soil Biol. Biochem. 2017, 113, 60–70. [Google Scholar] [CrossRef]
- Kimmel, K.; Furey, G.N.; Hobbie, S.E.; Isbell, F.; Tilman, D.; Reich, P.B. Diversity-dependent soil acidification under nitrogen enrichment constrains biomass productivity. Glob. Change Biol. 2020, 26, 6594–6603. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zheng, T.; Li, Y.; Zheng, X. A critical review of the central role of microbial regulation in the nitrogen biogeochemical process: New insights for controlling groundwater nitrogen contamination. J. Environ. Manag. 2023, 328, 116959. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chi, J.; Lu, X.; Cai, Y.; Jiang, H.; Zhang, Q.; Zhang, K. Fungal-bacterial composition and network complexity determine soil multifunctionality during ecological restoration. Catena 2023, 230, 107251. [Google Scholar] [CrossRef]
- Xue, R.; Wang, C.; Zhao, L.; Cao, J.; Liu, M.; Zhang, D. Agricultural intensification weakens soil multifunctionality by reducing fungal diversity. Appl. Soil Ecol. 2023, 189, 104900. [Google Scholar] [CrossRef]
- Zheng, Q.; Hu, Y.; Kosina, S.M.; Van Goethem, M.W.; Tringe, S.G.; Bowen, B.P.; Northen, T.R. Conservation of beneficial microbes between the rhizosphere and the cyanosphere. New Phytol. 2023, 240, 1246–1258. [Google Scholar] [CrossRef]
- Afridi, M.S.; Fakhar, A.; Kumar, A.; Ali, S.; Medeiros, F.H.; Muneer, M.A.; Ali, H.; Saleem, M. Harnessing microbial multitrophic interactions for rhizosphere microbiome engineering. Microbiol. Res. 2022, 265, 127199. [Google Scholar] [CrossRef] [PubMed]
- Dai, T.; Wen, D.; Bates, C.T.; Wu, L.; Guo, X.; Liu, S.; Su, Y.; Lei, J.; Zhou, J.; Yang, Y. Nutrient supply controls the linkage between species abundance and ecological interactions in marine bacterial communities. Nat. Commun. 2022, 13, 175. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Wu, X.; He, Y.; Li, Y.; Li, X.; Yu, X.; Shi, J. Mutual feedback mechanisms between functional traits and soil nutrients drive adaptive potential of tiger nuts (Cyperus esculentus L.) in marginal land. Plant Soil 2023, 495, 177–194. [Google Scholar] [CrossRef]
- Pascual, B.; Maroto, J.V.; López-Galarza, S.; Sanbautista, A.; Alagarda, J. Chufa (Cyperus esculentus L. var. sativus Boeck.): An unconventional crop. Studies related to applications and cultivation. Econ. Bot. 2000, 54, 439–448. [Google Scholar] [CrossRef]
- Wetters, S.; Häser, A.; Ehrlich, T.; Scheitle, C.; Nick, P. Tracing tiger nut (C. esculentus L.): Functional food from the colossal Cyperus genus. Eur. Food Res. Technol. 2023, 250, 225–238. [Google Scholar] [CrossRef]
- Pascual-Seva, N.; Pascual, B. Determination of crop coefficient for chufa crop (Cyperus esculentus L. var. sativus Boeck.) for sustainable irrigation scheduling. Sci. Total Environ. 2021, 768, 144975. [Google Scholar] [CrossRef]
- Scarabel, L.; Farinati, S.; Sattin, M. Occurrence of resistance to ALS inhibitors in European Cyperus esculentus L.: Characterisation and implications for management. Agronomy 2020, 10, 1133. [Google Scholar] [CrossRef]
- Ayeni, A.O. Hoop house and field evaluation of tigernut (Cyperus esculentus l. var. sativus boeck) selections in New Jersey, USA. Plants 2022, 11, 897. [Google Scholar] [CrossRef]
- Venturi, V.; Keel, C. Signaling in the rhizosphere. Trends Plant Sci. 2016, 21, 187–198. [Google Scholar] [CrossRef]
