Deep Learning-Enabled Dynamic Model for Nutrient Status Detection of Aquaponically Grown Plants
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Set-Up
2.2. Spectral Dataset Collection
2.3. Deep Autoencoder (DAE)
2.4. Genetic Algorithm (GA) and Sequential Forward Selection (SFS)-Based Feature Extraction
2.5. Long Short-Term Memory (LSTM)
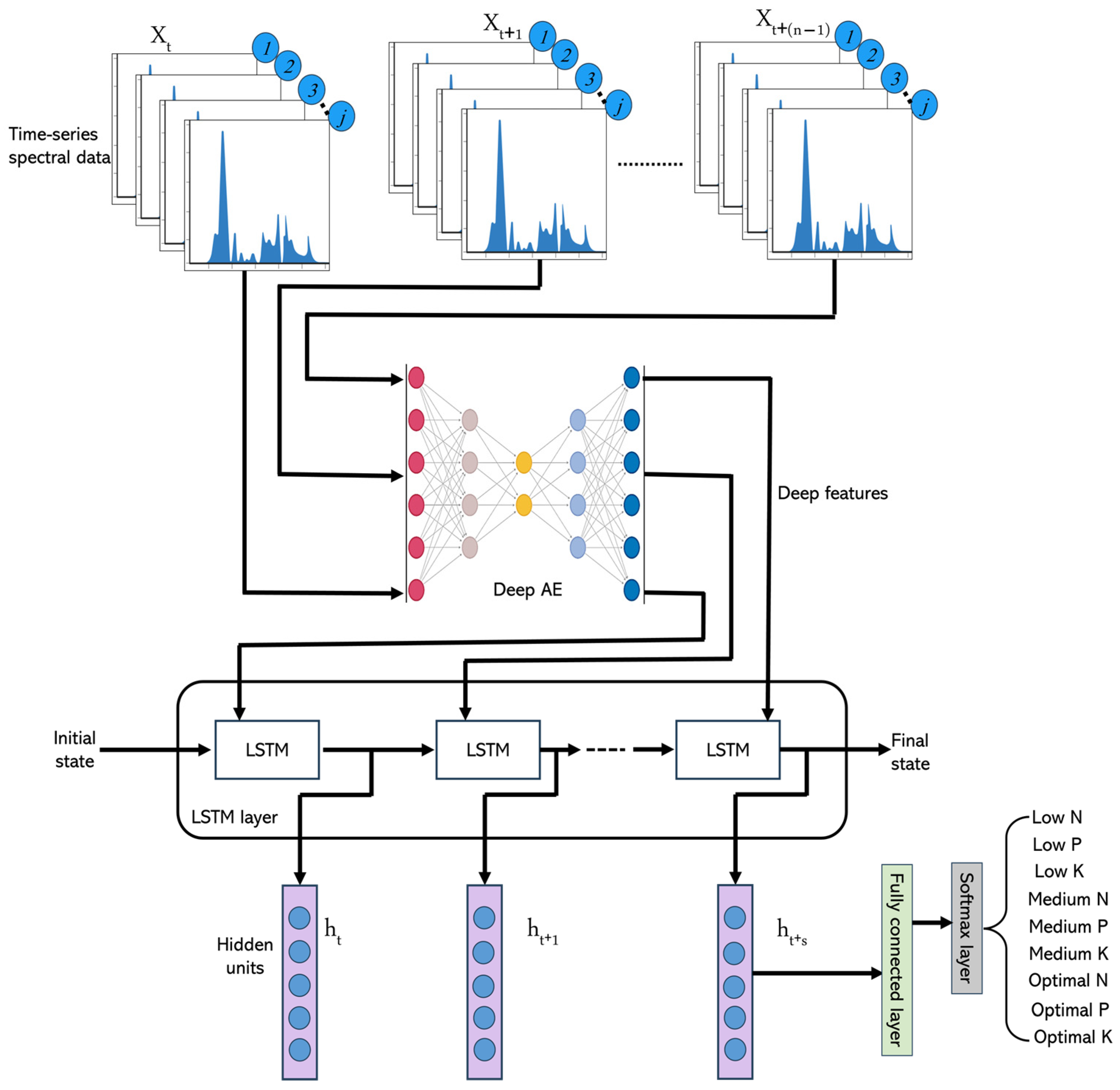
2.6. Deep Autoencoder–LSTM (DAE-LSTM)
2.7. Convolutional Neural Network (CNN)
2.8. Traditional Machine Learning
2.9. Performance Evaluation of the Proposed Framework
3. Results and Discussion
3.1. Evaluation of the Training Process
3.2. DAE vs. GA and SFS
3.3. LSTM vs. CNN and MCSVM
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eshkabilov, S.; Lee, A.; Sun, X.; Lee, C.W.; Simsek, H. Hyperspectral Imaging Techniques for Rapid Detection of Nutrient Content of Hydroponically Grown Lettuce Cultivars. Comput. Electron. Agric. 2021, 181, 105968. [Google Scholar] [CrossRef]
- Taha, M.F.; ElManawy, A.I.; Alshallash, K.S.; ElMasry, G.; Alharbi, K.; Zhou, L.; Liang, N.; Qiu, Z. Using Machine Learning for Nutrient Content Detection of Aquaponics-Grown Plants Based on Spectral Data. Sustainability 2022, 14, 12318. [Google Scholar] [CrossRef]
- Yang, T.; Kim, H.-J. Characterizing Nutrient Composition and Concentration in Tomato-, Basil-, and Lettuce-Based Aquaponic and Hydroponic Systems. Water 2020, 12, 1259. [Google Scholar] [CrossRef]
- Kovácsné Madar, Á.; Rubóczki, T.; Takácsné Hájos, M. Lettuce Production in Aquaponic and Hydroponic Systems. Acta Univ. Sapientiae Agric. Environ. 2019, 11, 51–59. [Google Scholar] [CrossRef]
- Sharaf-Eldin, M.A.; Elsayed, S.; Elmetwalli, A.H.; Yaseen, Z.M.; Moghanm, F.S.; Elbagory, M.; El-Nahrawy, S.; Omara, A.E.-D.; Tyler, A.N.; Elsherbiny, O. Using Optimized Three-Band Spectral Indices and a Machine Learning Model to Assess Squash Characteristics under Moisture and Potassium Deficiency Stress. Horticulturae 2023, 9, 79. [Google Scholar] [CrossRef]
