Silver and Hematite Nanoparticles Had a Limited Effect on the Bacterial Community Structure in Soil Cultivated with Phaseolus vulgaris L.
Abstract
:1. Introduction
- the growth and characteristics of common bean plants
- the diversity and taxonomic and putative functional profile of the soil bacterial community in cultivated and uncultivated soil.
2. Materials and Methods
2.1. Characterization of Nanoparticles
2.2. Seeds, Soil Sampling Site and Experimental Design
2.3. Measure of Morphological Parameters of the Plant and Mineral Content
2.4. DNA Extraction
2.5. PCR and DNA Sequencing
2.6. Bioinformatic Analysis
2.7. Statistical Analysis
3. Results
3.1. Nanoparticles’ Characteristics
3.2. Soil Characteristics
3.3. Effect of NPs on Bean Characteristics
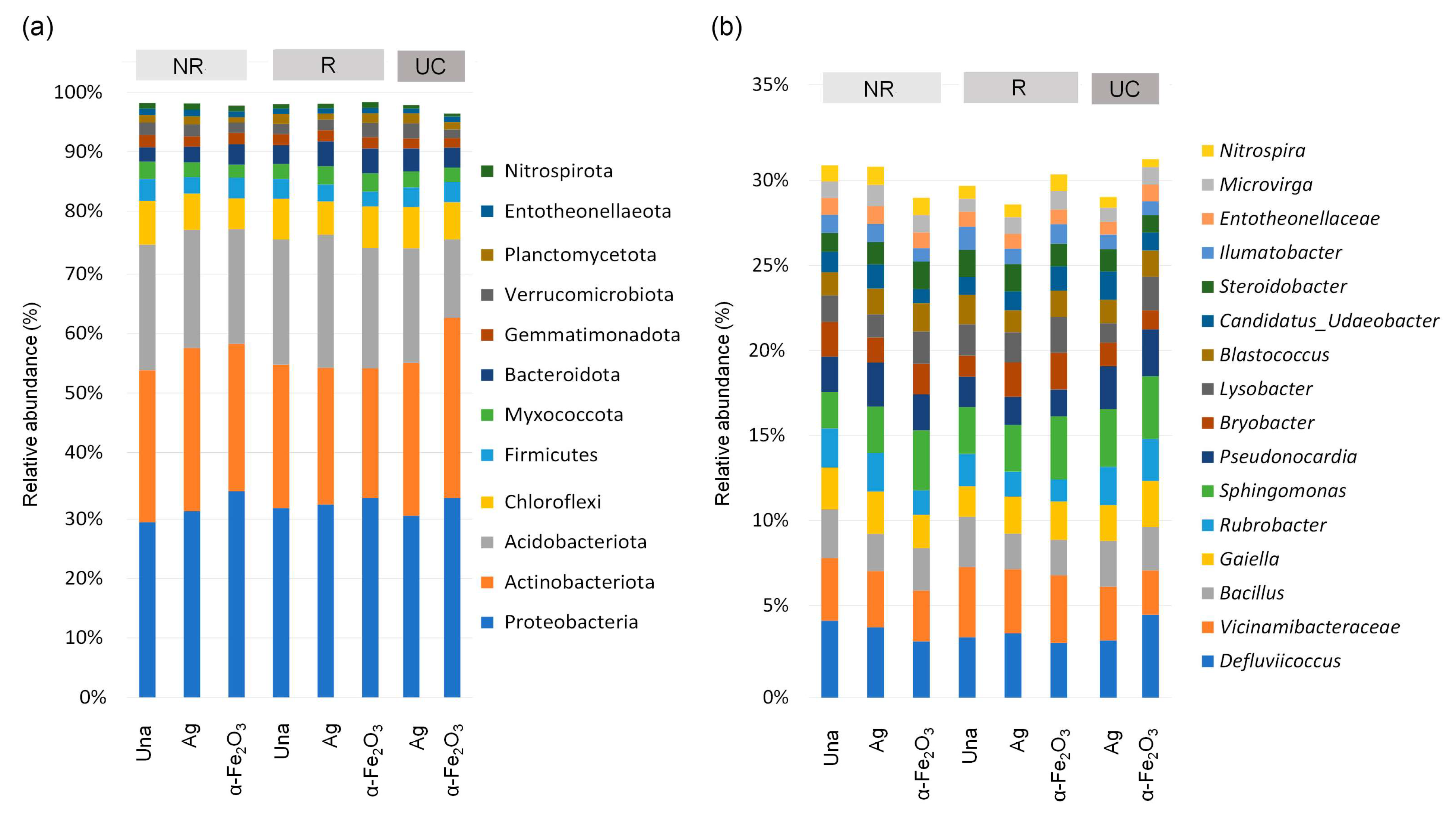
3.4. Alpha Diversity and Taxonomic Profile of the Bacterial Community in Soil
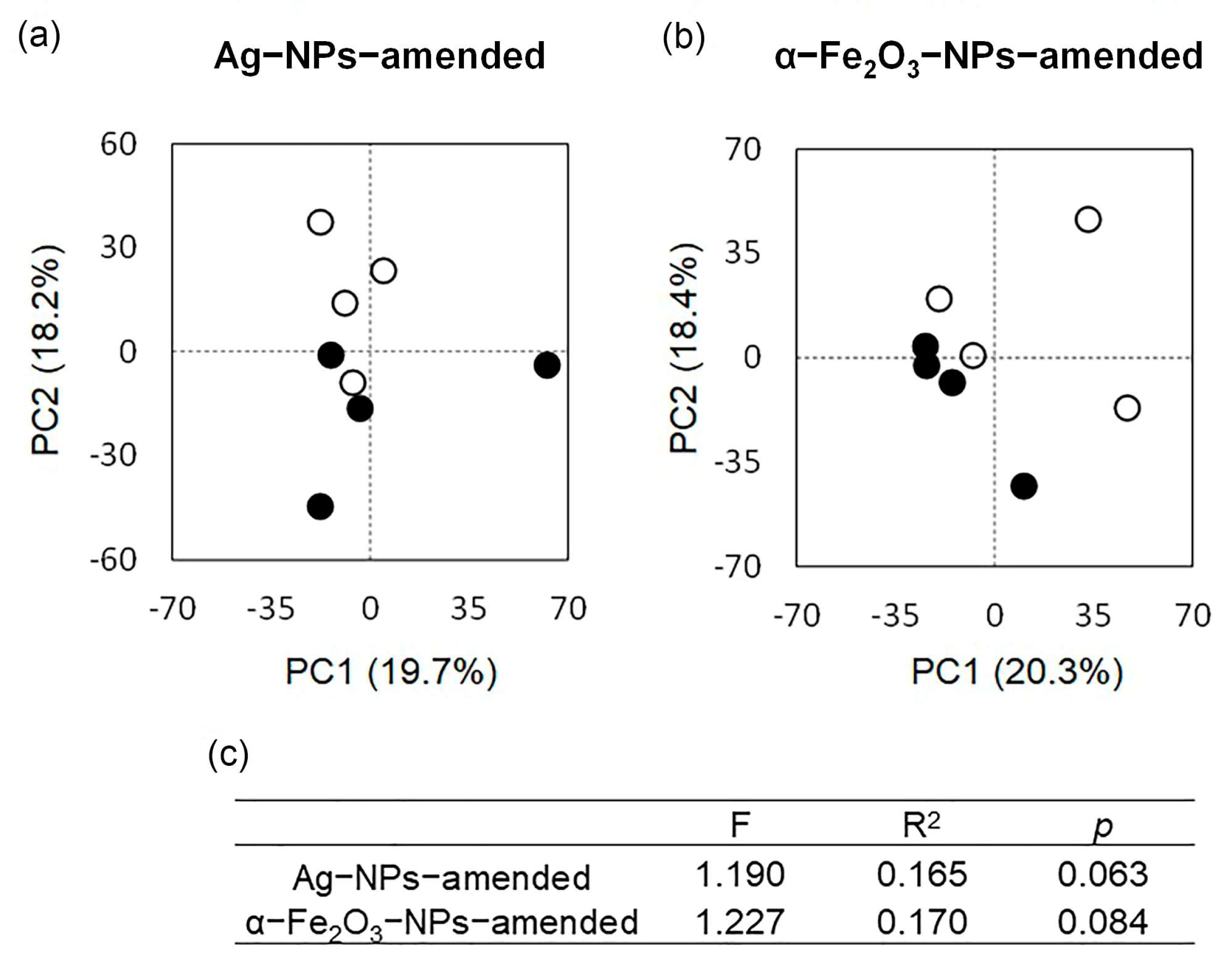
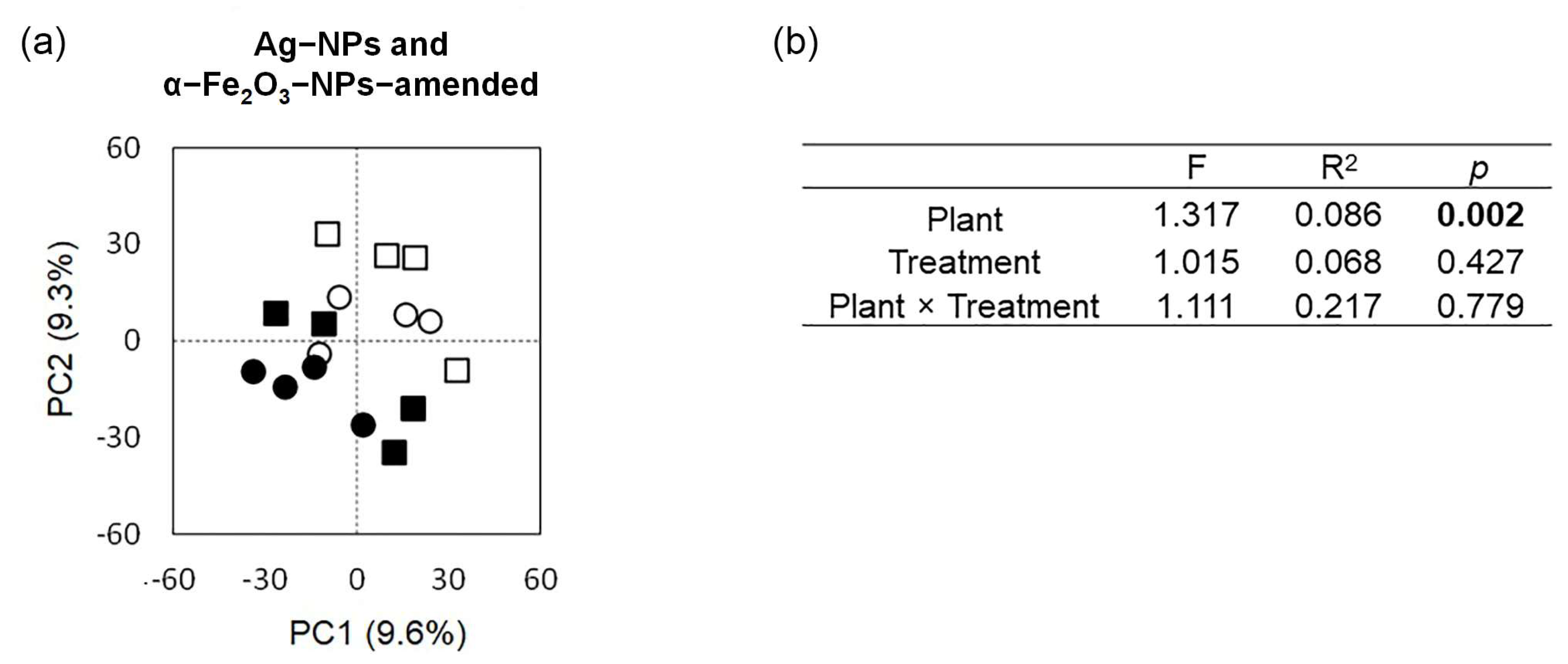
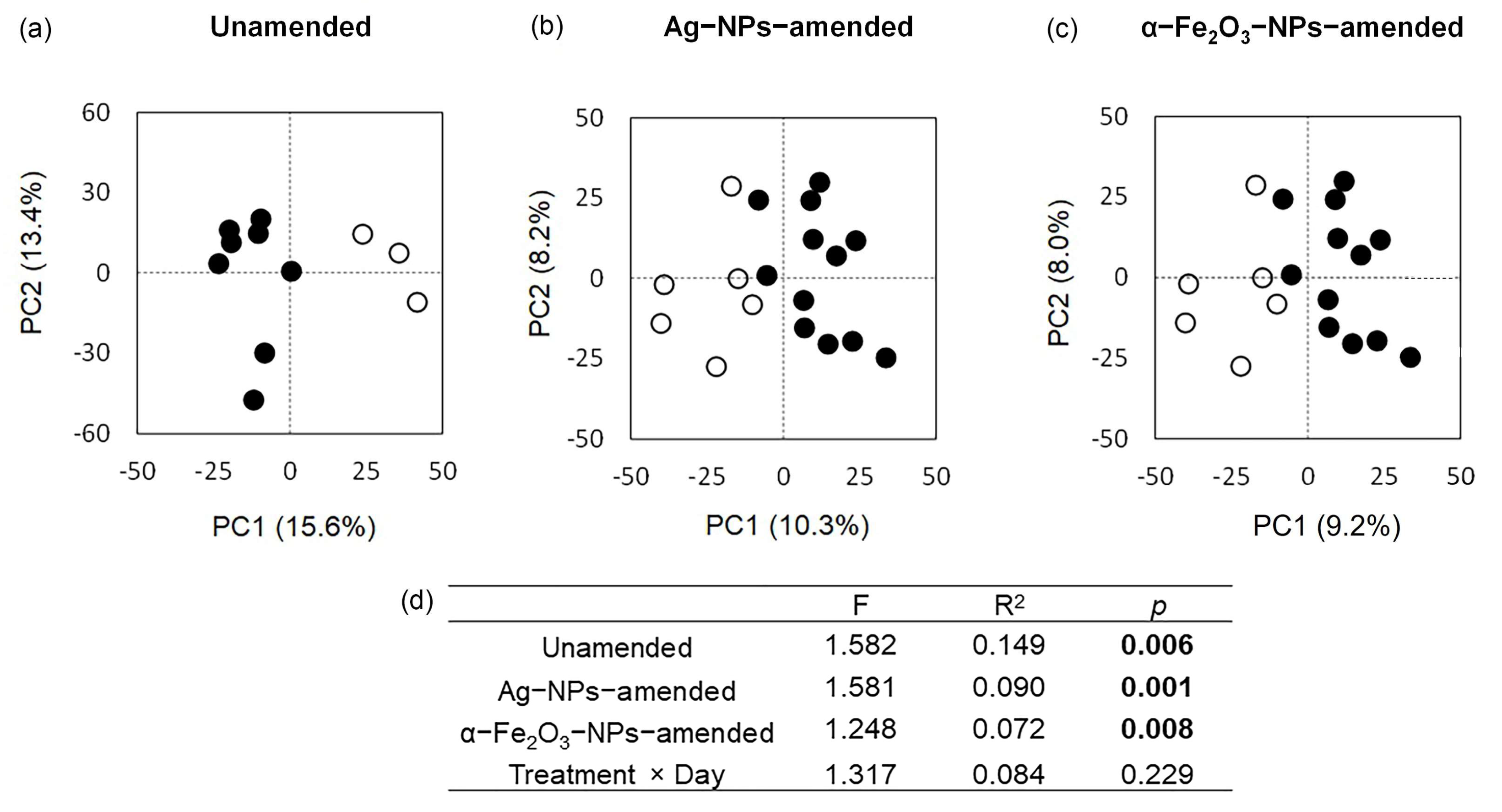
3.5. The Bacterial Community as Affected by Application of the Nanoparticles
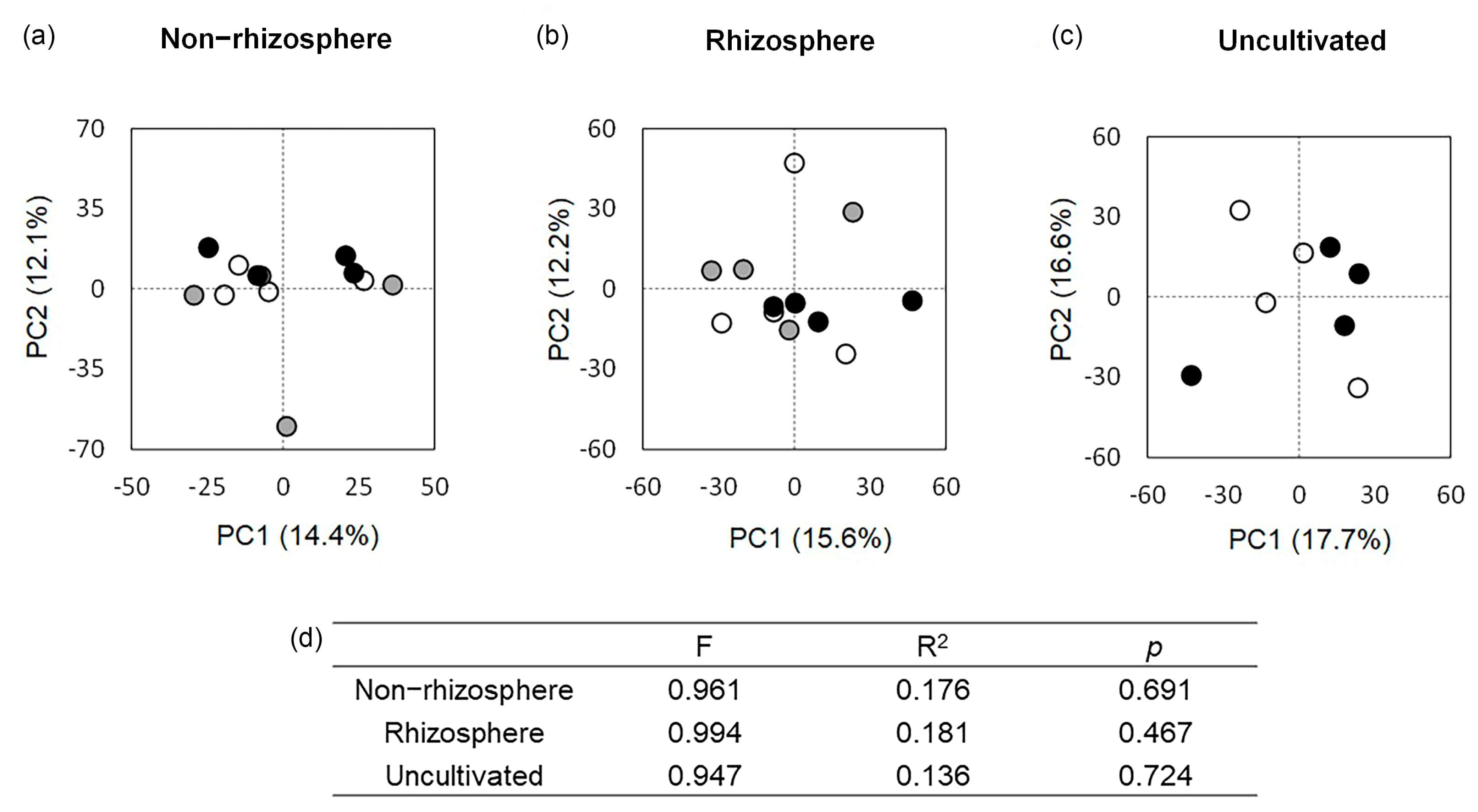
3.6. The Bacterial Community as Affected by Cultivation of the Bean Plants
3.7. Changes in the Bacterial Community over Time
