Multivariate Adaptive Regression Splines Enhance Genomic Prediction of Non-Additive Traits
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Simulation
2.2. Evaluated Scenarios
2.3. Phenotype Simulation
2.4. Genomic Best Linear Unbiased Predictor (GBLUP)
2.5. Multivariate Adaptive Regression Splines (MARS)
2.6. Linkage Disequilibrium
2.7. Assessing Methods
3. Results
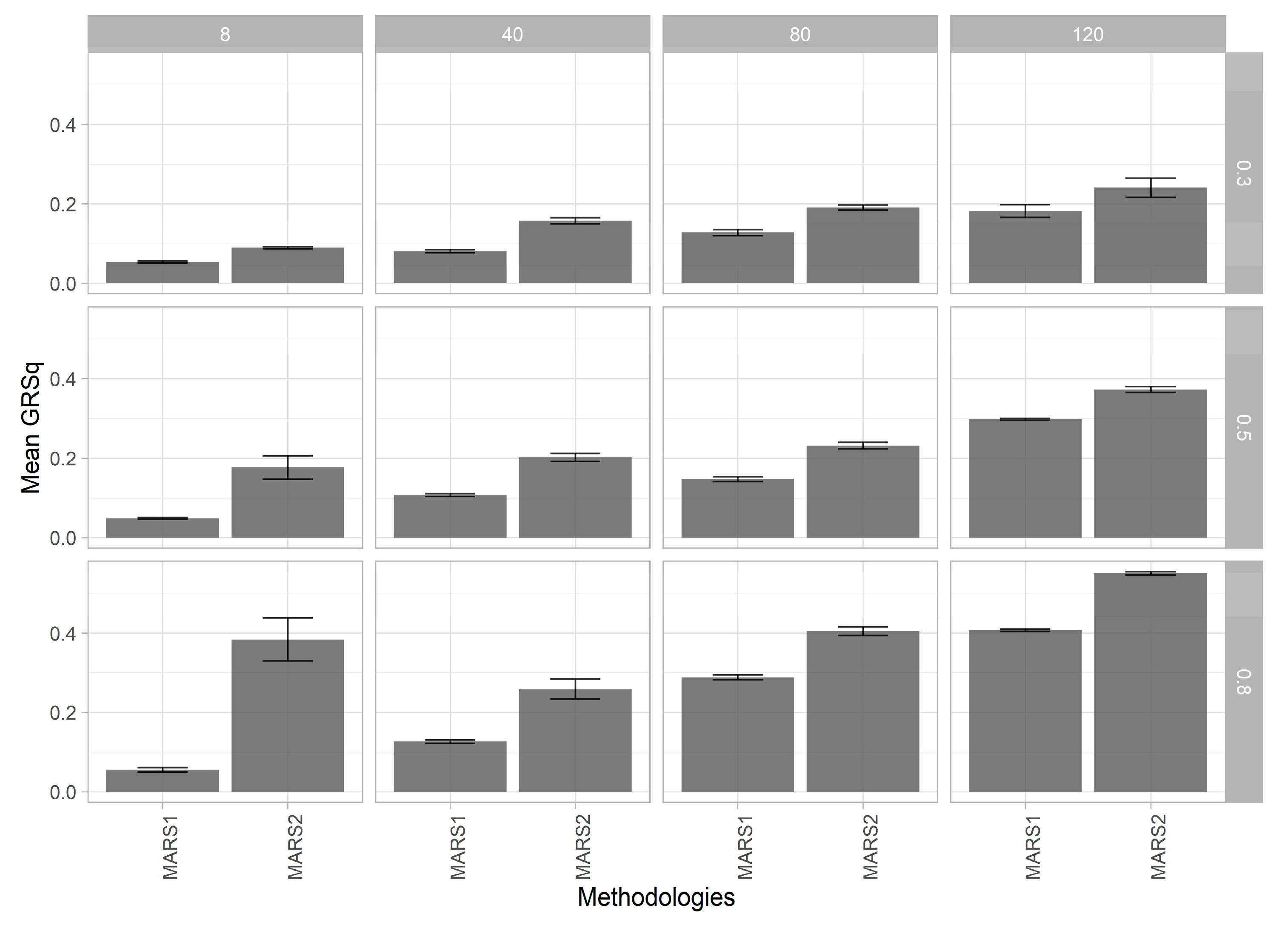
3.1. MARS Model Selection
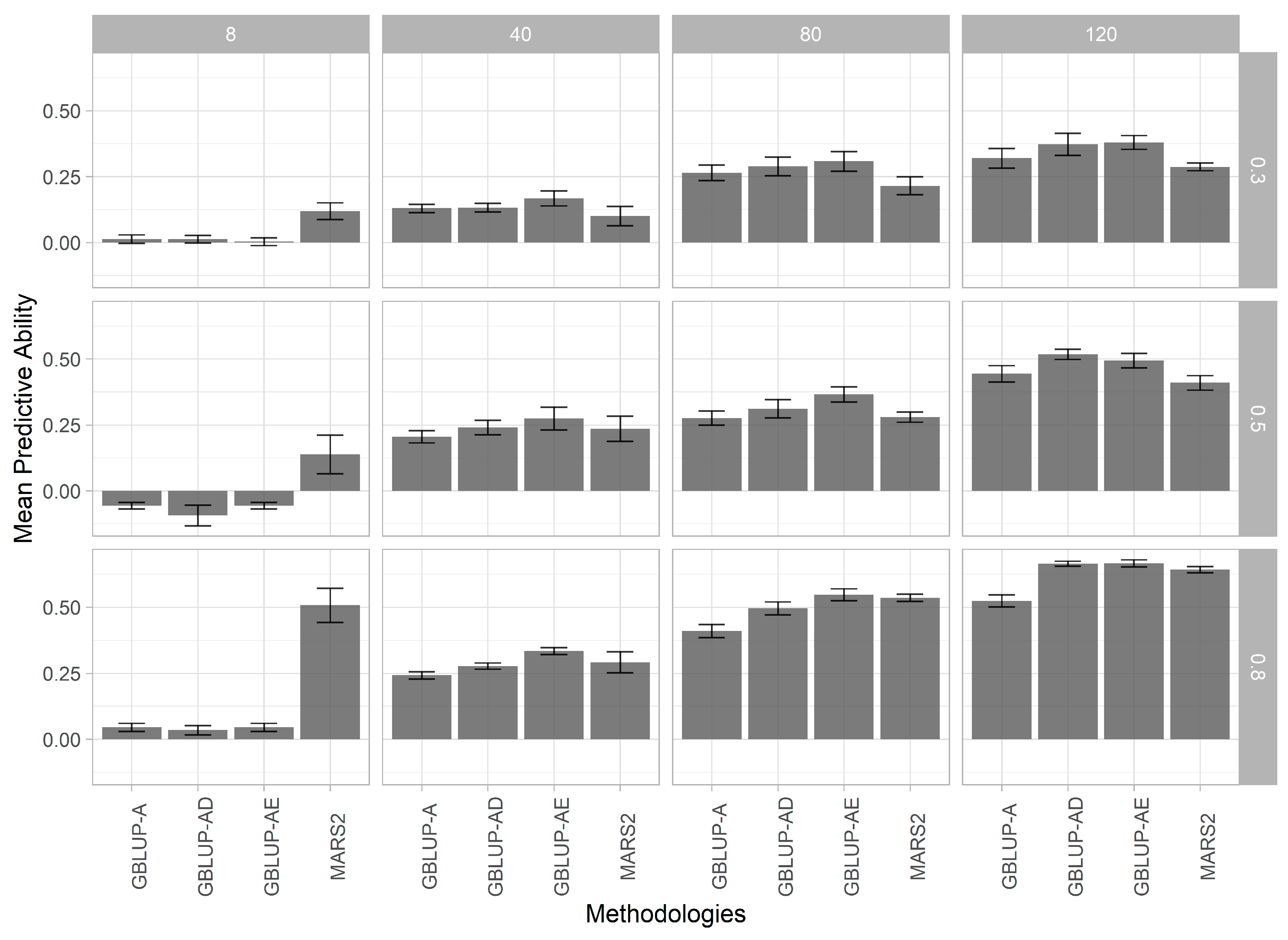
3.2. Genomic Prediction Models
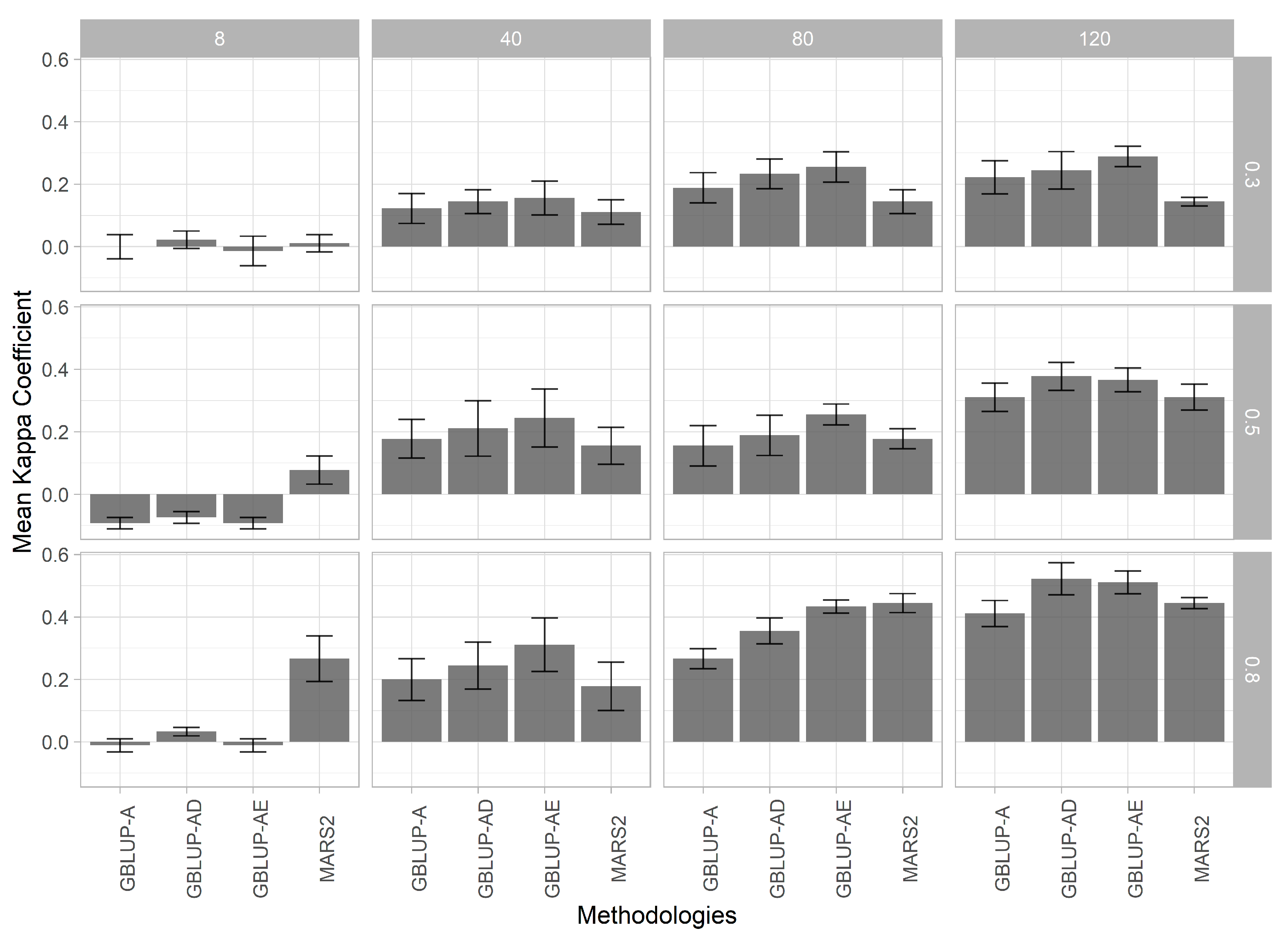
3.3. Cohen’s Kappa Coefficient of Agreement
3.4. Linkage Disequilibrium and Study of Trait Genetic Architecture
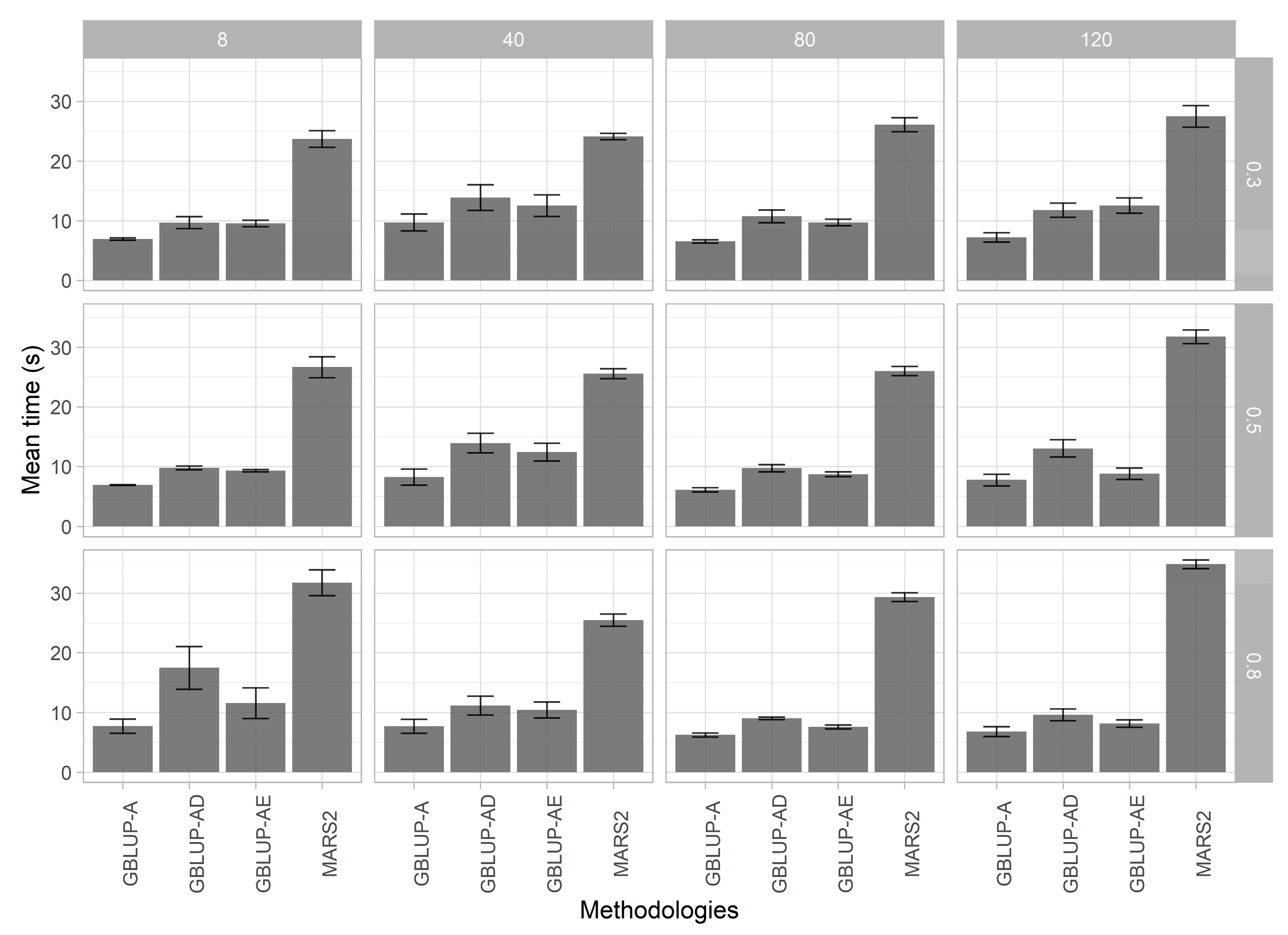
3.5. Computational Efficiency
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Mal, G.; Gautam, S.K.; Mukesh, M. Whole-Genome Selection in Livestock. In Advances in Animal Biotechnology; Singh, B., Mal, G., Gautam, S.K., Mukesh, M., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 349–364. ISBN 978-3-030-21309-1. [Google Scholar]