- Elsayed, S.; El-Hendawy, S.; Dewir, Y.H.; Schmidhalter, U.; Ibrahim, H.H.; Ibrahim, M.M.; Elsherbiny, O.; Farouk, M. Estimating the Leaf Water Status and Grain Yield of Wheat under Different Irrigation Regimes Using Optimized Two- and Three-Band Hyperspectral Indices and Multivariate Regression Models. Water 2021, 13, 2666. [Google Scholar] [CrossRef]
- Galieni, A.; D’Ascenzo, N.; Stagnari, F.; Pagnani, G.; Xie, Q.; Pisante, M. Past and Future of Plant Stress Detection: An Overview From Remote Sensing to Positron Emission Tomography. Front. Plant Sci. 2021, 11, 609155. [Google Scholar] [CrossRef]
- Rustioni, L.; Cola, G.; VanderWeide, J.; Murad, P.; Failla, O.; Sabbatini, P. Utilization of a Freeze-Thaw Treatment to Enhance Phenolic Ripening and Tannin Oxidation of Grape Seeds in Red (Vitis vinifera L.) Cultivars. Food Chem. 2018, 259, 139–146. [Google Scholar] [CrossRef]
- Saleh, A.H.; Elsayed, S.; Gad, M.; Elmetwalli, A.H.; Elsherbiny, O.; Hussein, H.; Moghanm, F.S.; Qazaq, A.S.; Eid, E.M.; El-Kholy, A.S.; et al. Utilization of Pollution Indices, Hyperspectral Reflectance Indices, and Data-Driven Multivariate Modelling to Assess the Bottom Sediment Quality of Lake Qaroun, Egypt. Water 2022, 14, 890. [Google Scholar] [CrossRef]
- Galal, H.; Elsayed, S.; Elsherbiny, O.; Allam, A.; Farouk, M. Using RGB Imaging, Optimized Three-Band Spectral Indices, and a Decision Tree Model to Assess Orange Fruit Quality. Agriculture 2022, 12, 1558. [Google Scholar] [CrossRef]
- Memon, M.S.; Chen, S.; Niu, Y.; Zhou, W.; Elsherbiny, O.; Liang, R.; Du, Z.; Guo, X. Evaluating the Efficacy of Sentinel-2B and Landsat-8 for Estimating and Mapping Wheat Straw Cover in Rice–Wheat Fields. Agronomy 2023, 13, 2691. [Google Scholar] [CrossRef]
- Liang, N.; Sun, S.; Zhou, L.; Zhao, N.; Taha, M.F.; He, Y.; Qiu, Z. High-Throughput Instance Segmentation and Shape Restoration of Overlapping Vegetable Seeds Based on Sim2real Method. Measurement 2023, 207, 112414. [Google Scholar] [CrossRef]
