Genome-Wide Identification and Expression Profiling of the BES1 Gene Family in Medicago sativa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Phylogenetic Analysis of MsBES1 Genes in M. sativa
2.2. Characteristics, Gene Structure, Conserved Motifs, and Cis-Elements of MsBES1s
2.3. Chromosomal Location, Subcellular Localization, and 3D Structure Prediction of MsBES1 Genes in M. sativa
2.4. Planting and Stress Treatment
2.5. Transcriptome Data Collection and Analysis
2.6. RNA Isolation and qRT-PCR Detection
2.7. RNA-Seq Analysis
3. Result
3.1. MsBES1 Gene Identification and Characterization
3.2. Phylogenetic Classification of MsBES1s in M. sativa
3.3. Conserved Motifs, Gene Structure, Cis-Regulatory Elements, and Three-Dimensional Structure Prediction of MsBES1s
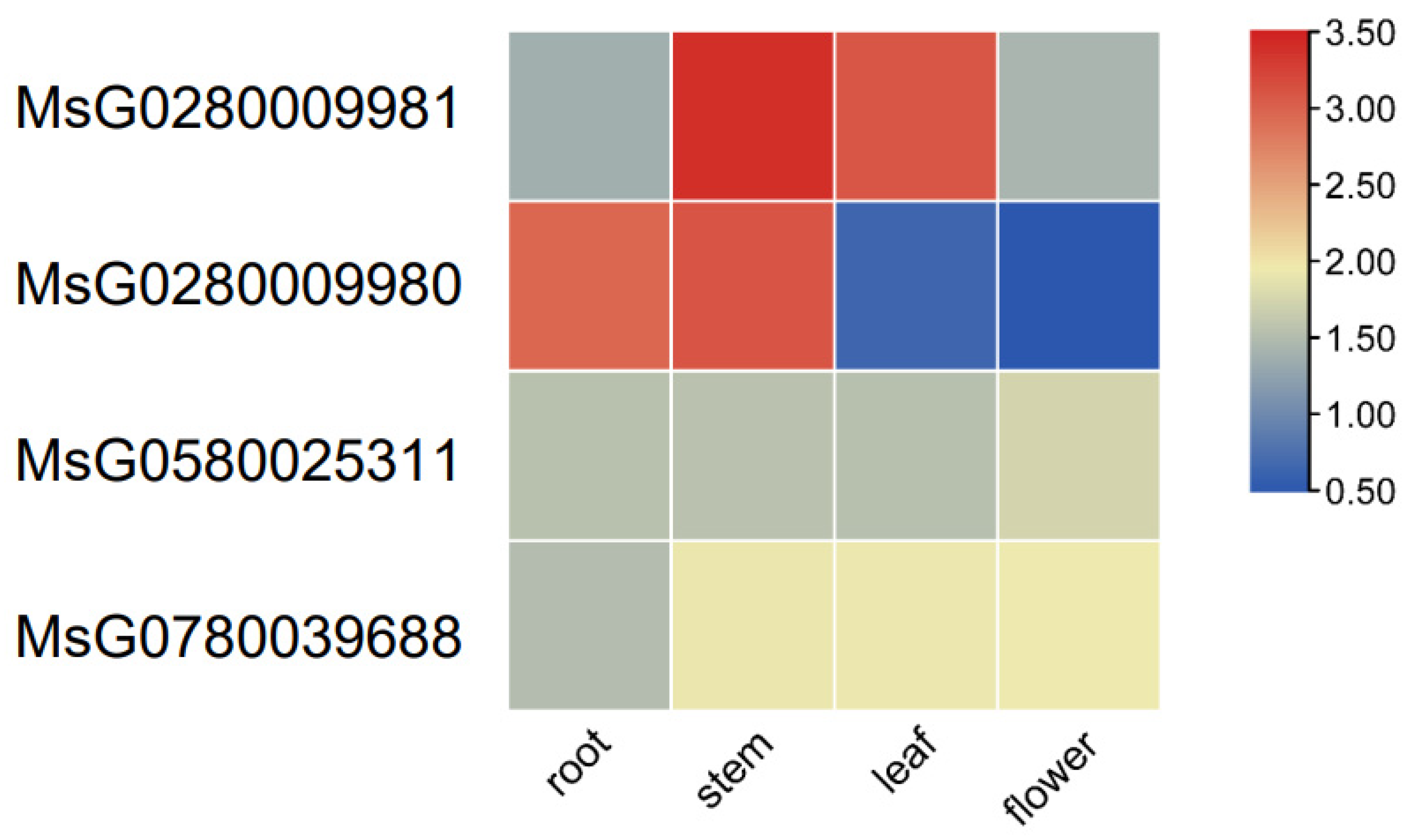
3.4. Transcriptome Analysis of MsBES1 Genes in Various Tissues
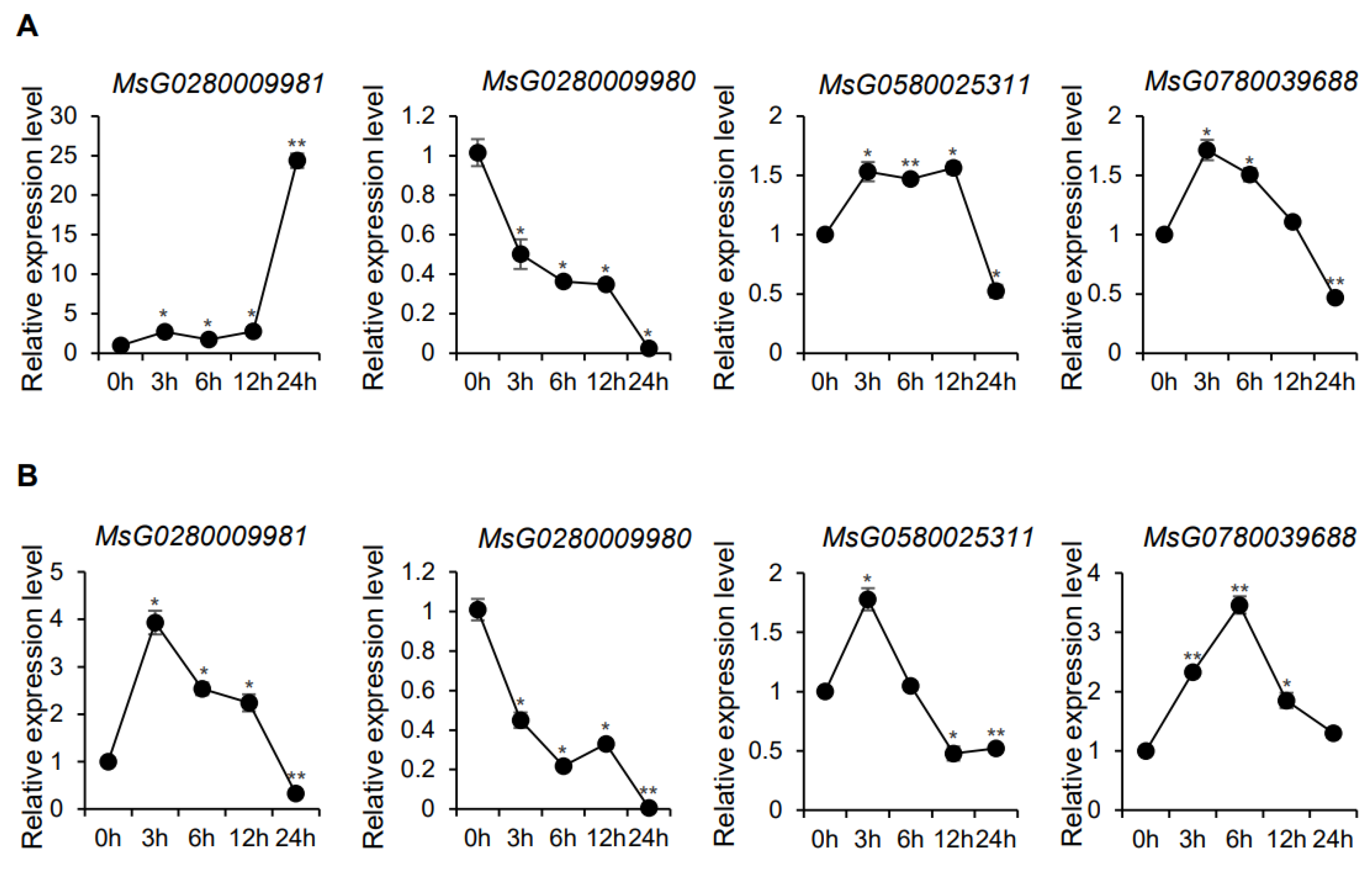
3.5. Expression of MsBES1 Genes under Different Abiotic Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wei, Z.Y.; Li, J. Brassinosteroids regulate root growth, development, and symbiosis. Mol. Plant 2016, 9, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Russinova, E. Brassinosteroid signalling. Curr. Biol. 2020, 30, R294–R298. [Google Scholar] [CrossRef] [PubMed]
- Oklestkova, J.; Rárová, L.; Kvasnica, M.; Strnad, M. Brassinosteroids: Synthesis and biological activities. Phytochem. Rev. 2015, 14, 1053–1072. [Google Scholar] [CrossRef]
- Saini, S.; Sharma, I.; Pati, P.K. Versatile roles of brassinosteroid in plants in the context of its homoeostasis, signaling and crosstalks. Front. Plant Sci. 2015, 6, 950. [Google Scholar] [CrossRef]
- Fridman, Y.; Savaldi-Goldstein, S. Brassinosteroids in growth control: How, when and where. Plant Sci. 2013, 209, 24–31. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef]
- Cao, X.; Wei, Y.; Shen, B.; Liu, L.; Mao, J. Interaction of the Transcription Factors BES1/BZR1 in Plant Growth and Stress Response. Int. J. Mol. Sci. 2024, 25, 6836. [Google Scholar] [CrossRef]
- Manghwar, H.; Hussain, A.; Ali, Q.; Liu, F. Brassinosteroids (BRs) Role in Plant Development and Coping with Different Stresses. Int. J. Mol. Sci. 2022, 23, 1012. [Google Scholar] [CrossRef]