4. Discussion
4.1. Plant Characteristics
4.2. Diversity and Structure of Soil Bacterial Community
4.2.1. Effect of Nanoparticles Application on the Structure and Taxonomic Composition of the Soil Bacterial Community
4.2.2. Effect of Cultivation of Common Bean Plants on the Structure and Taxonomic Composition of the Soil Bacterial Community
4.2.3. Effect of Time of Exposure
4.3. Putative Functional Profile of the Bacterial Community
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dimkpa, C.O. Can nanotechnology deliver the promised benefits without negatively impacting soil microbial life? J. Basic Microbiol. 2014, 54, 889–904. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Saeed, K.; Khan, I. Nanoparticles: Properties, applications and toxicities. Arab. J. Chem. 2019, 12, 908–931. [Google Scholar] [CrossRef]
- Ahmed, B.; Ameen, F.; Rizvi, A.; Ali, K.; Sonbol, H.; Zaidi, A.; Saghir Khan, M.; Musarrat, J. Destruction of cell topography, morphology, membrane, inhibition of respiration, biofilm formation, and bioactive molecule production by nanoparticles of Ag, ZnO, CuO, TiO2, and Al2O3 toward beneficial soil bacteria. ACS Omega 2020, 5, 7861–7876. [Google Scholar] [CrossRef] [PubMed]