- Liang, N.; Sun, S.; Yu, J.; Farag Taha, M.; He, Y.; Qiu, Z. Novel Segmentation Method and Measurement System for Various Grains with Complex Touching. Comput. Electron. Agric. 2022, 202, 107351. [Google Scholar] [CrossRef]
- Abdalla, A.; Cen, H.; Wan, L.; Mehmood, K.; He, Y. Nutrient Status Diagnosis of Infield Oilseed Rape via Deep Learning-Enabled Dynamic Model. IEEE Trans. Ind. Inf. 2021, 17, 4379–4389. [Google Scholar] [CrossRef]
- Wang, Y.; Li, T.; Chen, T.; Zhang, X.; Taha, M.F.; Yang, N.; Mao, H.; Shi, Q. Cucumber Downy Mildew Disease Prediction Using a CNN-LSTM Approach. Agriculture 2024, 14, 1155. [Google Scholar] [CrossRef]
- Yu, S.; Fan, J.; Lu, X.; Wen, W.; Shao, S.; Liang, D.; Yang, X.; Guo, X.; Zhao, C. Deep Learning Models Based on Hyperspectral Data and Time-Series Phenotypes for Predicting Quality Attributes in Lettuces under Water Stress. Comput. Electron. Agric. 2023, 211, 108034. [Google Scholar] [CrossRef]
- Zhu, Z.; Qi, G.; Lei, Y.; Jiang, D.; Mazur, N.; Liu, Y.; Wang, D.; Zhu, W. A Long Short-Term Memory Neural Network Based Simultaneous Quantitative Analysis of Multiple Tobacco Chemical Components by Near-Infrared Hyperspectroscopy Images. Chemosensors 2022, 10, 164. [Google Scholar] [CrossRef]
- Dandıl, E.; Karaca, S. Detection of Pseudo Brain Tumors via Stacked LSTM Neural Networks Using MR Spectroscopy Signals. Biocybern. Biomed. Eng. 2021, 41, 173–195. [Google Scholar] [CrossRef]
- Wang, P.; Guo, L.; Tian, Y.; Chen, J.; Huang, S.; Wang, C.; Bai, P.; Chen, D.; Zhu, W.; Yang, H.; et al. Discrimination of Blood Species Using Raman Spectroscopy Combined with a Recurrent Neural Network. OSA Contin. 2021, 4, 672. [Google Scholar] [CrossRef]
- Chen, C.; Zhu, W.; Steibel, J.; Siegford, J.; Han, J.; Norton, T. Classification of Drinking and Drinker-Playing in Pigs by a Video-Based Deep Learning Method. Biosyst. Eng. 2020, 196, 1–14. [Google Scholar] [CrossRef]
- Sheng, R.; Cheng, W.; Li, H.; Ali, S.; Akomeah Agyekum, A.; Chen, Q. Model Development for Soluble Solids and Lycopene Contents of Cherry Tomato at Different Temperatures Using Near-Infrared Spectroscopy. Postharvest Biol. Technol. 2019, 156, 110952. [Google Scholar] [CrossRef]
