Deciphering Codon Usage Patterns in the Mitochondrial Genome of the Oryza Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
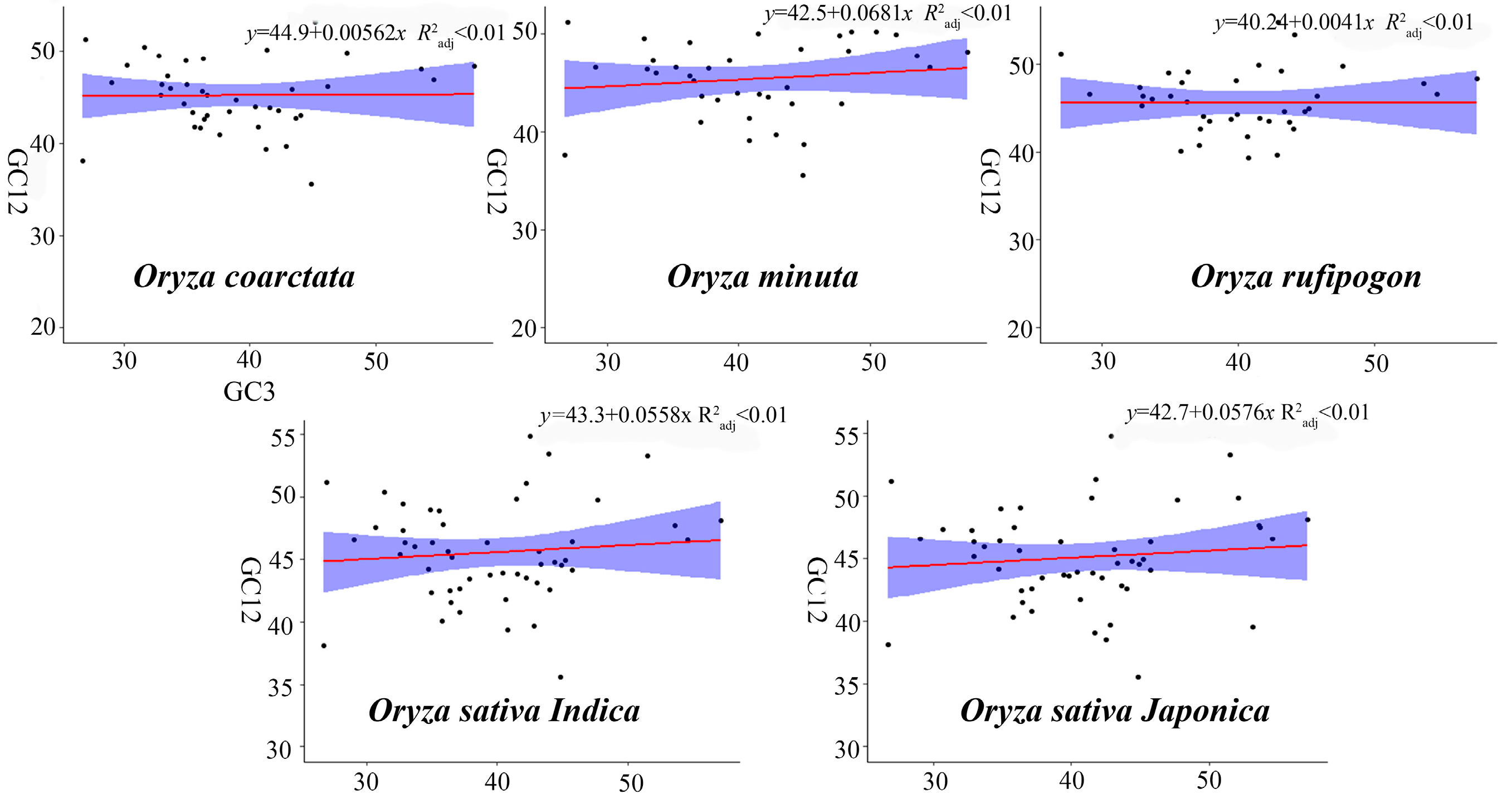
2.2. Analysis of Nucleotide Composition and Codon Bias Indices
2.3. Neutrality Plot and ENC Plot Analysis
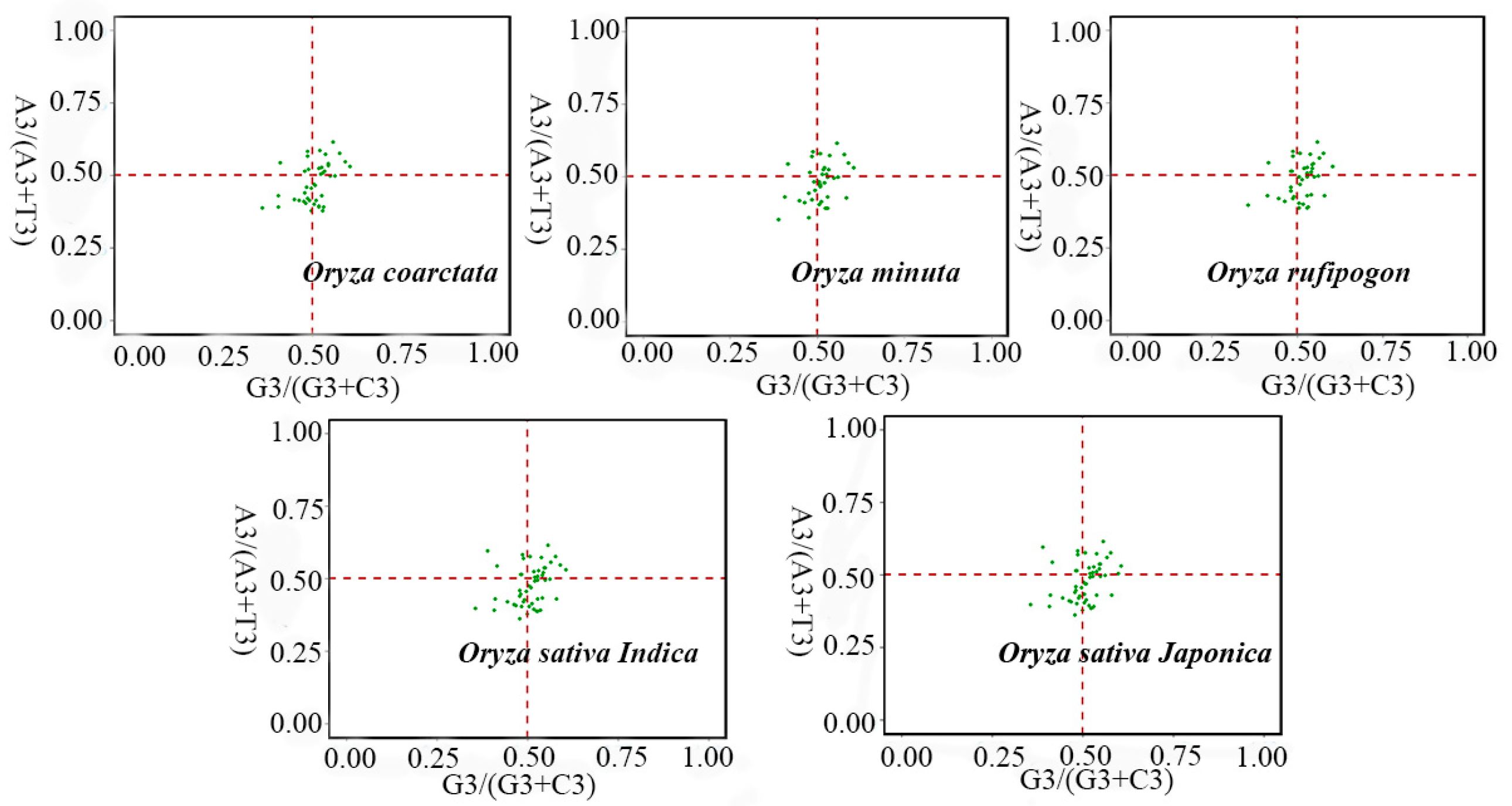
2.4. PR2-Bias Plot and Correlation Analysis
2.5. Phylogenetic Analysis
3. Results
3.1. Analysis of Codon Preference-Related Indices
3.2. Pattern of Codon Usage
3.3. Neutral Plot Analysis
3.4. PR2-Bias Plot Analysis
3.5. ENC Plot Analysis
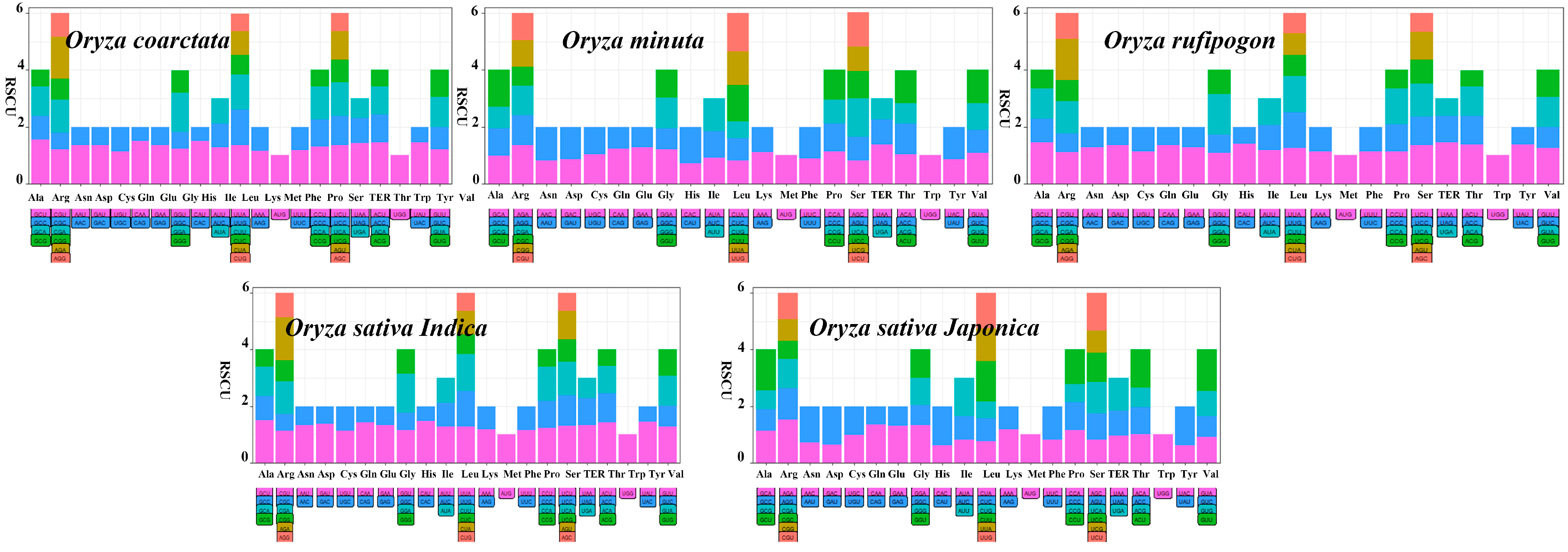
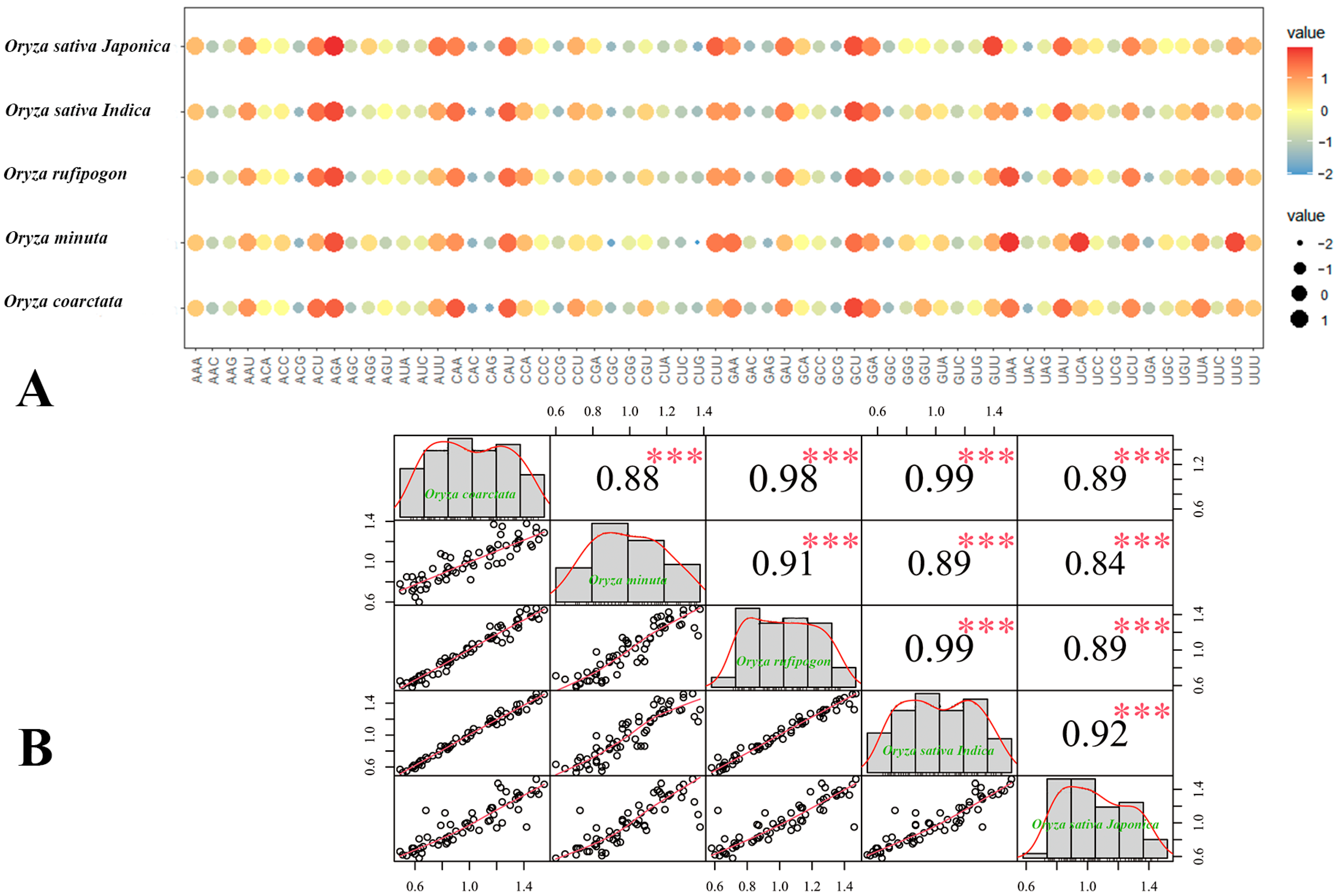
3.6. Application of RSCU
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Majeed, A.; Kaur, H.; Bhardwaj, P. Selection Constraints Determine Preference for a/u-Ending Codons in Taxus contorta. Genome 2020, 63, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Baeza, M.; Alcaíno, J.; Barahona, S.; Sepúlveda, D.; Cifuentes, V. Codon Usage and Codon Context Bias in Xanthophyllomyces dendrorhous. BMC Genom. 2015, 16, 293. [Google Scholar] [CrossRef] [PubMed]
- Dilucca, M.; Pavlopoulou, A.; Georgakilas, A.G.; Giansanti, A. Codon Usage Bias in Radioresistant Bacteria. Gene 2020, 742, 144554. [Google Scholar] [CrossRef] [PubMed]
- Vicario, S.; Moriyama, E.N.; Powell, J.R. Codon Usage in Twelve Species of Drosophila. BMC Evol. Biol. 2007, 7, 226. [Google Scholar] [CrossRef]