- Liu, X.; Wang, H.; Hu, X.; Li, K.; Liu, Z.; Wu, Y.; Huang, C. Improving Genomic Selection with Quantitative Trait Loci and Nonadditive Effects Revealed by Empirical Evidence in Maize. Front. Plant Sci. 2019, 10, 1129. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, C.O.; Wyatt, L.E.; Mazourek, M.R. Genomic Prediction and Selection for Fruit Traits in Winter Squash. G3 GenesGenomesGenetics 2020, 10, 3601–3610. [Google Scholar] [CrossRef]
- Varona, L.; Legarra, A.; Toro, M.A.; Vitezica, Z.G. Non-Additive Effects in Genomic Selection. Front. Genet. 2018, 9, 78. [Google Scholar] [CrossRef]
- Lebedev, V.G.; Lebedeva, T.N.; Chernodubov, A.I.; Shestibratov, K.A. Genomic Selection for Forest Tree Improvement: Methods, Achievements and Perspectives. Forests 2020, 11, 1190. [Google Scholar] [CrossRef]
- Martini, J.W.R.; Gao, N.; Cardoso, D.F.; Wimmer, V.; Erbe, M.; Cantet, R.J.C.; Simianer, H. Genomic Prediction with Epistasis Models: On the Marker-Coding-Dependent Performance of the Extended GBLUP and Properties of the Categorical Epistasis Model (CE). BMC Bioinform. 2017, 18, 3. [Google Scholar] [CrossRef]
- Calleja-Rodriguez, A.; Chen, Z.; Suontama, M.; Pan, J.; Wu, H.X. Genomic Predictions with Nonadditive Effects Improved Estimates of Additive Effects and Predictions of Total Genetic Values in Pinus Sylvestris. Front. Plant Sci. 2021, 12, 666820. [Google Scholar] [CrossRef] [PubMed]