- Simonin, M.; Richaume, A. Impact of engineered nanoparticles on the activity, abundance, and diversity of soil microbial communities: A review. Environ. Sci. Pollut. Res. 2015, 22, 13710–13723. [Google Scholar] [CrossRef]
- Beddow, J.; Stolpe, B.; Cole, P.; Lead, J.; Sapp, M.; Lyons, B.P.; Colbeck, I.; Whitby, C. Effects of engineered silver nanoparticles on the growth and activity of ecologically important microbes. Environ. Microbiol. Rep. 2014, 6, 448–458. [Google Scholar] [CrossRef]
- Chavan, S.; Nadanathangam, V. Effects of nanoparticles on plant growth-promoting bacteria in Indian agricultural soil. Agronomy 2019, 9, 140. [Google Scholar] [CrossRef]
- He, S.; Feng, Y.; Ren, H.; Zhang, Y.; Gu, N.; Lin, X. The impact of iron oxide magnetic nanoparticles on the soil bacterial community. J. Soils Sediments 2011, 11, 1408–1417. [Google Scholar] [CrossRef]
- Rui, M.; Ma, C.; Hao, Y.; Guo, J.; Rui, Y.; Tang, X.; Zhao, Q.; Fan, X.; Zhang, Z.; Hou, T.; et al. Iron oxide nanoparticles as a potential iron fertilizer for peanut (Arachis hypogaea). Front. Plant Sci. 2016, 7, 815. [Google Scholar] [CrossRef]
- Rajput, V.; Minkina, T.; Mazarji, M.; Shende, S.; Sushkova, S.; Mandzhieva, S.; Burachevskaya, M.; Chaplygin, V.; Singh, A.; Jatav, H. Accumulation of nanoparticles in the soil-plant systems and their effects on human health. Ann. Agric. Sci. 2020, 65, 137–143. [Google Scholar] [CrossRef]
