Genome-Wide Identification of the Glycine-Rich RNA-Binding Protein Genes and Their Expression Analysis upon Aspergillus flavus Infection in Groundnut (Arachis hypogaea)
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of GR-RBP Genes in Groundnut
2.2. Phylogenetic Analysis
2.3. Chromosomal Location, Synteny, and Evolutionary Analysis of GR-RBP Genes in Groundnut
2.4. Analysis of Intron-Exon Structure and Conserved Domains
2.5. Cis-Regulatory Element Analysis of Ah.GR-RBPs
2.6. Protein-Protein Interaction
2.7. Expression Analysis of GR-RBPs during A. flavus Infection
2.7.1. Plant Material
2.7.2. RNA Isolation and Expression Analysis
3. Results
3.1. Identification of GR-RBP Genes in Groundnut
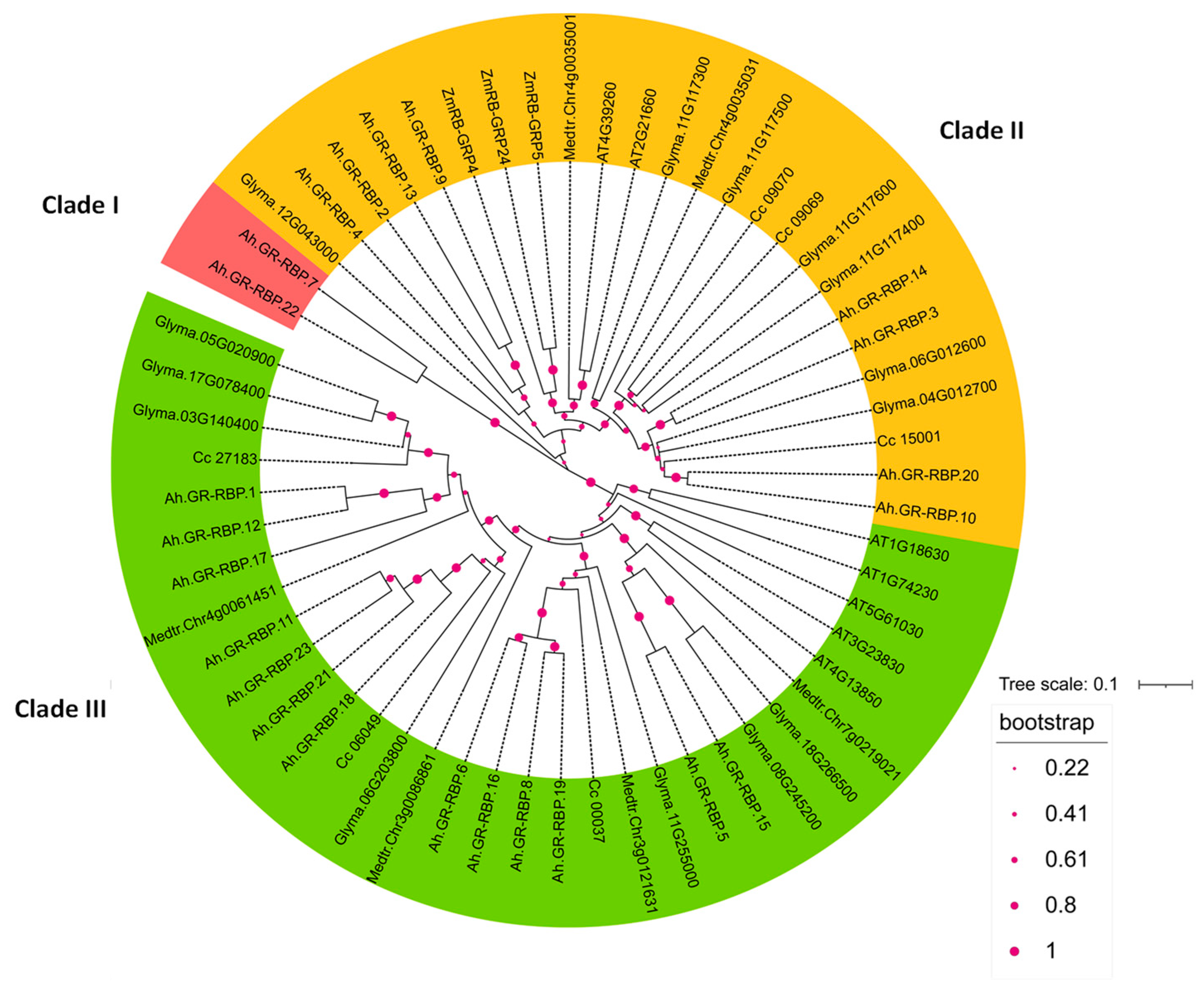
3.2. Phylogenetic Analysis of GR-RBPs in Groundnut
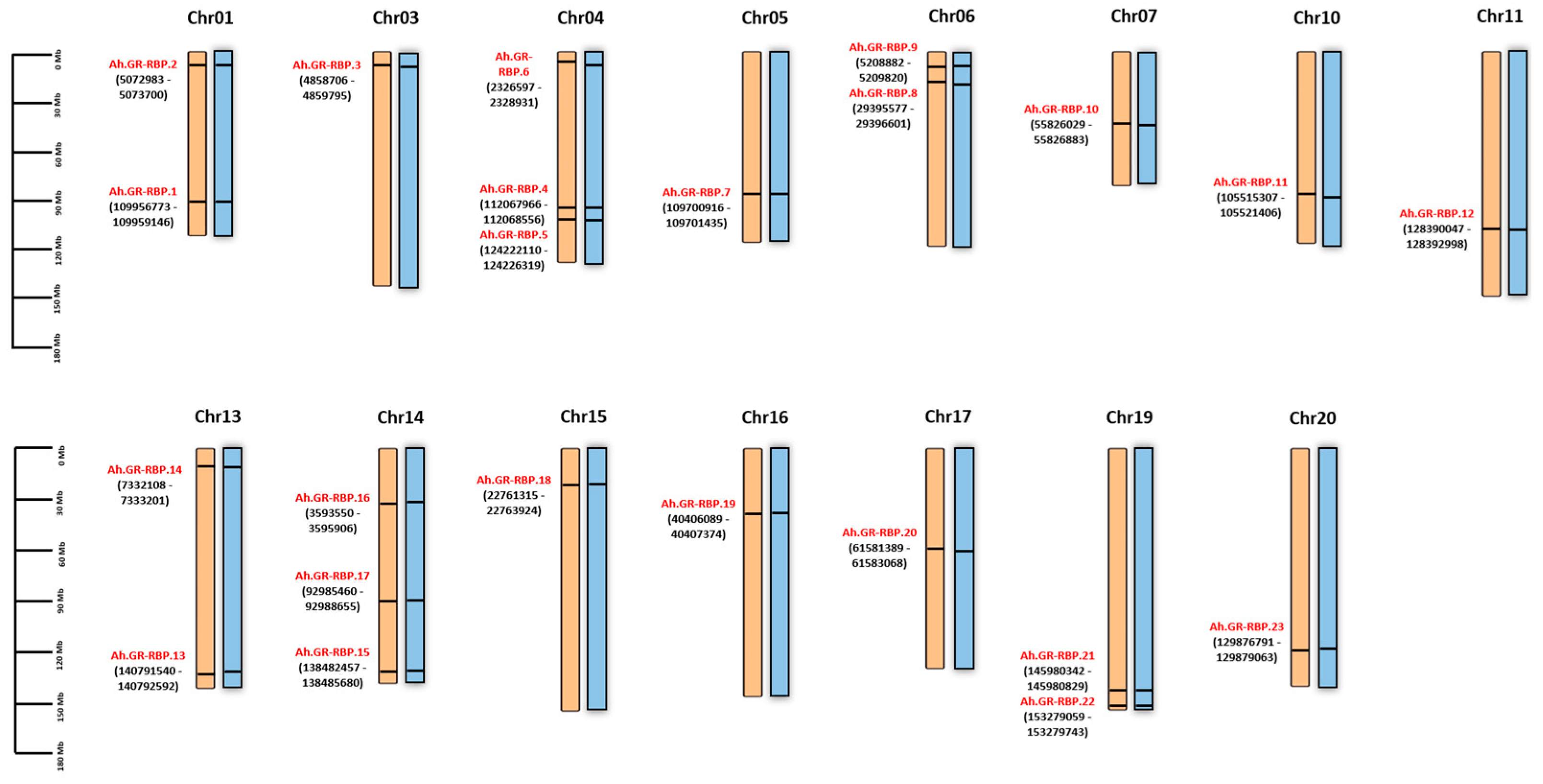
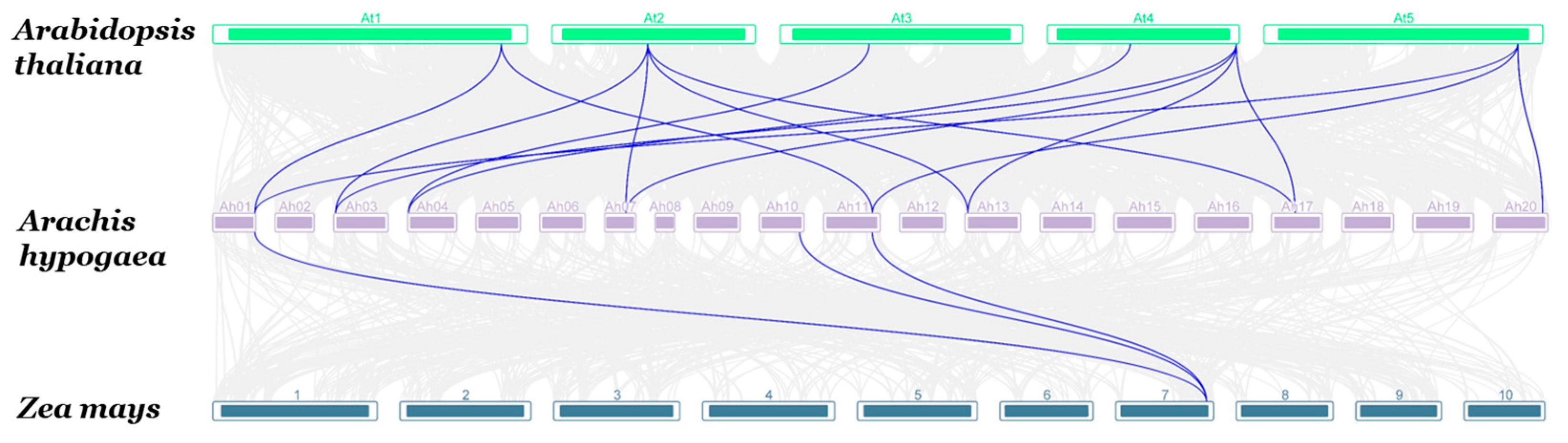
3.3. Chromosomal Location, Synteny, and Evolutionary Analysis of GR-RBP Genes in Groundnut
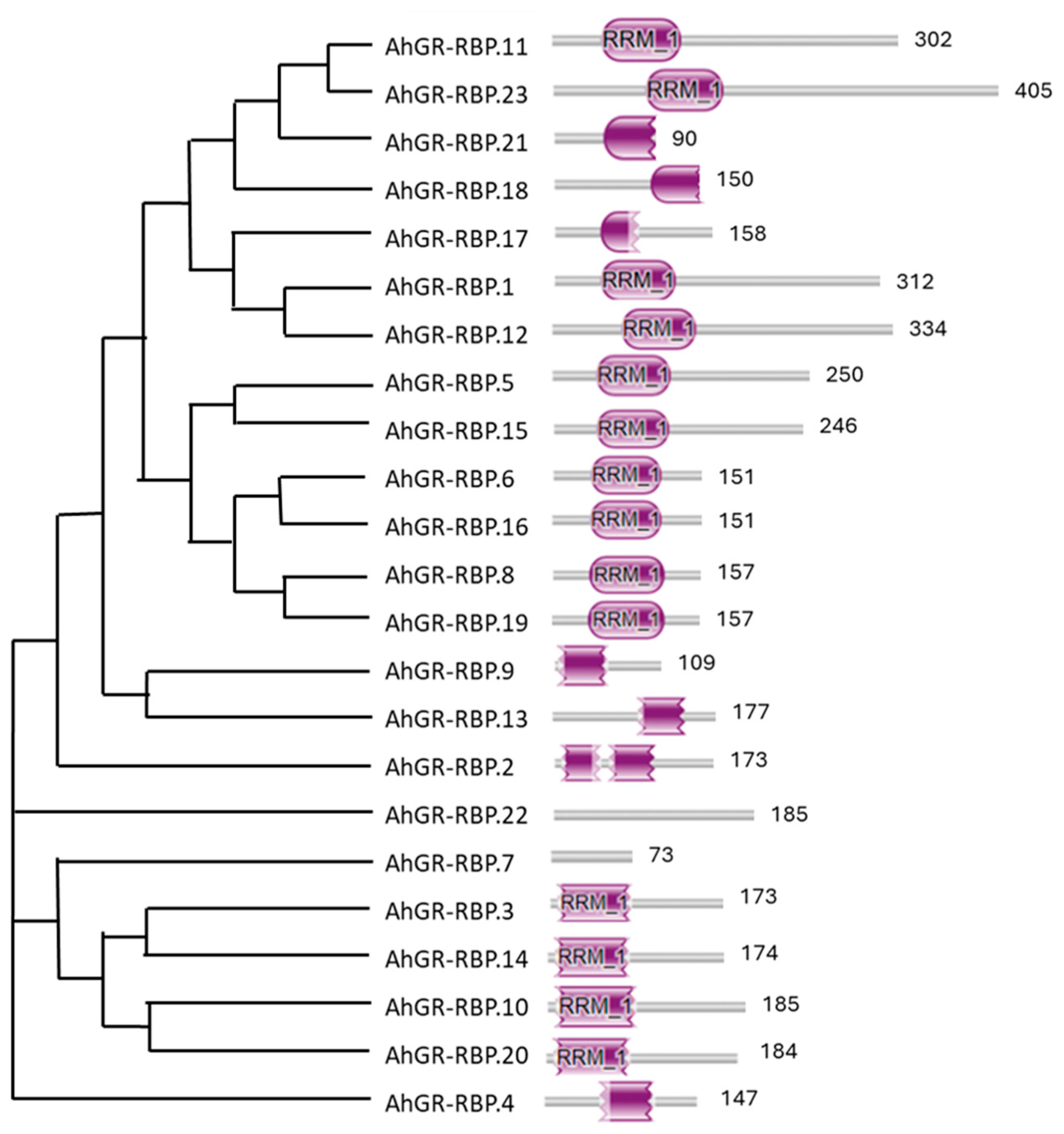
3.4. Analysis of Intron–Exon Structure and Conserved Domains
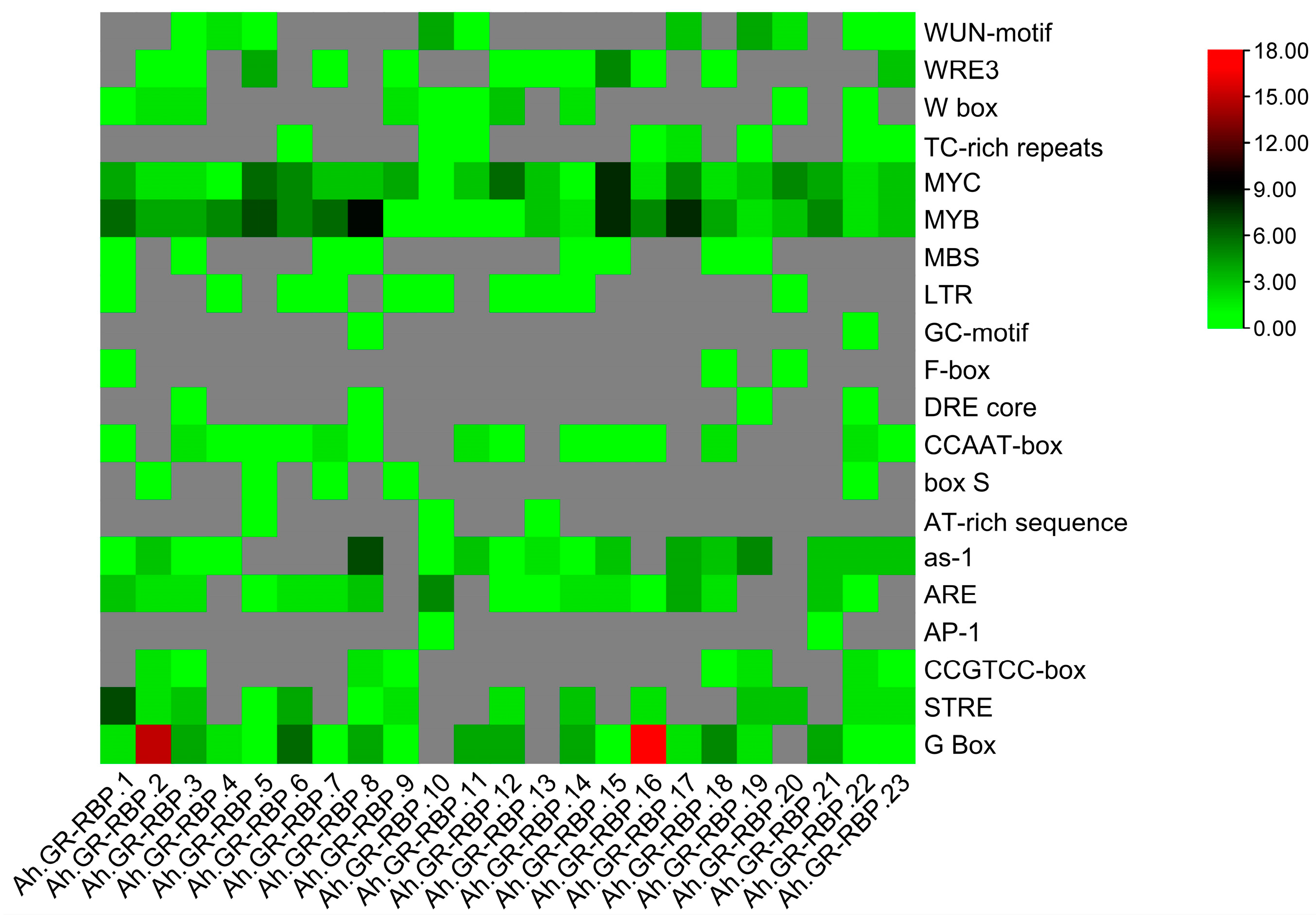
3.5. Cis-Regulatory Element Analysis of Ah.GR-RBPs
3.6. Protein–Protein Interaction of GR-RBPs in Groundnut
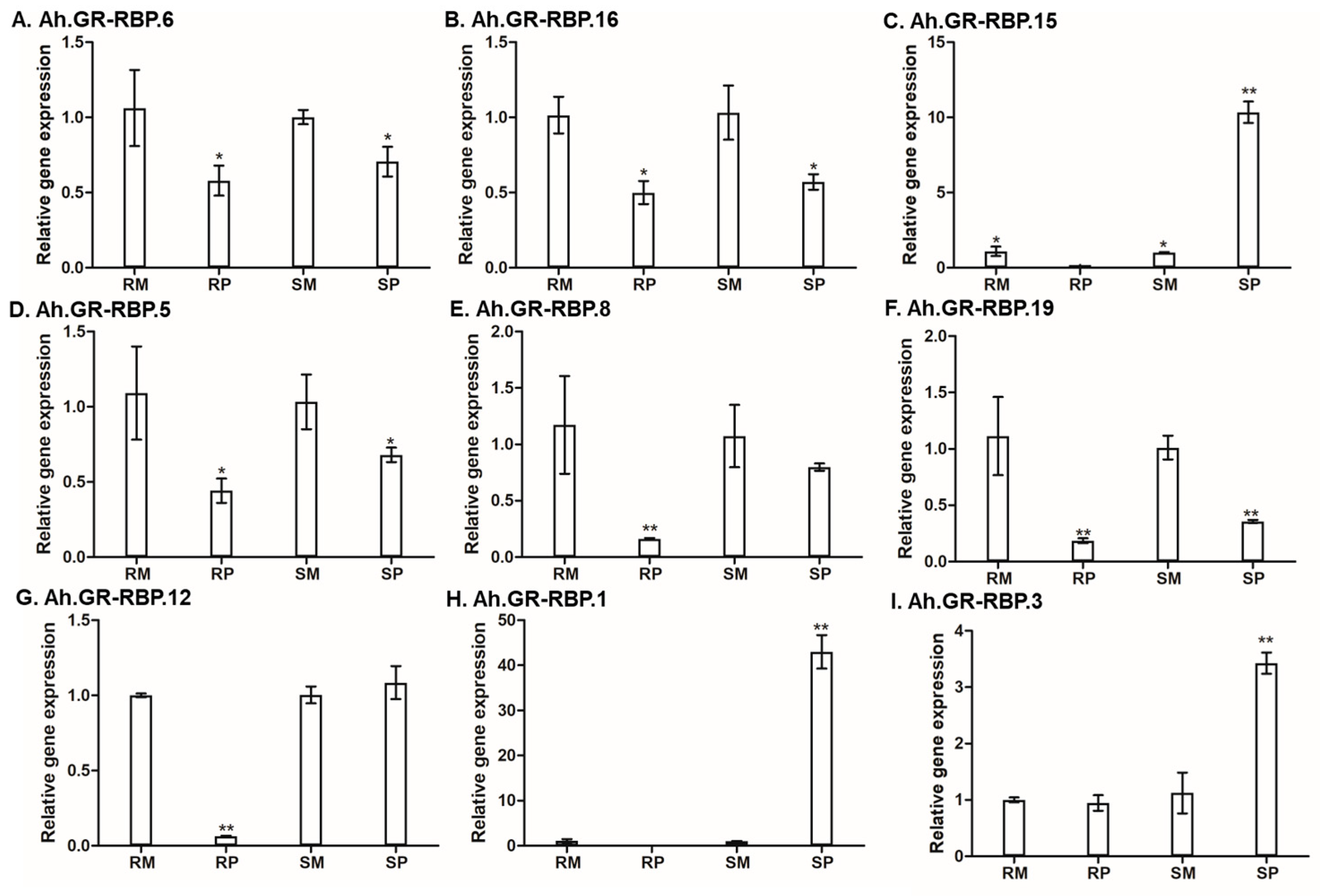
3.7. Expression Analysis of GR-RBPs during A. flavus Infection
4. Discussion
4.1. Identification and Diversity Analysis of GR-RBPs in Groundnut
4.2. Evolution of GR-RBP Genes in Groundnut
4.3. Regulation of GR-RBPs in Groundnut
4.4. Expression Analysis of GR-RBP Genes in Groundnut in Response to A. flavus Infection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nigam, S.N.; Waliyar, F.; Aruna, R.; Reddy, S.V.; Kumar, P.L.; Craufurd, P.Q.; Diallo, A.T.; Ntare, B.R.; Upadhyaya, H.D. Breeding Peanut for Resistance to Aflatoxin Contamination at ICRISAT. Peanut Sci. 2009, 36, 42–49. [Google Scholar] [CrossRef]