- Rüger, L.; Feng, K.; Dumack, K.; Freudenthal, J.; Chen, Y.; Sun, R.; Wilson, M.; Yu, P.; Sun, B.; Deng, Y. Assembly patterns of the rhizosphere microbiome along the longitudinal root axis of maize (Zea mays L.). Front. Microbiol. 2021, 12, 614501. [Google Scholar] [CrossRef]
- Qu, Q.; Wang, Z.; Gan, Q.; Liu, R.T.; Xu, H.W. Impact of drought on soil microbial biomass and extracellular enzyme activity. Front. Plant Sci. 2023, 14, 1221288. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.L.; Chen, Y.M.; Xu, W.J.; Liu, M. Effects of warming on the stoichiometry of soil microbial biomass and extracellular enzymes in an alpine shrubland. Geoderma 2023, 430, 116329. [Google Scholar] [CrossRef]
- Ke, W.S.; Li, C.X.; Zhu, F.; Luo, X.H.; Feng, J.P.; Li, X.; Jiang, Y.F.; Wu, C.; Hartley, W.; Xue, S.G. Effect of potentially toxic elements on soil multifunctionality at a lead smelting site. J. Hazard. Mater. 2023, 454, atr. [Google Scholar] [CrossRef] [PubMed]
- Sauvadet, M.; Saj, S.; Freschet, G.T.; Essobo, J.D.; Enock, S.; Becquer, T.; Tixier, P.; Harmand, J.M. Cocoa agroforest multifunctionality and soil fertility explained by shade tree litter traits. J. Appl. Ecol. 2020, 57, 476–487. [Google Scholar] [CrossRef]
- Byrnes, J.E.K.; Gamfeldt, L.; Isbell, F.; Lefcheck, J.S.; Griffin, J.N.; Hector, A.; Cardinale, B.J.; Hooper, D.U.; Dee, L.E.; Duffy, J.E. Investigating the relationship between biodiversity and ecosystem multifunctionality: Challenges and solutions. Methods Ecol. Evol. 2014, 5, 111–124. [Google Scholar] [CrossRef]
- Velmurugan, A.; Swarnam, T.P.; Jaisankar, I.; Swain, S.; Subramani, T. Vegetation-soil-microbial diversity influences ecosystem multifunctionality across different tropical coastal ecosystem types. Trop. Ecol. 2022, 63, 273–285. [Google Scholar] [CrossRef]
- Luo, J.; Liao, G.; Banerjee, S.; Gu, S.; Liang, J.; Guo, X.; Zhao, H.; Liang, Y.; Li, T. Long-term organic fertilization promotes the resilience of soil multifunctionality driven by bacterial communities. Soil Biol. 2023, 177, 108922. [Google Scholar] [CrossRef]
- Liu, Y.X.; Qin, Y.; Chen, T.; Lu, M.P.; Qian, X.B.; Guo, X.X.; Bai, Y. A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell 2021, 12, 315–330. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, 2584. [Google Scholar] [CrossRef]
- Edgar, R.C. UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv 2016. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Marsh, T.; Garrity, G.M.; et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009, 37, D141–D145. [Google Scholar] [CrossRef]
- Koljalg, U.; Larsson, K.H.; Abarenkov, K.; Nilsson, R.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Hoiland, K.; Kjoller, R.; Larsson, E.; et al. UNITE: A database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 2005, 166, 1063–1068. [Google Scholar] [CrossRef]
- Sun, Y.; Du, X.; Li, Y.; Han, X.; Fang, S.; Geisen, S.; Li, Q. Database and primer selections affect nematode community composition under different vegetations of Changbai Mountain. Soil Ecol. Lett. 2023, 5, 142–150. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Edgar, R. SINTAX: A simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv 2016. [Google Scholar] [CrossRef]
- Xu, S.D.; Liu, Y.X.; Cernava, T.; Wang, H.K.; Zhou, Y.Q.; Xia, T.; Cao, S.G.; Berg, G.; Shen, X.X.; Wen, Z.Y.; et al. Fusarium fruiting body microbiome member Pantoea agglomerans inhibits fungal pathogenesis by targeting lipid rafts. Nat. Microbiol. 2022, 7, 831–843. [Google Scholar] [CrossRef] [PubMed]