- González-Camacho, J.M.; Ornella, L.; Pérez-Rodríguez, P.; Gianola, D.; Dreisigacker, S.; Crossa, J. Applications of Machine Learning Methods to Genomic Selection in Breeding Wheat for Rust Resistance. Plant Genome 2018, 11, 170104. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Abid, M.A.; Rasheed, A.; Crossa, J.; Hearne, S.; Li, H. DNNGP, a Deep Neural Network-Based Method for Genomic Prediction Using Multi-Omics Data in Plants. Mol. Plant 2023, 16, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Coelho de Sousa, I.; Nascimento, M.; de Castro Sant’anna, I.; Caixeta, E.T.; Azevedo, C.F.; Cruz, C.D.; da Silva, F.L.; Alkimim, E.R.; Nascimento, A.C.C.; Serão, N.V.L. Marker Effects and Heritability Estimates Using Additive-Dominance Genomic Architectures via Artificial Neural Networks in Coffea Canephora. PLoS ONE 2022, 17, e0262055. [Google Scholar] [CrossRef]
- Montesinos-López, O.A.; Sivakumar, A.; Huerta Prado, G.I.; Salinas-Ruiz, J.; Agbona, A.; Ortiz Reyes, A.E.; Alnowibet, K.; Ortiz, R.; Montesinos-López, A.; Crossa, J. Exploring Data Augmentation Algorithm to Improve Genomic Prediction of Top-Ranking Cultivars. Algorithms 2024, 17, 260. [Google Scholar] [CrossRef]