- Pandey, M.K.; Kumar, R.; Pandey, A.K.; Soni, P.; Gangurde, S.S.; Sudini, H.K.; Fountain, J.C.; Liao, B.; Desmae, H.; Okori, P.; et al. Mitigating Aflatoxin Contamination in Groundnut through A Combination of Genetic Resistance and Post-Harvest Management Practices. Toxins 2019, 11, 315. [Google Scholar] [CrossRef] [PubMed]
- Govrin, E.M.; Levine, A. The Hypersensitive Response Facilitates Plant Infection by the Necrotrophic Pathogen Botrytis Cinerea. Curr. Biol. 2000, 10, 751–757. [Google Scholar] [CrossRef]
- Faris, J.D.; Zhang, Z.; Lu, H.; Lu, S.; Reddy, L.; Cloutier, S.; Fellers, J.P.; Meinhardt, S.W.; Rasmussen, J.B.; Xu, S.S.; et al. A Unique Wheat Disease Resistance-like Gene Governs Effector-Triggered Susceptibility to Necrotrophic Pathogens. Proc. Natl. Acad. Sci. USA 2010, 107, 13544–13549. [Google Scholar] [CrossRef]
- Kelley, R.Y.; Williams, W.P.; Mylroie, J.E.; Boykin, D.L.; Harper, J.W.; Windham, G.L.; Ankala, A.; Shan, X. Identification of Maize Genes Associated with Host Plant Resistance or Susceptibility to Aspergillus Flavus Infection and Aflatoxin Accumulation. PLoS ONE 2012, 7, e36892. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kim, W.Y.; Kwak, K.J.; Oh, S.H.; Han, Y.S.; Kang, H. Glycine-Rich RNA-Binding Proteins Are Functionally Conserved in Arabidopsis thaliana and Oryza sativa during Cold Adaptation Process. J. Exp. Bot. 2010, 61, 2317–2325. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, Y.; Xiao, H.; Zheng, Y.; Yue, B. Genome-wide Identification, Evolution, and Expression Analysis of RNA-binding Glycine-rich Protein Family in Maize. J. Integr. Plant Biol. 2014, 56, 1020–1031. [Google Scholar] [CrossRef]
- Tang, Y.; Huang, C.; Li, Y.; Wang, Y.; Zhang, C. Genome-Wide Identification, Phylogenetic Analysis, and Expression Profiling of Glycine-Rich RNA-Binding Protein (GRPs) Genes in Seeded and Seedless Grapes (Vitis vinifera). Physiol. Mol. Biol. Plants 2021, 27, 2231–2243. [Google Scholar] [CrossRef]