- Zhang, Y.; Shen, Z.; Meng, X.; Zhang, L.; Liu, Z.; Liu, M.; Zhang, F.; Zhao, J. Codon Usage Patterns across Seven Rosales Species. BMC Plant Biol. 2022, 22, 65. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.-H. The Codon Adaptation Index-a Measure of Directional Synonymous Codon Usage Bias, and Its Potential Applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef]
- Chakraborty, S.; Yengkhom, S.; Uddin, A. Analysis of Codon Usage Bias of Chloroplast Genes in Oryza Species: Codon Usage of Chloroplast Genes in Oryza Species. Planta 2020, 252, 67. [Google Scholar] [CrossRef]
- Saunders, R.; Deane, C.M. Synonymous Codon Usage Influences the Local Protein Structure Observed. Nucleic Acids Res. 2010, 38, 6719–6728. [Google Scholar] [CrossRef]
- Yang, J.; Ding, H.; Kan, X. Codon Usage Patterns and Evolution of HSP60 in Birds. Int. J. Biol. Macromol. 2021, 183, 1002–1012. [Google Scholar] [CrossRef]
- Geng, X.; Huang, N.; Zhu, Y.; Qin, L.; Hui, L. Codon Usage Bias Analysis of the Chloroplast Genome of Cassava. S. Afr. J. Bot. 2022, 151, 970–975. [Google Scholar] [CrossRef]
- Lewis, C.J.; Dixit, B.; Batiuk, E.; Hall, C.J.; O’Connor, M.S.; Boominathan, A. Codon Optimization Is an Essential Parameter for the Efficient Allotopic Expression of MtDNA Genes. Redox Biol. 2020, 30, 101429. [Google Scholar] [CrossRef] [PubMed]
- Dang, J.; Liu, Z.; Luo, X.; Jiang, Y.; Zhang, Z.; Abdullah, S.; Yusop, M.R. Codon Usage Characteristics and Evolutionary Analysis of Mitochondrial Genome of Winter Squash (Cucurbita maxima Duch.). J. Biobased Mater. Bioenergy 2024, 18, 428–443. [Google Scholar] [CrossRef]