- Feng, W.; Gao, P.; Wang, X. AI Breeder: Genomic Predictions for Crop Breeding. New Crops 2024, 1, 100010. [Google Scholar] [CrossRef]
- Wang, X.; Shi, S.; Wang, G.; Luo, W.; Wei, X.; Qiu, A.; Luo, F.; Ding, X. Using Machine Learning to Improve the Accuracy of Genomic Prediction of Reproduction Traits in Pigs. J. Anim. Sci. Biotechnol. 2022, 13, 60. [Google Scholar] [CrossRef] [PubMed]
- Azodi, C.B.; Bolger, E.; McCarren, A.; Roantree, M.; de los Campos, G.; Shiu, S.-H. Benchmarking Parametric and Machine Learning Models for Genomic Prediction of Complex Traits. G3 GenesGenomesGenetics 2019, 9, 3691–3702. [Google Scholar] [CrossRef]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; de los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y.; et al. Genomic Selection in Plant Breeding: Methods, Models, and Perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef] [PubMed]
- Montesinos López, O.A.; Montesinos López, A.; Crossa, J. Multivariate Statistical Machine Learning Methods for Genomic Prediction; Springer International Publishing: Cham, Switzerland, 2022; ISBN 978-3-030-89009-4. [Google Scholar]
- Zingaretti, L.M.; Gezan, S.A.; Ferrão, L.F.V.; Osorio, L.F.; Monfort, A.; Muñoz, P.R.; Whitaker, V.M.; Pérez-Enciso, M. Exploring Deep Learning for Complex Trait Genomic Prediction in Polyploid Outcrossing Species. Front. Plant Sci. 2020, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.H. Multivariate Adaptive Regression Splines. Ann. Stat. 1991, 19, 1–67. [Google Scholar] [CrossRef]
- Adnan, R.M.; Liang, Z.; Heddam, S.; Zounemat-Kermani, M.; Kisi, O.; Li, B. Least Square Support Vector Machine and Multivariate Adaptive Regression Splines for Streamflow Prediction in Mountainous Basin Using Hydro-Meteorological Data as Inputs. J. Hydrol. 2020, 586, 124371. [Google Scholar] [CrossRef]
- Cruz, C.D. Genes Software—Extended and Integrated with the R, Matlab and Selegen. Acta Sci. Agron. 2016, 38, 547–552. [Google Scholar] [CrossRef]
- Zhang, H.; Yin, L.; Wang, M.; Yuan, X.; Liu, X. Factors Affecting the Accuracy of Genomic Selection for Agricultural Economic Traits in Maize, Cattle, and Pig Populations. Front. Genet. 2019, 10, 189. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, C.F.; Nascimento, M.; Fontes, V.C.; Silva, F.F.E.; de Resende, M.D.V.; Cruz, C.D. GenomicLand: Software for Genome-Wide Association Studies and Genomic Prediction. Acta Sci. Agron. 2019, 41, e45361. [Google Scholar] [CrossRef]
- Huang, H.; Ji, X.; Xia, F.; Huang, S.; Shang, X.; Chen, H.; Zhang, M.; Dahlgren, R.A.; Mei, K. Multivariate Adaptive Regression Splines for Estimating Riverine Constituent Concentrations. Hydrol. Process. 2020, 34, 1213–1227. [Google Scholar] [CrossRef]
- Abdulelah Al-Sudani, Z.; Salih, S.Q.; Sharafati, A.; Yaseen, Z.M. Development of Multivariate Adaptive Regression Spline Integrated with Differential Evolution Model for Streamflow Simulation. J. Hydrol. 2019, 573, 1–12. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning; Springer Series in Statistics; Springer: New York, NY, USA, 2009; ISBN 978-0-387-84857-0. [Google Scholar]
- Milborrow, S.; Hastei, T.; Tibshirani, R.; Miller, A.; Lumley, T. Earth: Multivariate Adaptive Regression Splines. R Package Version 5.1.1. 2019. Available online: https://CRAN.R-project.org/package=earth (accessed on 11 March 2023).