- Lu, Y.; Sun, J.; Yang, Z.; Zhao, C.; Zhu, M.; Ma, D.; Dong, T.; Zhou, Z.; Liu, M.; Yang, D. Genome-Wide Identification and Expression Analysis of Glycine-Rich RNA-Binding Protein Family in Sweet Potato Wild Relative Ipomoea Trifida. Gene 2019, 686, 177–186. [Google Scholar] [CrossRef]
- Yang, W.; Yu, M.; Zou, C.; Lu, C.; Yu, D.; Cheng, H.; Jiang, P.; Feng, X.; Zhang, Y.; Wang, Q.; et al. Genome-Wide Comparative Analysis of RNA-Binding Glycine-Rich Protein Family Genes between Gossypium arboreum and Gossypium raimondii. PLoS ONE 2019, 14, e0218938. [Google Scholar] [CrossRef]
- Duan, M.; Zong, M.; Guo, N.; Han, S.; Wang, G.; Miao, L.; Liu, F. Comprehensive Genome-Wide Identification of the RNA-Binding Glycine-Rich Gene Family and Expression Profiling under Abiotic Stress in Brassica Oleracea. Plants 2023, 12, 3706. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.; Zhang, C.; Lu, Y.; Li, J.; Tang, H.; Ma, L.; Zhu, H. The Glycine-Rich RNA-Binding Protein Is a Vital Post-Transcriptional Regulator in Crops. Plants 2023, 12, 3504. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, S.J.; Jang, B.; Jung, C.-H.; Ahn, S.J.; Goh, C.-H.; Cho, K.; Han, O.; Kang, H. Functional Characterization of a Glycine-Rich RNA-Binding Protein 2 in Arabidopsis thaliana under Abiotic Stress Conditions. Plant J. 2007, 50, 439–451. [Google Scholar] [CrossRef]
- Kim, J.S.; Jung, H.J.; Lee, H.J.; Kim, K.A.; Goh, C.-H.; Woo, Y.; Oh, S.H.; Han, Y.S.; Kang, H. Glycine-Rich RNA-Binding Protein7 Affects Abiotic Stress Responses by Regulating Stomata Opening and Closing in Arabidopsis thaliana. Plant J. 2008, 55, 455–466. [Google Scholar] [CrossRef]
- Sahi, C.; Agarwal, M.; Singh, A.; Grover, A. Molecular Characterization of a Novel Isoform of Rice (Oryza sativa L.) Glycine Rich-RNA Binding Protein and Evidence for Its Involvement in High Temperature Stress Response. Plant Sci. 2007, 173, 144–155. [Google Scholar] [CrossRef]
- Kim, D.S.; Kim, N.H.; Hwang, B.K. Glycine-Rich Rna-Binding Protein1 Interacts with Receptor-like Cytoplasmic Protein Kinase1 and Suppresses Cell Death and Defense Responses in Pepper (Capsicum annuum). New Phytol. 2015, 205, 786–800. [Google Scholar] [CrossRef]