- Liao, X.; Zhao, Y.; Kong, X.; Khan, A.; Zhou, B.; Liu, D.; Kashif, M.H.; Chen, P.; Wang, H.; Zhou, R. Complete Sequence of Kenaf (Hibiscus cannabinus) Mitochondrial Genome and Comparative Analysis with the Mitochondrial Genomes of Other Plants. Sci. Rep. 2018, 8, 12714. [Google Scholar] [CrossRef] [PubMed]
- Castellana, S.; Vicario, S.; Saccone, C. Evolutionary Patterns of the Mitochondrial Genome in Metazoa: Exploring the Role of Mutation and Selection in Mitochondrial Protein–Coding Genes. Genome Biol. Evol. 2011, 3, 1067–1079. [Google Scholar] [CrossRef]
- Yan, D.; Tang, Y.; Hu, M.; Liu, F.; Zhang, D.; Fan, J. The Mitochondrial Genome of Frankliniella intonsa: Insights into the Evolution of Mitochondrial Genomes at Lower Taxonomic Levels in Thysanoptera. Genomics 2014, 104, 306–312. [Google Scholar] [CrossRef]
- Wu, J.; Chen, R.; Wan, Y.; Wang, S.; Ma, J. The Complete Mitochondrial Genome of Monochamus dubius Gahan (Coleoptera: Cerambycidae). Mitochondrial DNA B Resour. 2021, 6, 699–700. [Google Scholar] [CrossRef]
- Ran, J.-H.; Shen, T.-T.; Liu, W.-J.; Wang, P.-P.; Wang, X.-Q. Mitochondrial Introgression and Complex Biogeographic History of the Genus Picea. Mol. Phylogenet. Evol. 2015, 93, 63–76. [Google Scholar] [CrossRef]
- Cui, X.; Xu, S.M.; Mu, D.S.; Yang, Z.M. Genomic Analysis of Rice MicroRNA Promoters and Clusters. Gene 2009, 431, 61–66. [Google Scholar] [CrossRef]
- Wambugu, P.W.; Brozynska, M.; Furtado, A.; Waters, D.L.; Henry, R.J. Relationships of Wild and Domesticated Rices (Oryza AA Genome Species) Based upon Whole Chloroplast Genome Sequences. Sci. Rep. 2015, 5, 13957. [Google Scholar] [CrossRef]