- Qiao, S.; Sun, S.; Wang, L.; Wu, Z.; Li, C.; Li, X.; Wang, T.; Leng, L.; Tian, W.; Lu, T.; et al. The RLA1/SMOS1 Transcription Factor Functions with OsBZR1 to Regulate Brassinosteroid Signaling and Rice Architecture. Plant Cell 2017, 29, 292–309. [Google Scholar] [CrossRef]
- Ye, H.; Li, L.; Guo, H.; Yin, Y. MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in. Proc. Natl. Acad. Sci. USA 2012, 109, 20142–20147. [Google Scholar] [CrossRef]
- Gampala, S.S.; Kim, T.W.; He, J.X.; Tang, W.; Deng, Z.; Bai, M.Y.; Guan, S.; Lalonde, S.; Sun, Y.; Gendron, J.M.; et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 2007, 13, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Ma, X.; Li, C.; Hu, J.; Yang, Q.; Wang, T.; Wang, L.; Wang, J.; Guo, D.; Ge, W.; et al. Comprehensive analyses of the BES1 gene family in Brassica napus and examination of their evolutionary pattern in representative species. BMC Genom. 2018, 19, 346. [Google Scholar] [CrossRef]
- Ryu, H.; Cho, H.; Bae, W.; Hwang, I. Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of. Nat. Commun. 2014, 5, 4138. [Google Scholar] [CrossRef]
- Yu, X.; Li, L.; Zola, J.; Aluru, M.; Ye, H.; Foudree, A.; Guo, H.; Anderson, S.; Aluru, S.; Liu, P.; et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in. Plant J. 2011, 65, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.M.; Vukašinović, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional Regulators of Plant Growth, Development, and Stress Responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef]
- Lv, M.; Li, J. Molecular Mechanisms of Brassinosteroid-Mediated Responses to Changing Environments in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 2737. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.F.; Lu, J.; Yu, J.W.; Zhang, C.Q.; He, J.X.; Liu, Q.Q. The brassinosteroid-regulated transcription factors BZR1/BES1 function as a coordinator in multisignal-regulated plant growth. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 561–571. [Google Scholar] [CrossRef]
- González-García, M.P.; Vilarrasa-Blasi, J.; Zhiponova, M.; Divol, F.; Mora-García, S.; Russinova, E.; Caño-Delgado, A.I. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 2011, 138, 849–859. [Google Scholar] [CrossRef]