- Lee, H.J.; Kim, J.S.; Yoo, S.J.; Kang, E.Y.; Han, S.H.; Yang, K.-Y.; Kim, Y.C.; McSpadden Gardener, B.; Kang, H. Different Roles of Glycine-Rich RNA-Binding Protein7 in Plant Defense against Pectobacterium Carotovorum, Botrytis Cinerea, and Tobacco Mosaic Viruses. Plant Physiol. Biochem. 2012, 60, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Springer Protocols Handbooks; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. ISBN 978-1-59259-890-8. [Google Scholar]
- Thumuluri, V.; Almagro Armenteros, J.J.; Johansen, A.R.; Nielsen, H.; Winther, O. DeepLoc 2.0: Multi-Label Subcellular Localization Prediction Using Protein Language Models. Nucleic Acids Res. 2022, 50, W228–W234. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Chao, J.; Li, Z.; Sun, Y.; Aluko, O.O.; Wu, X.; Wang, Q.; Liu, G. MG2C: A User-Friendly Online Tool for Drawing Genetic Maps. Mol. Hortic. 2021, 1, 16. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER Web Server: 2018 Update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P. STRING V10: Protein–Protein Interaction Networks, Integrated over the Tree of Life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Commey, L.; Tengey, T.K.; Cobos, C.J.; Dampanaboina, L.; Dhillon, K.K.; Pandey, M.K.; Sudini, H.K.; Falalou, H.; Varshney, R.K.; Burow, M.D.; et al. Peanut Seed Coat Acts as a Physical and Biochemical Barrier against Aspergillus Flavus Infection. J. Fungi 2021, 7, 1000. [Google Scholar] [CrossRef] [PubMed]
- Prasad, K.; Yogendra, K.; Sanivarapu, H.; Rajasekaran, K.; Cary, J.W.; Sharma, K.K.; Bhatnagar-Mathur, P. Multiplexed Host-Induced Gene Silencing of Aspergillus Flavus Genes Confers Aflatoxin Resistance in Groundnut. Toxins 2023, 15, 319. [Google Scholar] [CrossRef]
- Arias, R.S.; Dang, P.M.; Sobolev, V.S. RNAi-Mediated Control of Aflatoxins in Peanut: Method to Analyze Mycotoxin Production and Transgene Expression in the Peanut/Aspergillus Pathosystem. J. Vis. Exp. 2015, e53398. [Google Scholar] [CrossRef]