- Gadirov, H.; Tkachev, G.; Ertl, T.; Frey, S. Evaluation and Selection of Autoencoders for Expressive Dimensionality Reduction of Spatial Ensembles; Springer: Berlin/Heidelberg, Germany, 2021; pp. 222–234. [Google Scholar]
- Palm, H.W.; Knaus, U.; Appelbaum, S.; Strauch, S.M.; Kotzen, B. Coupled Aquaponics Systems. In Aquaponics Food Production Systems; Springer International Publishing: Cham, Switzerland, 2019; pp. 163–199. [Google Scholar]
- Ahmed, Z.F.R.; Alnuaimi, A.K.H.; Askri, A.; Tzortzakis, N. Evaluation of Lettuce (Lactuca sativa L.) Production under Hydroponic System: Nutrient Solution Derived from Fish Waste vs. Inorganic Nutrient Solution. Horticulturae 2021, 7, 292. [Google Scholar] [CrossRef]
- Pineda-Pineda, J.; Miranda-Velázquez, I.; Rodríguez-Pérez, J.E.; Ramírez-Arias, J.A.; Pérez-Gómez, E.A.; García-Antonio, I.N.; Morales-Parada, J.J. Nutrimental Balance in Aquaponic Lettuce Production. Acta Hortic. 2017, 1093–1100. [Google Scholar] [CrossRef]
- Goddek, S.; Delaide, B.; Mankasingh, U.; Ragnarsdottir, K.; Jijakli, H.; Thorarinsdottir, R. Challenges of Sustainable and Commercial Aquaponics. Sustainability 2015, 7, 4199–4224. [Google Scholar] [CrossRef]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the Dimensionality of Data with Neural Networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef]
- Han, J.; Tao, J.; Wang, C. FlowNet: A Deep Learning Framework for Clustering and Selection of Streamlines and Stream Surfaces. IEEE Trans. Vis. Comput. Graph. 2019, 26, 1732–1744. [Google Scholar] [CrossRef]
- Guo, X.; Liu, X.; Zhu, E.; Yin, J. Deep Clustering with Convolutional Autoencoders; Springer: Berlin/Heidelberg, Germany, 2017; pp. 373–382. [Google Scholar]
- Fang, H. Retrieving Leaf Area Index Using a Genetic Algorithm with a Canopy Radiative Transfer Model. Remote Sens. Environ. 2003, 85, 257–270. [Google Scholar] [CrossRef]