- Yang, R.; Liu, P.; Zhang, T.; Dong, H.; Jing, Y.; Yang, Z.; Tang, S.; Zhang, Y.; Lv, M.; Liu, J.; et al. Plant-specific BLISTER interacts with kinase BIN2 and Brassinazole Resistant1 during skotomorphogenesis. Plant Physiol. 2023, 193, 1580–1596. [Google Scholar] [CrossRef]
- Zhao, N.; Zhao, M.; Tian, Y.; Wang, Y.; Han, C.; Fan, M.; Guo, H.; Bai, M.Y. Interaction between BZR1 and EIN3 mediates signalling crosstalk between brassinosteroids and ethylene. New Phytol. 2021, 232, 2308–2323. [Google Scholar] [CrossRef]
- Wang, L.; Tian, Y.; Shi, W.; Yu, P.; Hu, Y.; Lv, J.; Fu, C.; Fan, M.; Bai, M.Y. The miR396-GRFs Module Mediates the Prevention of Photo-oxidative Damage by Brassinosteroids during Seedling De-Etiolation in Arabidopsis. Plant Cell 2020, 32, 2525–2542. [Google Scholar] [CrossRef]
- Ravindran, N.; Ramachandran, H.; Job, N.; Yadav, A.; Vaishak, K.P.; Datta, S. B-box protein BBX32 integrates light and brassinosteroid signals to inhibit cotyledon opening. Plant Physiol. 2021, 187, 446–461. [Google Scholar] [CrossRef]
- Ji, Y.; Qu, Y.; Jiang, Z.; Yan, J.; Chu, J.; Xu, M.; Su, X.; Yuan, H.; Wang, A. The mechanism for brassinosteroids suppressing climacteric fruit ripening. Plant Physiol. 2021, 185, 1875–1893. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, B.; Xu, Y.; Li, H.; Li, S.; Zhang, D.; Mao, Z.; Guo, S.; Yang, C.; Weng, Y.; et al. The cyclophilin CYP20-2 modulates the conformation of Brassinazole-Resistant1, which binds the promoter of Flowering Locus D to regulate flowering in Arabidopsis. Plant Cell 2013, 25, 2504–2521. [Google Scholar] [CrossRef]
- Tian, X.; He, M.; Mei, E.; Zhang, B.; Tang, J.; Xu, M.; Liu, J.; Li, X.; Wang, Z.; Tang, W.; et al. WRKY53 integrates classic brassinosteroid signaling and the mitogen-activated protein kinase pathway to regulate rice architecture and seed size. Plant Cell 2021, 33, 2753–2775. [Google Scholar] [CrossRef]
- Chen, J.; Nolan, T.M.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54, and WRKY70 Transcription Factors Are Involved in Brassinosteroid-Regulated Plant Growth and Drought Responses. Plant Cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef]