- He, M.; Cui, S.; Yang, X.; Mu, G.; Chen, H.; Liu, L. Selection of Suitable Reference Genes for Abiotic Stress-Responsive Gene Expression Studies in Peanut by Real-Time Quantitative PCR. Electron. J. Biotechnol. 2017, 28, 76–86. [Google Scholar] [CrossRef]
- Patel, P.; Panchal, H. Primary, Secondary and Tertiary Structural Analysis of Disease Resistance Protein RGA4 of ZEA Maize Using Bioinformatics Tools. Int. J. Adv. Res. Comput. Commun. Eng. 2016, 5, 7–12. [Google Scholar]
- Mangeon, A.; Magioli, C.; Menezes-Salgueiro, A.D.; Cardeal, V.; De Oliveira, C.; Galvão, V.C.; Margis, R.; Engler, G.; Sachetto-Martins, G. AtGRP5, a Vacuole-Located Glycine-Rich Protein Involved in Cell Elongation. Planta 2009, 230, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Nakaminami, K.; Sasaki, K.; Kajita, S.; Takeda, H.; Karlson, D.; Ohgi, K.; Imai, R. Heat Stable ssDNA/RNA-Binding Activity of a Wheat Cold Shock Domain Protein. FEBS Lett. 2005, 579, 4887–4891. [Google Scholar] [CrossRef]
- Brady, K.P.; Darvill, A.G.; Albersheim, P. Activation of a Tobacco Glycine-rich Protein Gene by a Fungal Glucan Preparation. Plant J. 1993, 4, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Chaikam, V.; Karlson, D. Functional Characterization of Two Cold Shock Domain Proteins from Oryza sativa. Plant Cell Environ. 2008, 31, 995–1006. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Karlson, D.T. Overexpression of AtCSP4 Affects Late Stages of Embryo Development in Arabidopsis. J. Exp. Bot. 2011, 62, 2079–2091. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, R.; Liang, D.; Ma, F.; Shu, H. Molecular Characterization and Expression Analysis of a Glycine-Rich RNA-Binding Protein Gene from Malus hupehensis Rehd. Mol. Biol. Rep. 2012, 39, 4145–4153. [Google Scholar] [CrossRef] [PubMed]