- McCouch, S.R.; Teytelman, L.; Xu, Y.; Lobos, K.B.; Clare, K.; Walton, M.; Fu, B.; Maghirang, R.; Li, Z.; Xing, Y. Development and Mapping of 2240 New SSR Markers for Rice (Oryza sativa L.). DNA Res. 2002, 9, 199–207. [Google Scholar] [CrossRef]
- Hu, H.; Dong, B.; Fan, X.; Wang, M.; Wang, T.; Liu, Q. Mutational Bias and Natural Selection Driving the Synonymous Codon Usage of Single-Exon Genes in Rice (Oryza sativa L.). Rice 2023, 16, 11. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Pan, Z.; Gao, S.; He, Y.; Xia, Q.; Jin, Y.; Yao, H. Analysis of Synonymous Codon Usage of Chloroplast Genome in Porphyra umbilicalis. Genes Genom. 2019, 41, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Gan, Z.; Zhang, F.; Yi, X.; Zhang, J.; Wan, X. Analysis of Codon Usage Patterns in Citrus Based on Coding Sequence Data. BMC Genom. 2020, 21, 234. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Lu, Y.; Song, Y.; Jing, L. Analysis of Codon Usage Bias of WRKY Transcription Factors in Helianthus annuus. BMC Genom. Data 2022, 23, 46. [Google Scholar] [CrossRef]
- Liu, H.; Lu, Y.; Lan, B.; Xu, J. Codon Usage by Chloroplast Gene Is Bias in Hemiptelea davidii. J. Genet. 2020, 99, 8. [Google Scholar] [CrossRef]
- Li, X.; Song, H.; Kuang, Y.; Chen, S.; Tian, P.; Li, C.; Nan, Z. Genome-Wide Analysis of Codon Usage Bias in Epichloë festucae. Int. J. Mol. Sci. 2016, 17, 1138. [Google Scholar] [CrossRef]
- Li, G.; Zhang, L.; Xue, P. Codon Usage Pattern and Genetic Diversity in Chloroplast Genomes of Panicum Species. Gene 2021, 802, 145866. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT Version 5: Improvement in Accuracy of Multiple Sequence Alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Lanfear, R. PartitionFinder v1.1.0 and PartitionFinderProtein v1.1.0. Tutoriais Manuais 2012, 29, 1695–1701. [Google Scholar]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian Inference of Phylogenetic Trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, Y.; Luo, L.; Wang, Y.-P.; Dong, Z.-M.; Sun, S.-H.; Qiu, L.-J. Analysis of Nuclear Gene Codon Bias on Soybean Genome and Transcriptome. Acta Agron. Sin. 2011, 37, 965–974. [Google Scholar] [CrossRef]
- Yang, X.; Qin, L.; Jiang, X. Analysis of Codon Usage Bias in the Genome of Trichoderma reesei. Sci. Technol. Food Ind. 2022, 43, 141–149. [Google Scholar] [CrossRef]
- Wu, D.; Xie, L.; Sun, Y.; Huang, Y.; Jia, L.; Dong, C.; Shen, E.; Ye, C.-Y.; Qian, Q.; Fan, L. A Syntelog-Based Pan-Genome Provides Insights into Rice Domestication and de-Domestication. Genome Biol. 2023, 24, 179. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-C.; Hickey, D.A. Rapid Divergence of Codon Usage Patterns within the Rice Genome. BMC Evol. Biol. 2007, 7, S6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Xu, W.; Lu, X.; Wang, L. Analysis of Codon Usage Bias of Chloroplast Genomes in Gynostemma Species. Physiol. Mol. Biol. Plants 2021, 27, 2727–2737. [Google Scholar] [CrossRef]
- Han, B.; Wang, C.; Tang, Z.; Ren, Y.; Li, Y.; Zhang, D.; Dong, Y.; Zhao, X. Genome-Wide Analysis of Microsatellite Markers Based on Sequenced Database in Chinese Spring Wheat (Triticum aestivum L.). PLoS ONE 2015, 10, e0141540. [Google Scholar] [CrossRef]
- Zervas, A.; Petersen, G.; Seberg, O. Mitochondrial Genome Evolution in Parasitic Plants. BMC Evol. Biol. 2019, 19, 87. [Google Scholar] [CrossRef]