- Albertos, P.; Dündar, G.; Schenk, P.; Carrera, S.; Cavelius, P.; Sieberer, T.; Poppenberger, B. Transcription factor BES1 interacts with HSFA1 to promote heat stress resistance of plants. EMBO J. 2022, 41, e108664. [Google Scholar] [CrossRef]
- Li, H.; Ye, K.; Shi, Y.; Cheng, J.; Zhang, X.; Yang, S. BZR1 Positively Regulates Freezing Tolerance via CBF-Dependent and CBF-Independent Pathways in Arabidopsis. Mol. Plant 2017, 10, 545–559. [Google Scholar] [CrossRef]
- Cao, X.; Khaliq, A.; Lu, S.; Xie, M.; Ma, Z.; Mao, J.; Chen, B. Genome-wide identification and characterization of the BES1 gene family in apple (Malus domestica). Plant Biol. 2020, 22, 723–733. [Google Scholar] [CrossRef]
- Su, D.; Xiang, W.; Wen, L.; Lu, W.; Shi, Y.; Liu, Y.; Li, Z. Genome-wide identification, characterization and expression analysis of BES1 gene family in tomato. BMC Plant Biol. 2021, 21, 161. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Leng, F.; Ma, C.; Zhang, C.; Wang, S. Genome-wide identification, characterization and gene expression of BES1 transcription factor family in grapevine (Vitis vinifera L.). Sci. Rep. 2023, 13, 240. [Google Scholar] [CrossRef]
- Cheng, M.; Yuan, H.; Wang, R.; Wang, W.; Zhang, L.; Fan, F.; Li, S. Identification and characterization of BES1 genes involved in grain size development of Oryza sativa L. Int. J. Biol. Macromol. 2023, 253 Pt 6, 127327. [Google Scholar] [CrossRef]
- Yan, Q.; Wang, L.; Li, X. GmBEHL1, a BES1/BZR1 family protein, negatively regulates soybean nodulation. Sci. Rep. 2018, 8, 7614. [Google Scholar] [CrossRef]
- Park, C.R.; Nguyen, V.T.; Min, J.H.; Sang, H.; Lim, G.H.; Kim, C.S. Isolation and Functional Characterization of Soybean BES1/BZR1 Homolog 3-Like 1 (GmBEH3L1) Associated with Dehydration Sensitivity and Brassinosteroid Signaling in Arabidopsis thaliana. Plants 2022, 11, 2565. [Google Scholar] [CrossRef]
- Liao, K.; Peng, Y.J.; Yuan, L.B.; Dai, Y.S.; Chen, Q.F.; Yu, L.J.; Bai, M.Y.; Zhang, W.Q.; Xie, L.J.; Xiao, S. Brassinosteroids Antagonize Jasmonate-Activated Plant Defense Responses through BRI1-EMS-SUPPRESSOR1 (BES1). Plant Physiol. 2020, 182, 1066–1082. [Google Scholar] [CrossRef]
- Oh, E.; Zhu, J.Y.; Wang, Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012, 14, 802–809. [Google Scholar] [CrossRef]