- Muthusamy, M.; Kim, J.-H.; Kim, J.A.; Lee, S.-I. Plant RNA Binding Proteins as Critical Modulators in Drought, High Salinity, Heat, and Cold Stress Responses: An Updated Overview. Int. J. Mol. Sci. 2021, 22, 6731. [Google Scholar] [CrossRef]
- Condit, C.M.; Meagher, R.B. Expression of a Gene Encoding a Glycine-Rich Protein in Petunia. Mol. Cell. Biol. 1987, 7, 4273–4279. [Google Scholar] [CrossRef] [PubMed]
- Fusaro, A.F.; Bocca, S.N.; Ramos, R.L.B.; Barrôco, R.M.; Magioli, C.; Jorge, V.C.; Coutinho, T.C.; Rangel-Lima, C.M.; De Rycke, R.; Inzé, D.; et al. AtGRP2, a Cold-Induced Nucleo-Cytoplasmic RNA-Binding Protein, Has a Role in Flower and Seed Development. Planta 2007, 225, 1339–1351. [Google Scholar] [CrossRef]
- Vermel, M.; Guermann, B.; Delage, L.; Grienenberger, J.-M.; Maréchal-Drouard, L.; Gualberto, J.M. A Family of RRM-Type RNA-Binding Proteins Specific to Plant Mitochondria. Proc. Natl. Acad. Sci. USA 2002, 99, 5866–5871. [Google Scholar] [CrossRef] [PubMed]
- Dunn, M.A.; Brown, K.; Lightowlers, R.; Hughes, M.A. A Low-Temperature-Responsive Gene from Barley Encodes a Protein with Single-Stranded Nucleic Acid-Binding Activity Which Is Phosphorylated in Vitro. Plant Mol. Biol. 1996, 30, 947–959. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Sugita, M.; Sugiura, M. cDNA Structure, Expression and Nucleic Acid-Binding Properties of Three RNA-Binding Proteins in Tobacco: Occurence of Tissue-Specific Alternative Splicing. Nucleic Acids Res. 1993, 21, 3981–3987. [Google Scholar] [CrossRef] [PubMed]
- Ludevid, M.D.; Freire, M.A.; Gómez, J.; Burd, C.G.; Albericio, F.; Giralt, E.; Dreyfuss, G.; Pagès, M. RNA Binding Characteristics of a 16 kDa Glycine-rich Protein from Maize. Plant J. 1992, 2, 999–1003. [Google Scholar] [CrossRef]
- Hanano, S.; Sugita, M.; Sugiura, M. Isolation of a Novel RNA-Binding Protein and Its Association with a Large Ribonucleoprotein Particle Present in the Nucleoplasm of Tobacco Cells. Plant Mol. Biol. 1996, 31, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Hernández-Hernández, T.; Martínez-Castilla, L.P.; Alvarez-Buylla, E.R. Functional Diversification of B MADS-Box Homeotic Regulators of Flower Development: Adaptive Evolution in Protein–Protein Interaction Domains after Major Gene Duplication Events. Mol. Biol. Evol. 2007, 24, 465–481. [Google Scholar] [CrossRef]