- Guan, D.-L.; Qian, Z.-Q.; Ma, L.-B.; Bai, Y.; Xu, S.-Q. Different Mitogenomic Codon Usage Patterns between Damselflies and Dragonflies and Nine Complete Mitogenomes for Odonates. Sci. Rep. 2019, 9, 678. [Google Scholar] [CrossRef]
- Muyle, A.; Serres-Giardi, L.; Ressayre, A.; Escobar, J.; Glémin, S. GC-Biased Gene Conversion and Selection Affect GC Content in the Oryza Genus (Rice). Mol. Biol. Evol. 2011, 28, 2695–2706. [Google Scholar] [CrossRef]
- Patil, S.S.; Indrabalan, U.B.; Suresh, K.P.; Shome, B.R. Analysis of Codon Usage Bias of Classical Swine Fever Virus. Vet. World 2021, 14, 1450. [Google Scholar] [CrossRef]
- Hussain, S.; Rasool, S.T. Analysis of Synonymous Codon Usage in Zika Virus. Acta Trop. 2017, 173, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cai, Q.; Wang, Y.; Li, M.; Wang, C.; Wang, Z.; Jiao, C.; Xu, C.; Wang, H.; Zhang, Z. Comparative Analysis of Codon Bias in the Chloroplast Genomes of Theaceae Species. Front. Genet. 2022, 13, 824610. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Sun, M.H.; Jiang, Z.S.; Shu, H.R.; Zhang, S.Z. Genome-Wide Analysis of the Synonymous Codon Usage Patterns in Apple. J. Integr. Agric. 2016, 15, 983–991. [Google Scholar] [CrossRef]
- Barbhuiya, R.I.; Uddin, A.; Chakraborty, S. Codon Usage Pattern and Its Influencing Factors for Mitochondrial CO Genes among Different Classes of Arthropoda Codon Usage Pattern and Its Influencing Factors for Mitochondrial CO Genes. Mitochondrial DNA Part A 2020, 31, 313–326. [Google Scholar] [CrossRef]