- Jia, C.; Zhao, S.; Bao, T.; Zhao, P.; Peng, K.; Guo, Q.; Gao, X.; Qin, J. Tomato BZR/BES transcription factor SlBZR1 positively regulates BR signaling and salt stress tolerance in tomato and Arabidopsis thaliana. Plant Sci. 2021, 302, 110719. [Google Scholar] [CrossRef]
- Sun, F.; Ding, L.; Feng, W.; Cao, Y.; Lu, F.; Yang, Q.; Li, W.; Lu, Y.; Shabek, N.; Fu, F.; et al. Maize transcription factor ZmBES1/BZR1-5 positively regulates kernel size. J. Exp. Bot. 2021, 72, 1714–1726. [Google Scholar] [CrossRef]
- Mecchia, M.A.; García-Hourquet, M.; Lozano-Elena, F.; Planas-Riverola, A.; Blasco-Escamez, D.; Marquès-Bueno, M.; Mora-García, S.; Caño-Delgado, A.I. The BES1/BZR1-family transcription factor MpBES1 regulates cell division and differentiation in Marchantia polymorpha. Curr. Biol. 2021, 31, 4860–4869.e8. [Google Scholar] [CrossRef]
- Lorenzo, C.D.; García-Gagliardi, P.; Antonietti, M.S.; Sánchez-Lamas, M.; Mancini, E.; Dezar, C.A.; Vazquez, M.; Watson, G.; Yanovsky, M.J.; Cerdán, P.D. Improvement of alfalfa forage quality and management through the down-regulation of MsFTa1. Plant Biotechnol. J. 2020, 18, 944–954. [Google Scholar] [CrossRef]
- Wang, Q.; Hu, J.; Lou, T.; Li, Y.; Shi, Y.; Hu, H. Integrated agronomic, physiological, microstructure, and whole-transcriptome analyses reveal the role of biomass accumulation and quality formation during Se biofortification in alfalfa. Front. Plant Sci. 2023, 14, 1198847. [Google Scholar] [CrossRef]
- Shen, C.; Du, H.; Chen, Z.; Lu, H.; Zhu, F.; Chen, H.; Meng, X.; Liu, Q.; Liu, P.; Zheng, L.; et al. The Chromosome-Level Genome Sequence of the Autotetraploid Alfalfa and Resequencing of Core Germplasms Provide Genomic Resources for Alfalfa Research. Mol. Plant 2020, 13, 1250–1261. [Google Scholar] [CrossRef]
- Sun, S.; Ma, W.; Jia, Z.; Ou, C.; Li, M.; Mao, P. Genomic Identification and Expression Profiling of Lesion Simulating Disease Genes in Alfalfa (Medicago sativa) Elucidate Their Responsiveness to Seed Vigor. Antioxidants 2023, 12, 1768. [Google Scholar] [CrossRef]
- Yu, A.; Jiang, X.; Sun, Y.; Hu, Q.; Zhu, X.; Kang, J.; Chen, L.; Liu, L.; Hao, L.; Yang, Q.; et al. Genome-wide identification, characterization, and expression analysis of UDP-glycosyltransferase genes associated with secondary metabolism in alfalfa (Medicago sativa L.). Front Plant Sci. 2022, 13, 1001206. [Google Scholar] [CrossRef]