- Park, J.; Kim, J. Defining Heatwave Thresholds Using an Inductive Machine Learning Approach. PLoS ONE 2018, 13, e0206872. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Covarrubias-Pazaran, G. Genome-Assisted Prediction of Quantitative Traits Using the R Package Sommer. PLoS ONE 2016, 11, e0156744. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, W.S.; Devlin, S.J. Locally Weighted Regression: An Approach to Regression Analysis by Local Fitting. J. Am. Stat. Assoc. 1988, 83, 596–610. [Google Scholar] [CrossRef]
- Vos, P.G.; Paulo, M.J.; Voorrips, R.E.; Visser, R.G.F.; van Eck, H.J.; van Eeuwijk, F.A. Evaluation of LD Decay and Various LD-Decay Estimators in Simulated and SNP-Array Data of Tetraploid Potato. Theor. Appl. Genet. 2017, 130, 123–135. [Google Scholar] [CrossRef]
- Otyama, P.I.; Wilkey, A.; Kulkarni, R.; Assefa, T.; Chu, Y.; Clevenger, J.; O’Connor, D.J.; Wright, G.C.; Dezern, S.W.; MacDonald, G.E.; et al. Evaluation of Linkage Disequilibrium, Population Structure, and Genetic Diversity in the U.S. Peanut Mini Core Collection. BMC Genom. 2019, 20, 481. [Google Scholar] [CrossRef]
- Jannink, J.-L.; Lorenz, A.J.; Iwata, H. Genomic Selection in Plant Breeding: From Theory to Practice. Brief. Funct. Genom. 2010, 9, 166–177. [Google Scholar] [CrossRef]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- McHugh, M.L. Interrater Reliability: The Kappa Statistic. Biochem. Medica 2012, 22, 276–282. [Google Scholar] [CrossRef]
- Toro, M.A.; Varona, L. A Note on Mate Allocation for Dominance Handling in Genomic Selection. Genet. Sel. Evol. 2010, 42, 33. [Google Scholar] [CrossRef]
- Henderson, C.R. Best Linear Unbiased Prediction of Nonadditive Genetic Merits in Noninbred Populations. J. Anim. Sci. 1985, 60, 111–117. [Google Scholar] [CrossRef]
- Mackay, T.F. Q&A: Genetic Analysis of Quantitative Traits. J. Biol. 2009, 8, 23. [Google Scholar] [CrossRef]
- Barbosa, I.D.P.; da Silva, M.J.; da Costa, W.G.; de Castro Sant’Anna, I.; Nascimento, M.; Cruz, C.D. Genome-Enabled Prediction through Machine Learning Methods Considering Different Levels of Trait Complexity. Crop Sci. 2021, 61, 1890–1902. [Google Scholar] [CrossRef]
- Li, D.H.W.; Chen, W.; Li, S.; Lou, S. Estimation of Hourly Global Solar Radiation Using Multivariate Adaptive Regression Spline (MARS)—A Case Study of Hong Kong. Energy 2019, 186, 115857. [Google Scholar] [CrossRef]
- Phillips, P.C. Epistasis—The Essential Role of Gene Interactions in the Structure and Evolution of Genetic Systems. Nat. Rev. Genet. 2008, 9, 855–867. [Google Scholar] [CrossRef]
- Holland, J.B. Genetic Architecture of Complex Traits in Plants. Curr. Opin. Plant Biol. 2007, 10, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Hayes, B.J.; Pryce, J.; Chamberlain, A.J.; Bowman, P.J.; Goddard, M.E. Genetic Architecture of Complex Traits and Accuracy of Genomic Prediction: Coat Colour, Milk-Fat Percentage, and Type in Holstein Cattle as Contrasting Model Traits. PLOS Genet. 2010, 6, e1001139. [Google Scholar] [CrossRef] [PubMed]