- Chase, M.W.; Christenhusz, M.J.M.; Fay, M.F.; Byng, J.W.; Judd, W.S.; Soltis, D.E.; Mabberley, D.J.; Sennikov, A.N.; Soltis, P.S.; Stevens, P.F. An Update of the Angiosperm Phylogeny Group Classification for the Orders and Families of Flowering Plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S. A Comprehensive Evolutionary Classification of Proteins Encoded in Complete Eukaryotic Genomes. Genome Biol. 2004, 5, R7. [Google Scholar] [CrossRef]
- Gernandt, D.S.; López, G.G.; García, S.O.; Liston, A. Phylogeny and Classification of Pinus. Taxon 2005, 54, 29–42. [Google Scholar] [CrossRef]
- Wang, L.; Xing, H.; Yuan, Y.; Wang, X.; Saeed, M.; Tao, J.; Feng, W.; Zhang, G.; Song, X.; Sun, X. Genome-Wide Analysis of Codon Usage Bias in Four Sequenced Cotton Species. PLoS ONE 2018, 13, e0194372. [Google Scholar] [CrossRef]
- Niu, Y.; Luo, Y.; Wang, C.; Liao, W. Deciphering Codon Usage Patterns in Genome of Cucumis sativus in Comparison with Nine Species of Cucurbitaceae. Agronomy 2021, 11, 2289. [Google Scholar] [CrossRef]
- Newman, Z.R.; Young, J.M.; Ingolia, N.T.; Barton, G.M. Differences in Codon Bias and GC Content Contribute to the Balanced Expression of TLR7 and TLR9. Proc. Natl. Acad. Sci. USA 2016, 113, E1362–E1371. [Google Scholar] [CrossRef]

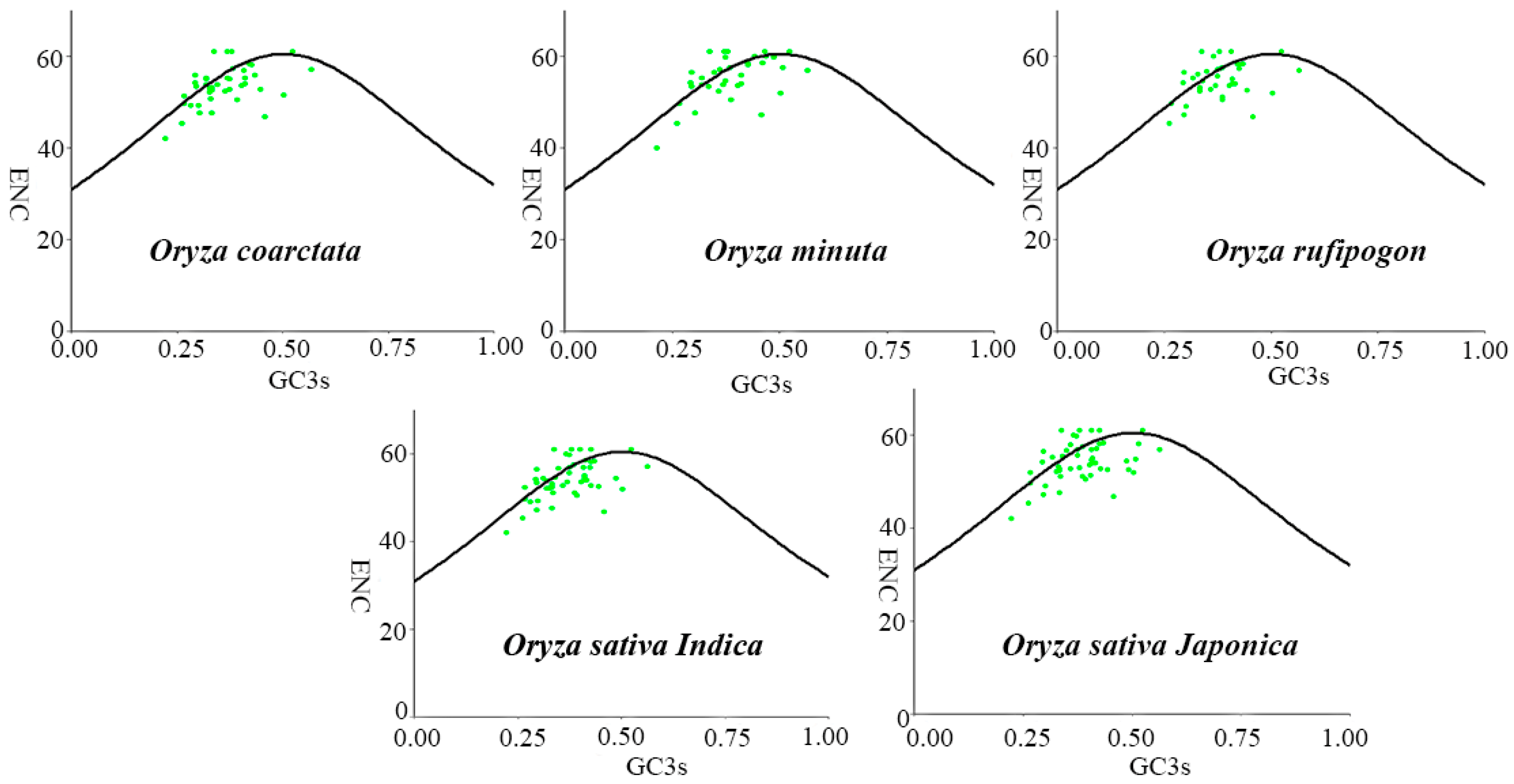
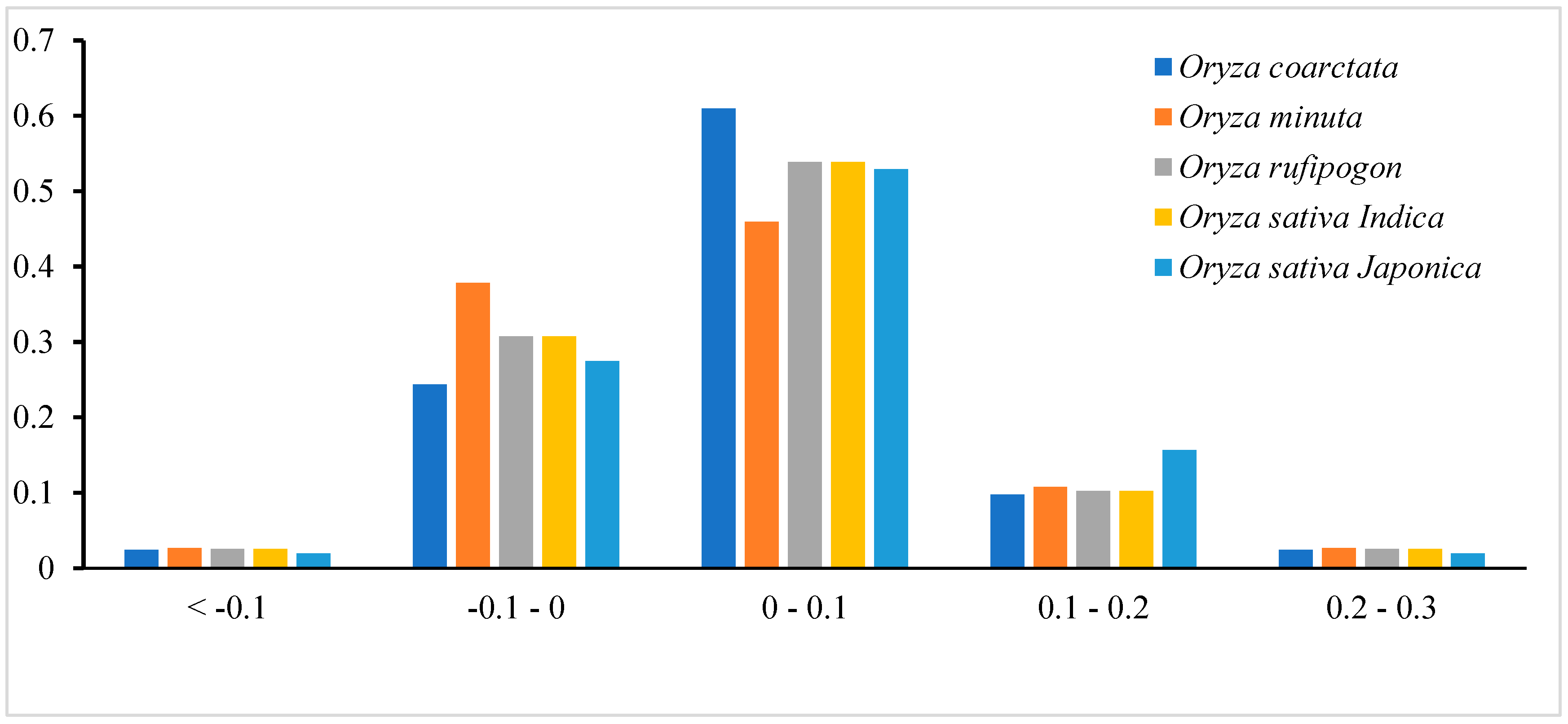

| Species | T3s | A3s | G3s | C3s | GC3s | GC | ENC |
|---|---|---|---|---|---|---|---|
| Oryza coarctata | 0.402 | 0.231 | 0.362 | 0.226 | 0.362 | 0.431 | 53.521 |
| Oryza minuta | 0.379 | 0.241 | 0.359 | 0.247 | 0.386 | 0.441 | 55.120 |