- Du, W.; Yang, J.; Li, Q.; He, C.; Pang, Y. Identification and Characterization of Abiotic Stress-Responsive NF-YB Family Genes in Medicago. Int. J. Mol. Sci. 2022, 23, 6906. [Google Scholar] [CrossRef]
- Wagh, K.; Stavreva, D.A.; Upadhyaya, A.; Hager, G.L. Transcription Factor Dynamics: One Molecule at a Time. Annu. Rev. Cell Dev. Biol. 2023, 39, 277–305. [Google Scholar] [CrossRef]
- Diao, P.; Chen, C.; Zhang, Y.; Meng, Q.; Lv, W.; Ma, N. The role of NAC transcription factor in plant cold response. Plant Signal Behav. 2020, 15, 1785668. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; López-Vidriero, I.; Carrasco, J.L.; Godoy, M.; Vera, P.; Solano, R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA 2014, 111, 2367–2372. [Google Scholar] [CrossRef]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple Functions of MYB Transcription Factors in Abiotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef]
- Edwards, H.E.; Gorelick, D.A. The evolution and structure/function of bHLH-PAS transcription factor family. Biochem. Soc. Trans. 2022, 50, 1227–1243. [Google Scholar] [CrossRef]
- Wang, H.; Chen, W.; Xu, Z.; Chen, M.; Yu, D. Functions of WRKYs in plant growth and development. Trends Plant Sci. 2023, 28, 630–645. [Google Scholar] [CrossRef]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Shi, H.; Li, X.; Lv, M.; Li, J. BES1/BZR1 Family Transcription Factors Regulate Plant Development via Brassinosteroid-Dependent and Independent Pathways. Int. J. Mol. Sci. 2022, 23, 10149. [Google Scholar] [CrossRef]
- Wu, Q.; Tong, C.; Chen, Z.; Huang, S.; Zhao, X.; Hong, H.; Li, J.; Feng, M.; Wang, H.; Xu, M.; et al. NLRs derepress MED10b-and MED7-mediated repression of jasmonate-dependent transcription to activate immunity. Proc. Natl. Acad. Sci. USA. 2023, 120, e2302226120. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, Z.; Wu, Q.; Cai, Y.; Zhang, Y.; Zhao, R.; Yan, J.; Qian, X.; Li, J.; Zhu, M.; et al. The Sw-5b NLR nucleotide-binding domain plays a role in oligomerization, and its self-association is important for activation of cell death signaling. J. Exp. Bot. 2021, 72, 6581–6595. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, K.; Xu, X.; Yang, Y.; Yan, S.; Wei, P.; Liu, H.; Xu, J.; Xiao, F.; Zhou, H.; et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018, 9, 4475. [Google Scholar] [CrossRef]