- Nagasubramanian, K.; Jones, S.; Sarkar, S.; Singh, A.K.; Singh, A.; Ganapathysubramanian, B. Hyperspectral Band Selection Using Genetic Algorithm and Support Vector Machines for Early Identification of Charcoal Rot Disease in Soybean Stems. Plant Methods 2018, 14, 86. [Google Scholar] [CrossRef]
- Zhang, W.; Li, X.; Zhao, L. Band Priority Index: A Feature Selection Framework for Hyperspectral Imagery. Remote Sens. 2018, 10, 1095. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Elsherbiny, O.; Elaraby, A.; Alahmadi, M.; Hamdan, M.; Gao, J. Rapid Grapevine Health Diagnosis Based on Digital Imaging and Deep Learning. Plants 2024, 13, 135. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Ding, Y.; Wang, X.; Lu, W.; Sun, Y.; Yu, H. Nondestructive Determination of Nitrogen, Phosphorus and Potassium Contents in Greenhouse Tomato Plants Based on Multispectral Three-Dimensional Imaging. Sensors 2019, 19, 5295. [Google Scholar] [CrossRef] [PubMed]
- Pacumbaba, R.O.; Beyl, C.A. Changes in Hyperspectral Reflectance Signatures of Lettuce Leaves in Response to Macronutrient Deficiencies. Adv. Space Res. 2011, 48, 32–42. [Google Scholar] [CrossRef]
- Herrmann, I.; Karnieli, A.; Bonfil, D.J.; Cohen, Y.; Alchanatis, V. SWIR-Based Spectral Indices for Assessing Nitrogen Content in Potato Fields. Int. J. Remote Sens. 2010, 31, 5127–5143. [Google Scholar] [CrossRef]
- Petkovski, A.; Shehu, V. Anomaly Detection on Univariate Sensing Time Series Data for Smart Aquaculture Using Deep Learning. SEEU Rev. 2023, 18, 1–16. [Google Scholar] [CrossRef]
- Wang, X.; Tian, S.; Yu, L.; Lv, X.; Zhang, Z. Rapid Screening of Hepatitis B Using Raman Spectroscopy and Long Short-Term Memory Neural Network. Lasers Med. Sci. 2020, 35, 1791–1799. [Google Scholar] [CrossRef]
- Wang, D.; Tian, F.; Yang, S.X.; Zhu, Z.; Jiang, D.; Cai, B. Improved Deep CNN with Parameter Initialization for Data Analysis of Near-Infrared Spectroscopy Sensors. Sensors 2020, 20, 874. [Google Scholar] [CrossRef]