- Ameen, F.; Alsamhary, K.; Alabdullatif, J.A.; ALNadhari, S. A review on metal-based nanoparticles and their toxicity to beneficial soil bacteria and fungi. Ecotoxicol. Environ. Saf. 2021, 213, 112027. [Google Scholar] [CrossRef]
- Wang, J.; Shu, K.; Zhang, L.; Si, Y. Effects of silver nanoparticles on soil microbial communities and bacterial nitrification in suburban vegetable soils. Pedosphere 2017, 27, 482–490. [Google Scholar] [CrossRef]
- Sillen, W.M.; Thijs, S.; Abbamondi, G.R.; Janssen, J.; Weyens, N.; White, J.C.; Vangronsveld, J. Effects of silver nanoparticles on soil microorganisms and maize biomass are linked in the rhizosphere. Soil Biol. Biochem. 2015, 91, 14–22. [Google Scholar] [CrossRef]
- Karami Mehrian, S.; Heidari, R.; Rahmani, F.; Najafi, S. Effect of chemical synthesis silver nanoparticles on germination indices and seedlings growth in seven varieties of Lycopersicon esculentum Mill (tomato) plants. J. Clust. Sci. 2016, 27, 327–340. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, L.; Si, Y.; Shu, K. Size-dependent cytotoxicity of silver nanoparticles to Azotobacter vinelandii: Growth inhibition, cell injury, oxidative stress and internalization. PLoS ONE 2018, 13, e0209020. [Google Scholar] [CrossRef]