- Kaushal, M. Insights Into Microbially Induced Salt Tolerance and Endurance Mechanisms (STEM) in Plants. Front. Microbiol. 2020, 11, 1518. [Google Scholar] [CrossRef]
- Raza, A.; Mubarik, M.S.; Sharif, R.; Habib, M.; Jabeen, W.; Zhang, C.; Chen, H.; Chen, Z.H.; Siddique, K.H.M.; Zhuang, W.; et al. Developing drought-smart, ready-to-grow future crops. Plant Genome. 2023, 16, e20279. [Google Scholar] [CrossRef]
- Klemens, P.A.W.; Patzke, K.; Trentmann, O.; Poschet, G.; Büttner, M.; Schulz, A.; Marten, I.; Hedrich, R.; Neuhaus, H.E. Overexpression of a proton-coupled vacuolar glucose exporter impairs freezing tolerance and seed germination. New Phytol. 2014, 202, 188–197. [Google Scholar] [CrossRef]
- Yu, H.Q.; Feng, W.; Sun, F.; Zhang, Y.Y.; Qu, J.T.; Liu, B.; Lu, F.; Yang, L.; Fu, F.; Li, W. Cloning and characterization of BES1/BZR1 transcription factor genes in maize. Plant Growth Regul. 2018, 86, 235–249. [Google Scholar] [CrossRef]
- Tian, Y.; Zhao, N.; Wang, M.; Zhou, W.; Guo, J.; Han, C.; Zhou, C.; Wang, W.; Wu, S.; Tang, W.; et al. Integrated regulation of periclinal cell division by transcriptional module of BZR1-SHR in Arabidopsis thaliana roots. New Phytol. 2022, 233, 795–808. [Google Scholar] [CrossRef]
- Cui, Y.; Cao, Q.; Li, Y.; He, M.; Liu, X. Advances in cis-element- and natural variation-mediated transcriptional regulation and applications in gene editing of major crops. J. Exp. Bot. 2023, 74, 5441–5457. [Google Scholar] [CrossRef]
- Feng, W.; Liu, Y.; Cao, Y.; Zhao, Y.; Zhang, H.; Sun, F.; Yang, Q.; Li, W.; Lu, Y.; Zhang, X.; et al. Maize ZmBES1/BZR1-3 and -9 Transcription Factors Negatively Regulate Drought Tolerance in Transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 6025. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell. Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Wang, J.; Wu, D.; Wang, Y.; Xie, D. Jasmonate action in plant defense against insects. J. Exp. Bot. 2019, 70, 3391–3400. [Google Scholar] [CrossRef]