- Lau, C.P.Y.; Ma, W.; Law, K.Y.; Lacambra, M.D.; Wong, K.C.; Lee, C.W.; Lee, O.K.; Dou, Q.; Kumta, S.M. Development of Deep Learning Algorithms to Discriminate Giant Cell Tumors of Bone from Adjacent Normal Tissues by Confocal Raman Spectroscopy. Analyst 2022, 147, 1425–1439. [Google Scholar] [CrossRef]
- Yu, X.; Lu, H.; Liu, Q. Deep-Learning-Based Regression Model and Hyperspectral Imaging for Rapid Detection of Nitrogen Concentration in Oilseed Rape (Brassica napus L.) Leaf. Chemom. Intell. Lab. Syst. 2018, 172, 188–193. [Google Scholar] [CrossRef]
- Zhou, X.; Sun, J.; Tian, Y.; Lu, B.; Hang, Y.; Chen, Q. Hyperspectral Technique Combined with Deep Learning Algorithm for Detection of Compound Heavy Metals in Lettuce. Food Chem. 2020, 321, 126503. [Google Scholar] [CrossRef]
- Liao, F.; Feng, X.; Li, Z.; Wang, D.; Xu, C.; Chu, G.; Ma, H.; Yao, Q.; Chen, S. A Hybrid CNN-LSTM Model for Diagnosing Rice Nutrient Levels at the Rice Panicle Initiation Stage. J. Integr. Agric. 2024, 23, 711–723. [Google Scholar] [CrossRef]
- Belward, A.S.; Skøien, J.O. Who Launched What, When and Why; Trends in Global Land-Cover Observation Capacity from Civilian Earth Observation Satellites. ISPRS J. Photogramm. Remote Sens. 2015, 103, 115–128. [Google Scholar] [CrossRef]

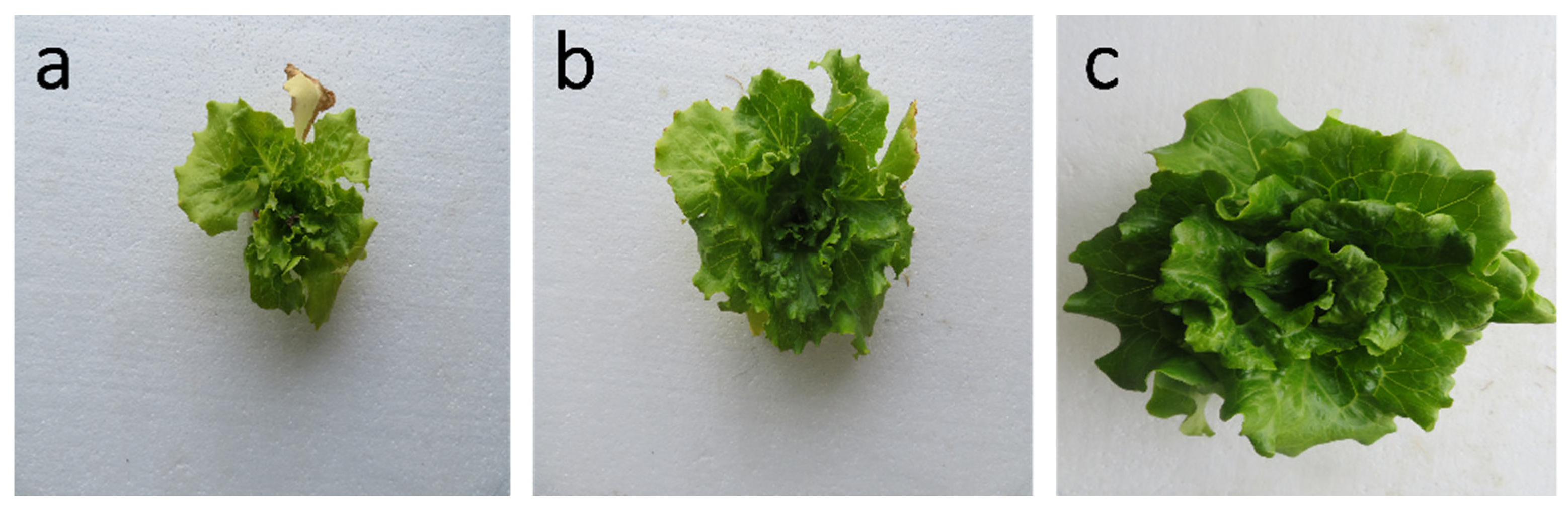
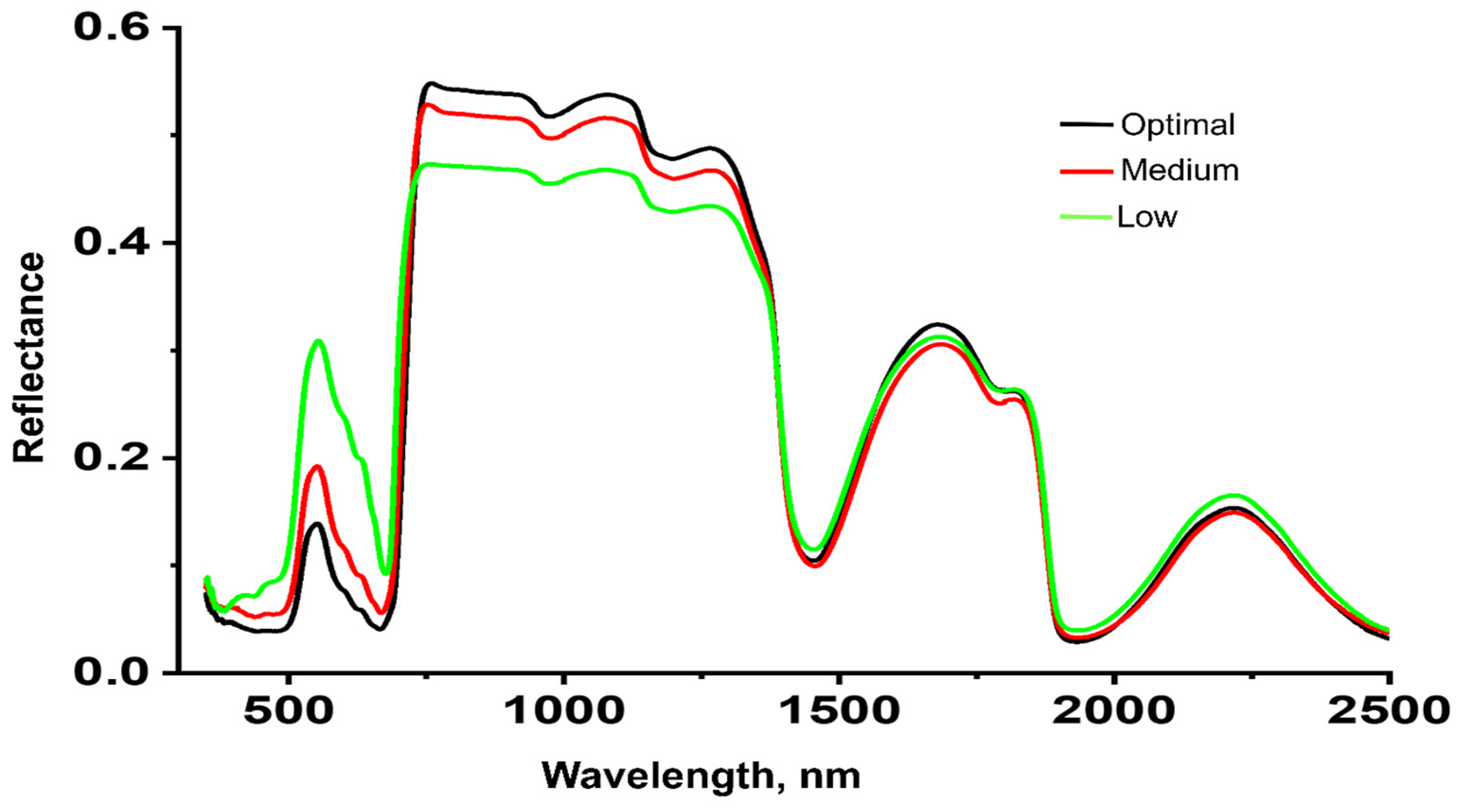





| Nutrient | Nutrient Concentration at Each Level, g/L | ||
|---|---|---|---|
| Low | Medium | Optimal | |
| N | 20 | 23 | 28 |
| P | 23 | 28 | 36 |
| K | 0.6 | 1.1 | 2.4 |
| No. | Acquisition Date | Number of Measurements | Total |
|---|---|---|---|
| 1 | 15 November 2023 | 900 | 8100 |
| 2 | 22 November 2023 | 900 | |
| 3 | 3 December 2023 | 900 | |
| 4 | 10 December 2023 | 900 | |
| 5 | 16 December 2023 | 900 | |
| 6 | 23 December 2023 | 900 | |
| 7 | 28 December 2023 | 900 | |
| 8 | 4 January 2024 | 900 | |
| 9 | 8 January 2024 | 900 |
| Layer | Type | Input Size | Layer | Type | Input Size |
|---|---|---|---|---|---|
| 0 | Input data | 1 × 120 × 1 | 8 | Droput | 1 × 30 × 64 |
| 1 | Conv2D | 1 × 120 × 1 | 9 | Conv2D | 1 × 30 × 64 |