- He, S.; Feng, Y.; Ni, J.; Sun, Y.; Xue, L.; Feng, Y.; Yu, Y.; Lin, X.; Yang, L. Different responses of soil microbial metabolic activity to silver and iron oxide nanoparticles. Chemosphere 2016, 147, 195–202. [Google Scholar] [CrossRef]
- Mahmud, K.; Missaoui, A.; Lee, K.; Ghimire, B.; Presley, H.W.; Makaju, S. Rhizosphere microbiome manipulation for sustainable crop production. Curr. Plant Biol. 2021, 27, 100210. [Google Scholar] [CrossRef]
- Priya, P.; Aneesh, B.; Harikrishnan, K. Genomics as a potential tool to unravel the rhizosphere microbiome interactions on plant health. J. Microbiol. Methods 2021, 185, 106215. [Google Scholar] [CrossRef]
- Thomas, G.W. Soil pH and Soil Acidity. In Methods of Soil Analysis: Chemical Methods; Sparks, D.L., Ed.; American Society of Agronomy: Madison, WI, USA, 1996; pp. 475–490. [Google Scholar]
- Rhoades, J.D.; Mantghi, N.A.; Shause, P.J.; Alves, W. Estimating soil salinity from saturate soilpaste electrical conductivity. Soil Sci. Soc. Am. J. 1989, 53, 428–433. [Google Scholar] [CrossRef]