- Oren, A.; Garrity, G.M. Valid publication of the names of forty-two phyla of prokaryotes. J Int. J. Syst. Evol. Microbiol. 2021, 71, 005056. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.S.; Zou, Y.; Zhang, J.L.; Peres-Neto, P.R. Generalizing hierarchical and variation partitioning in multiple regression and canonical analyses using the rdacca.hp R package. Methods Ecol. Evol. 2022, 13, 782–788. [Google Scholar] [CrossRef]
- Dong, Y.; Gao, M.; Qiu, W.; Song, Z. Effect of microplastics and arsenic on nutrients and microorganisms in rice rhizosphere soil. Ecotoxicol. Environ. Saf. 2021, 211, 111899. [Google Scholar] [CrossRef] [PubMed]
- Preece, C.; Penuelas, J. A return to the wild: Root exudates and food security. Trends Plant Sci. 2020, 25, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, S.; Semenov, M.V.; Yao, F.; Ye, J.; Bu, R.; Ma, R.; Lin, J.; Kurganova, I.; Wang, X. Temperature sensitivity of SOM decomposition is linked with a K-selected microbial community. Glob. Change Biol. 2021, 27, 2763–2779. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Wilhelm, R.C.; Muñoz-Ucros, J.; Weikl, F.; Pritsch, K.; Goebel, M.; Buckley, D.H.; Bauerle, T.L. The effects of mixed-species root zones on the resistance of soil bacteria and fungi to long-term experimental and natural reductions in soil moisture. Sci. Total Environ. 2023, 873, 162266. [Google Scholar] [CrossRef]
- Li, C.; Li, X.; Min, K.; Liu, T.; Li, D.; Xu, J.; Zhao, Y.; Li, H.; Chen, H.; Hu, F. Copiotrophic taxa in pig manure mitigate nitrogen limitation of soil microbial communities. Chemosphere 2022, 301, 134812. [Google Scholar] [CrossRef]
- Duan, Y.; Wang, X.; Wang, L.; Lian, J.; Wang, W.; Wu, F.; Li, Y.; Li, Y. Biogeographic patterns of soil microbe communities in the deserts of the Hexi Corridor, northern China. Catena 2022, 211, 106026. [Google Scholar] [CrossRef]
- Horton, D.J.; Theis, K.R.; Uzarski, D.G.; Learman, D.R. Microbial community structure and microbial networks correspond to nutrient gradients within coastal wetlands of the Laurentian Great Lakes. FEMS Microbiol. Ecol. 2019, 95, fiz033. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Liu, G.; Zhang, C.; Wang, G. Bacterial richness is negatively related to potential soil multifunctionality in a degraded alpine meadow. Ecol. Indic. 2021, 121, 106996. [Google Scholar] [CrossRef]
- Li, J.; Delgado-Baquerizo, M.; Wang, J.-T.; Hu, H.-W.; Cai, Z.-J.; Zhu, Y.-N.; Singh, B.K. Fungal richness contributes to multifunctionality in boreal forest soil. Soil Biol. Biochem. 2019, 136, 107526. [Google Scholar] [CrossRef]
- Hu, W.; Ran, J.; Dong, L.; Du, Q.; Ji, M.; Yao, S.; Sun, Y.; Gong, C.; Hou, Q.; Gong, H. Aridity-driven shift in biodiversity–soil multifunctionality relationships. Nat. Commun. 2021, 12, 5350. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-F.; Chen, P.; Wang, F.-H.; Han, W.-X.; Qiao, M.; Dong, W.-X.; Hu, C.-S.; Zhu, D.; Chu, H.-Y.; Zhu, Y.-G. The ecological clusters of soil organisms drive the ecosystem multifunctionality under long-term fertilization. Environ. Int. 2022, 161, 107133. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Xu, X.; Zhang, F.; Si, H.; Li, L.; Mao, G. The Preliminary Research on Shifts in Maize Rhizosphere Soil Microbial Communities and Symbiotic Networks under Different Fertilizer Sources. Agronomy 2023, 13, 2111. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).