- Doyle, J.J.; Flagel, L.E.; Paterson, A.H.; Rapp, R.A.; Soltis, D.E.; Soltis, P.S.; Wendel, J.F. Evolutionary Genetics of Genome Merger and Doubling in Plants. Annu. Rev. Genet. 2008, 42, 443–461. [Google Scholar] [CrossRef]
- Jiang, Y.; Zeng, B.; Zhao, H.; Zhang, M.; Xie, S.; Lai, J. Genome-wide Transcription Factor Gene Prediction and Their Expressional Tissue-Specificities in Maize. J. Integr. Plant Biol. 2012, 54, 616–630. [Google Scholar] [CrossRef]
- Gokul, B.S.; Gohil, D.S.; Roy Choudhury, S. Genome-Wide Identification, Evolutionary and Expression Analysis of the Cyclin-Dependent Kinase Gene Family in Peanut. BMC Plant Biol. 2023, 23, 43. [Google Scholar] [CrossRef]
- Kaur, A.; Pati, P.K.; Pati, A.M.; Nagpal, A.K. In-Silico Analysis of Cis-Acting Regulatory Elements of Pathogenesis-Related Proteins of Arabidopsis thaliana and Oryza sativa. PLoS ONE 2017, 12, e0184523. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Gangwar, R.; Zahra, S.; Poddar, N.; Singh, A.; Kumar, S. Genome-Wide Characterization and Comparative Analysis of the OSCA Gene Family and Identification of Its Potential Stress-Responsive Members in Legumes. Sci. Rep. 2023, 13, 5914. [Google Scholar] [CrossRef] [PubMed]
- Césari, S.; Kanzaki, H.; Fujiwara, T.; Bernoux, M.; Chalvon, V.; Kawano, Y.; Shimamoto, K.; Dodds, P.; Terauchi, R.; Kroj, T. The NB-LRR Proteins RGA4 and RGA5 Interact Functionally and Physically to Confer Disease Resistance. EMBO J. 2014, 33, 1941–1959. [Google Scholar] [CrossRef] [PubMed]
- Lorković, Z.J. Role of Plant RNA-Binding Proteins in Development, Stress Response and Genome Organization. Trends Plant Sci. 2009, 14, 229–236. [Google Scholar] [CrossRef]