- Barreto, C.A.V.; das Graças Dias, K.O.; de Sousa, I.C.; Azevedo, C.F.; Nascimento, A.C.C.; Guimarães, L.J.M.; Guimarães, C.T.; Pastina, M.M.; Nascimento, M. Genomic Prediction in Multi-Environment Trials in Maize Using Statistical and Machine Learning Methods. Sci. Rep. 2024, 14, 1062. [Google Scholar] [CrossRef]
- Mathew, B.; Léon, J.; Sannemann, W.; Sillanpää, M.J. Detection of Epistasis for Flowering Time Using Bayesian Multilocus Estimation in a Barley MAGIC Population. Genetics 2018, 208, 525–536. [Google Scholar] [CrossRef]
- Nascimento, M.; Nascimento, A.C.C.; Azevedo, C.F.; de Oliveira, A.C.B.; Caixeta, E.T.; Jarquin, D. Enhancing Genomic Prediction with Stacking Ensemble Learning in Arabica Coffee. Front. Plant Sci. 2024, 15, 1373318. [Google Scholar] [CrossRef]
- De Sousa, I.C.; Barreto, C.A.V.; Caixeta, E.T.; Nascimento, A.C.C.; Azevedo, C.F.; Alkimim, E.R.; Nascimento, M. The Trade-off between Density Marker Panels Size and Predictive Ability of Genomic Prediction for Agronomic Traits in Coffea Canephora. Euphytica 2024, 220, 46. [Google Scholar] [CrossRef]

| Heritability (h2) | Number of Loci (QTLs) | |||
|---|---|---|---|---|
| 8 | 40 | 80 | 120 | |
| 0.3 | S1 | S4 | S7 | S10 |
| 0.5 | S2 | S5 | S8 | S11 |
| 0.8 | S3 | S6 | S9 | S12 |
| Heritability (h2) | Number of Loci (QTLs) | |||
|---|---|---|---|---|
| 8 | 40 | 80 | 120 | |
| 0.3 | 14 | 12 | 16 | 17 |
| 0.5 | 25 | 16 | 14 | 18 |
| 0.8 | 22 | 18 | 18 | 17 |
| Number of QTLs | h2 | MARS2 | GBLUP-A | GBLUP-AD | GBLUP-AE |
|---|---|---|---|---|---|
| 8 | 0.3 | 0.569 | 0.782 | 0.881 | 0.782 |
| 0.5 | 0.534 | 0.831 | 0.831 | 0.831 | |
| 0.8 | 0.625 | 0.804 | 0.707 | 0.804 | |
| 40 | 0.3 | 0.636 | 0.612 | 0.919 | 0.727 |
| 0.5 | 0.621 | 0.945 | 0.760 | 0.605 | |
| 0.8 | 0.587 | 0.762 | 0.930 | 0.605 | |
| 80 | 0.3 | 0.569 | 0.859 | 0.817 | 0.699 |
| 0.5 | 0.468 | 0.762 | 0.628 | 0.533 | |
| 0.8 | 0.601 | 0.929 | 0.961 | 0.606 | |
| 120 | 0.3 | 0.624 | 0.896 | 0.751 | 0.527 |
| 0.5 | 0.523 | 0.866 | 0.543 | 0.639 | |
| 0.8 | 0.492 | 0.868 | 0.837 | 0.621 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Oliveira Celeri, M.; da Costa, W.G.; Nascimento, A.C.C.; Azevedo, C.F.; Cruz, C.D.; Sagae, V.S.; Nascimento, M. Multivariate Adaptive Regression Splines Enhance Genomic Prediction of Non-Additive Traits. Agronomy 2024, 14, 2234. https://doi.org/10.3390/agronomy14102234
de Oliveira Celeri M, da Costa WG, Nascimento ACC, Azevedo CF, Cruz CD, Sagae VS, Nascimento M. Multivariate Adaptive Regression Splines Enhance Genomic Prediction of Non-Additive Traits. Agronomy. 2024; 14(10):2234. https://doi.org/10.3390/agronomy14102234
Chicago/Turabian Stylede Oliveira Celeri, Maurício, Weverton Gomes da Costa, Ana Carolina Campana Nascimento, Camila Ferreira Azevedo, Cosme Damião Cruz, Vitor Seiti Sagae, and Moysés Nascimento. 2024. "Multivariate Adaptive Regression Splines Enhance Genomic Prediction of Non-Additive Traits" Agronomy 14, no. 10: 2234. https://doi.org/10.3390/agronomy14102234
APA Stylede Oliveira Celeri, M., da Costa, W. G., Nascimento, A. C. C., Azevedo, C. F., Cruz, C. D., Sagae, V. S., & Nascimento, M. (2024). Multivariate Adaptive Regression Splines Enhance Genomic Prediction of Non-Additive Traits. Agronomy, 14(10), 2234. https://doi.org/10.3390/agronomy14102234