- Xu, J.; Lu, X.; Liu, Y.; Lan, W.; Wei, Z.; Yu, W.; Li, C. Interaction between ABA and NO in plants under abiotic stresses and its regulatory mechanisms. Front. Plant Sci. 2024, 15, 1330948. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, J.; Gao, Y.; Wang, X.; Wang, R.; Huang, H.; Zhang, Y.; Zhao, L.; Wang, P. Research on drought stress in Medicago sativa L. from 1998 to 2023: A bibliometric analysis. Front Plant Sci. 2024, 15, 1406256. [Google Scholar] [CrossRef]
- Ma, L.; Li, X.; Zhang, J.; Yi, D.; Li, F.; Wen, H.; Liu, W.; Wang, X. MsWRKY33 increases alfalfa (Medicago sativa L.) salt stress tolerance through altering the ROS scavenger via activating MsERF5 transcription. Plant Cell Environ. 2023, 46, 3887–3901. [Google Scholar] [CrossRef]


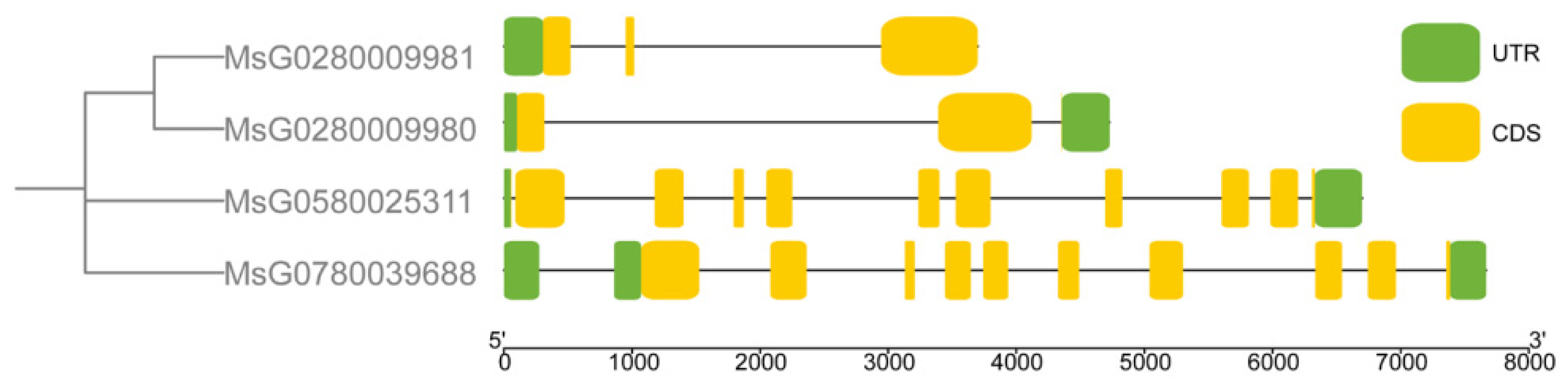
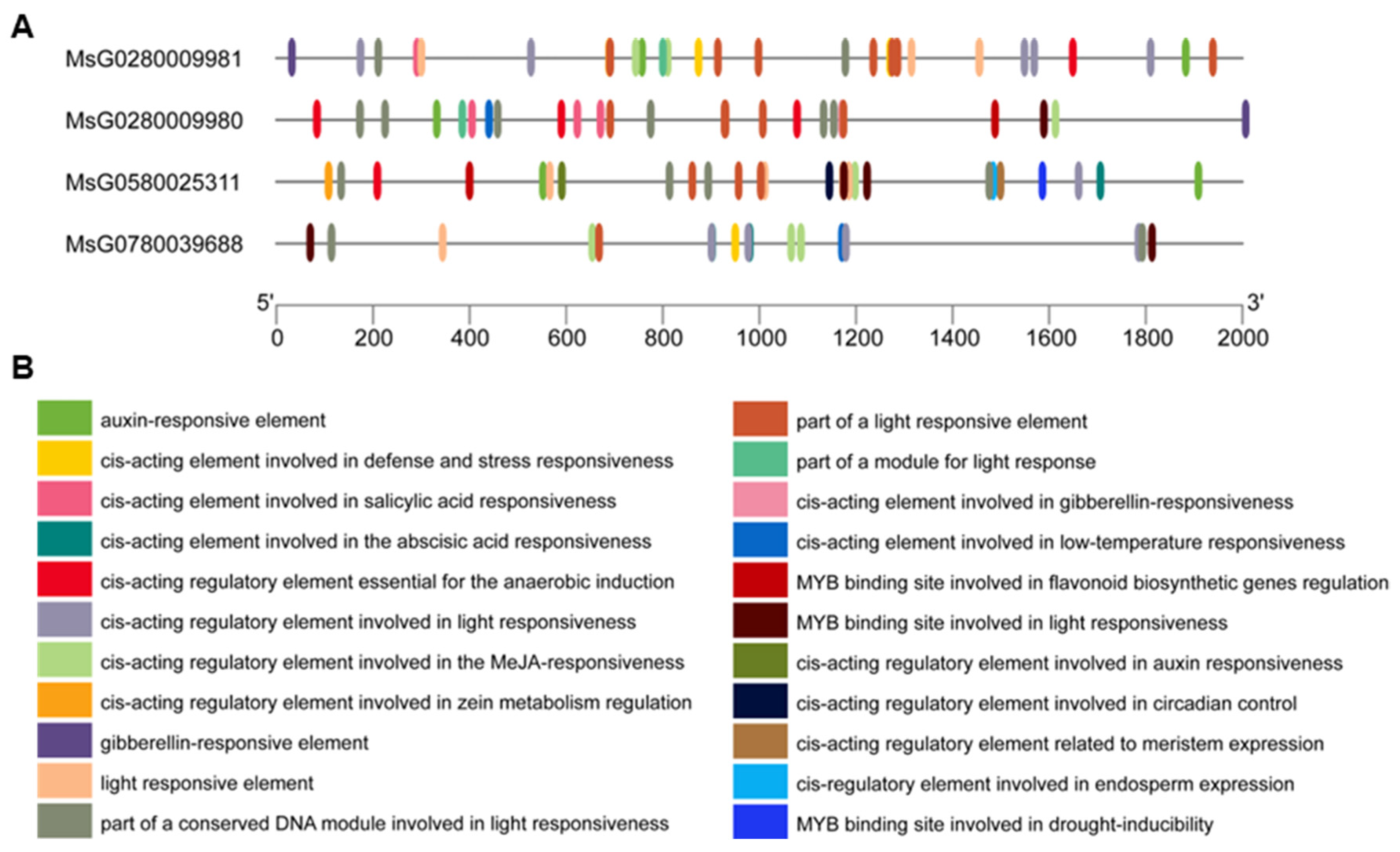
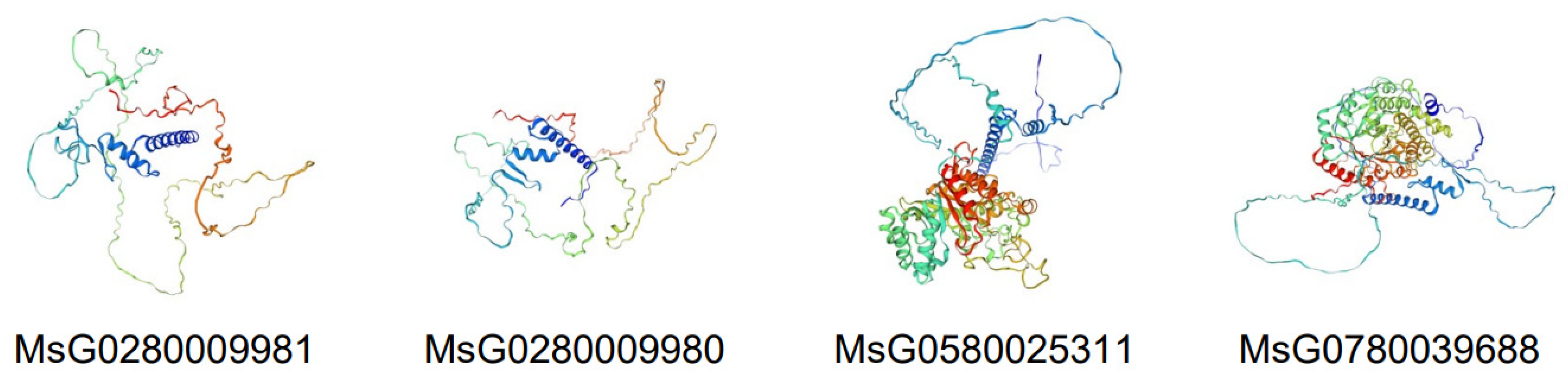
| Gene ID | CDS Length (bp) | AA | PI | Mw(kDa) | Locations on Chromosome | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| MsG0280009981 | 1038 | 345 | 8.59 | 37.316 | Chr2: 62,283,844–62,287,544 | −0.633 | Nucleus |
| MsG0280009980 | 951 | 316 | 8.65 | 34.141 | Chr2: 62,273,143–62,277,871 | −0.628 | Nucleus |
| MsG0580025311 | 1923 | 640 | 5.98 | 72.661 | Chr5: 16,822,237–16,828,937 | −0.424 | Nucleus |
| MsG0780039688 | 2100 | 699 | 5.31 | 78.120 | Chr7: 67,576,707–67,584,373 | −0.451 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Chen, F.; Jia, R.; Qin, Y.; Zhang, Y.; Lin, K. Genome-Wide Identification and Expression Profiling of the BES1 Gene Family in Medicago sativa. Agronomy 2024, 14, 2287. https://doi.org/10.3390/agronomy14102287
Chen Z, Chen F, Jia R, Qin Y, Zhang Y, Lin K. Genome-Wide Identification and Expression Profiling of the BES1 Gene Family in Medicago sativa. Agronomy. 2024; 14(10):2287. https://doi.org/10.3390/agronomy14102287
Chicago/Turabian StyleChen, Zhengqiang, Fangqi Chen, Ruifang Jia, Yaxuan Qin, Yuanyuan Zhang, and Kejian Lin. 2024. "Genome-Wide Identification and Expression Profiling of the BES1 Gene Family in Medicago sativa" Agronomy 14, no. 10: 2287. https://doi.org/10.3390/agronomy14102287
APA StyleChen, Z., Chen, F., Jia, R., Qin, Y., Zhang, Y., & Lin, K. (2024). Genome-Wide Identification and Expression Profiling of the BES1 Gene Family in Medicago sativa. Agronomy, 14(10), 2287. https://doi.org/10.3390/agronomy14102287