- Lorković, Z.J.; Barta, A. Genome Analysis: RNA Recognition Motif (RRM) and K Homology (KH) Domain RNA-Binding Proteins from the Flowering Plant Arabidopsis thaliana. Nucleic Acids Res. 2002, 30, 623–635. [Google Scholar] [CrossRef]
- Xu, T.; Lee, H.J.; Sy, N.D.; Kang, H. Wheat (Triticum aestivum) Zinc Finger-Containing Glycine-Rich RNA-Binding Protein TaRZ1 Affects Plant Growth and Defense Response in Arabidopsis thaliana. Plant Growth Regul. 2015, 76, 243–250. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Guo, M.; Jeong, B.; Tian, F.; Elthon, T.E.; Cerny, R.L.; Staiger, D.; Alfano, J.R. A Type III Effector ADP-Ribosylates RNA-Binding Proteins and Quells Plant Immunity. Nature 2007, 447, 284–288. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jose, A.M.; Tejaswi, A.; Kokiladevi, E.; Sharma, N.; Yogendra, K. Genome-Wide Identification of the Glycine-Rich RNA-Binding Protein Genes and Their Expression Analysis upon Aspergillus flavus Infection in Groundnut (Arachis hypogaea). Agronomy 2024, 14, 165. https://doi.org/10.3390/agronomy14010165
Jose AM, Tejaswi A, Kokiladevi E, Sharma N, Yogendra K. Genome-Wide Identification of the Glycine-Rich RNA-Binding Protein Genes and Their Expression Analysis upon Aspergillus flavus Infection in Groundnut (Arachis hypogaea). Agronomy. 2024; 14(1):165. https://doi.org/10.3390/agronomy14010165
Chicago/Turabian StyleJose, Alin M., Avuthu Tejaswi, Eswaran Kokiladevi, Niharika Sharma, and Kalenahalli Yogendra. 2024. "Genome-Wide Identification of the Glycine-Rich RNA-Binding Protein Genes and Their Expression Analysis upon Aspergillus flavus Infection in Groundnut (Arachis hypogaea)" Agronomy 14, no. 1: 165. https://doi.org/10.3390/agronomy14010165
APA StyleJose, A. M., Tejaswi, A., Kokiladevi, E., Sharma, N., & Yogendra, K. (2024). Genome-Wide Identification of the Glycine-Rich RNA-Binding Protein Genes and Their Expression Analysis upon Aspergillus flavus Infection in Groundnut (Arachis hypogaea). Agronomy, 14(1), 165. https://doi.org/10.3390/agronomy14010165