| 2 | LeakyReLU | 1 × 120 × 32 | 10 | LeakyReLU | 1 × 30 × 128 |
| 3 | Maxpooling2D | 1 × 120 × 32 | 11 | Maxpooling2D | 1 × 30 × 128 |
| 4 | Droput | 1 × 60 × 32 | 12 | Droput | 1 × 15 × 128 |
| 5 | Conv2D | 1 × 60 × 32 | 13 | Dense | 1 × 15 × 128 |
| 6 | LeakyReLU | 1 × 60 × 64 | 14 | Flatten | 1 × 15 × 128 |
| 7 | Maxpooling2D | 1 × 60 × 64 | 15 | Dense | 1 × 15 × 128 |
| Model | Method | Classification Performance Metrics, % | |||
|---|---|---|---|---|---|
| Acc | Pr | Re | Fm | ||
| LSTM | DAE | 94.7 | 94.8 | 94.7 | 94.7 |
| GA | 73.4 | 74.1 | 73.4 | 72.1 | |
| SFS | 80.4 | 81.5 | 81.4 | 80.5 | |
| CNN | DAE | 83.3 | 83.2 | 83.3 | 83.3 |
| GA | 55.8 | 56.3 | 55.9 | 56.09 | |
| SFS | 78.9 | 80.9 | 78.9 | 78.9 | |
| MCSVM | GA | 72.2 | 72.1 | 72.2 | 72.2 |
| SFS | 70.7 | 70.3 | 70.7 | 70.7 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taha, M.F.; Mao, H.; Mousa, S.; Zhou, L.; Wang, Y.; Elmasry, G.; Al-Rejaie, S.; Elwakeel, A.E.; Wei, Y.; Qiu, Z. Deep Learning-Enabled Dynamic Model for Nutrient Status Detection of Aquaponically Grown Plants. Agronomy 2024, 14, 2290. https://doi.org/10.3390/agronomy14102290
Taha MF, Mao H, Mousa S, Zhou L, Wang Y, Elmasry G, Al-Rejaie S, Elwakeel AE, Wei Y, Qiu Z. Deep Learning-Enabled Dynamic Model for Nutrient Status Detection of Aquaponically Grown Plants. Agronomy. 2024; 14(10):2290. https://doi.org/10.3390/agronomy14102290
Chicago/Turabian StyleTaha, Mohamed Farag, Hanping Mao, Samar Mousa, Lei Zhou, Yafei Wang, Gamal Elmasry, Salim Al-Rejaie, Abdallah Elshawadfy Elwakeel, Yazhou Wei, and Zhengjun Qiu. 2024. "Deep Learning-Enabled Dynamic Model for Nutrient Status Detection of Aquaponically Grown Plants" Agronomy 14, no. 10: 2290. https://doi.org/10.3390/agronomy14102290
APA StyleTaha, M. F., Mao, H., Mousa, S., Zhou, L., Wang, Y., Elmasry, G., Al-Rejaie, S., Elwakeel, A. E., Wei, Y., & Qiu, Z. (2024). Deep Learning-Enabled Dynamic Model for Nutrient Status Detection of Aquaponically Grown Plants. Agronomy, 14(10), 2290. https://doi.org/10.3390/agronomy14102290