| Oryza rufipogon | 0.386 | 0.239 | 0.360 | 0.242 | 0.380 | 0.440 | 54.853 |
| Oryza sativa Indica | 0.394 | 0.234 | 0.361 | 0.233 | 0.370 | 0.437 | 54.111 |
| Oryza sativa Japonica | 0.387 | 0.242 | 0.353 | 0.242 | 0.383 | 0.439 | 54.283 |
| GC1 | GC2 | GC12 | |
|---|---|---|---|
| GC2 | 0.257 ** | ||
| GC12 | 0.805 ** | 0.780 ** | |
| GC3 | −0.064 | 0.185 ** | 0.072 |
| Species | RSCU > 1.0 | The Highest Frequency Codons | The Lowest Frequency Codons |
|---|---|---|---|
| Oryza coarctata | 30 | GCU (1.53) | CAG (0.49) |
| Oryza minuta | 29 | UAA (1.38) | CUG (0.60) |
| Oryza rufipogon | 31 | AGA (1.47) | ACG (0.58) |
| Oryza sativa Indica | 30 | AGA (1.52) | CAC (0.53) |
| Oryza sativa Japonica | 27 | AGA (1.52) | CUG (0.58) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Ma, Y.; Yu, H.; Han, Y.; Yu, T. Deciphering Codon Usage Patterns in the Mitochondrial Genome of the Oryza Species. Agronomy 2024, 14, 2722. https://doi.org/10.3390/agronomy14112722
Zhang Y, Ma Y, Yu H, Han Y, Yu T. Deciphering Codon Usage Patterns in the Mitochondrial Genome of the Oryza Species. Agronomy. 2024; 14(11):2722. https://doi.org/10.3390/agronomy14112722
Chicago/Turabian StyleZhang, Yuyang, Yunqi Ma, Huanxi Yu, Yu Han, and Tao Yu. 2024. "Deciphering Codon Usage Patterns in the Mitochondrial Genome of the Oryza Species" Agronomy 14, no. 11: 2722. https://doi.org/10.3390/agronomy14112722
APA StyleZhang, Y., Ma, Y., Yu, H., Han, Y., & Yu, T. (2024). Deciphering Codon Usage Patterns in the Mitochondrial Genome of the Oryza Species. Agronomy, 14(11), 2722. https://doi.org/10.3390/agronomy14112722