- Gee, G.W.; Bauder, J.W. Particle size analysis. In Methods of Soil Analysis: Physical and Mineralogical Methods; Klute, A., Ed.; American Society of Agronomy: Madison, WI, USA, 1986; Volume 1, pp. 383–411. [Google Scholar]
- Bremner, J.M. Nitrogen-Total. In Methods of Soil Analysis: Chemical Methods Part 3; Sparks, D.L., Ed.; Soil Science Society of America Inc., American Society of Agronomy: Madison, WI, USA, 1996; pp. 1085–1122. [Google Scholar]
- Hurlbert, S.H. Pseudoreplication and the design of ecological field experiments. Ecol. Monogr. 1984, 54, 187–211. [Google Scholar] [CrossRef]
- Hoffman, C.; Winston, F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 1987, 57, 267–272. [Google Scholar] [CrossRef]
- Sambrook, J.; Rusell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Valenzuela-Encinas, C.; Neria-González, I.; Alcántara-Hernández, R.J.; Enríquez-Aragón, J.A.; Estrada-Alvarado, I.; Hernández-Rodríguez, C.; Dendooven, L.; Marsch, R. Phylogenetic analysis of the archaeal community in an alkaline-saline soil of the former lake Texcoco (Mexico). Extremophiles 2008, 12, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Ceja-Navarro, J.A.; Rivera-Orduna, F.N.; Patiño-Zúñiga, L.; Vila-Sanjurjo, A.; Crossa, J.; Govaerts, B.; Dendooven, L. Phylogenetic and multivariate analyses to determine the effects of different tillage and residue management practices on soil bacterial communities. Appl. Environ. Microbiol. 2010, 76, 3685–3691. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2021. [Google Scholar]
- Chao, A.; Chiu, C.H.; Jost, L. Unifying species diversity, phylogenetic diversity, functional diversity, and related similarity and differentiation measures through hill numbers. Annu. Rev. Ecol. Evol. Syst. 2014, 45, 297–324. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Fernandes, A.D.; Reid, J.N.; Macklaim, J.M.; McMurrough, T.A.; Edgell, D.R.; Gloor, G.B. Unifying the analysis of high-throughput sequencing datasets: Characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2014, 2, 15. [Google Scholar] [CrossRef]
- Husson, F.; Josse, J.; Le, S.; Mazet, J. Multivariate Exploratory Analysis and Data Mining. Package: FactoMineR. 2020. Available online: https://CRAN.R-project.org/package=FactoMineR (accessed on 15 January 2022).
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.B.; Friendly, M.; McGlinn, D.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5-7. 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 15 January 2022).
- Anderson, M.J. Distance-based tests for homogeneity of multivariate dispersions. Biometrics 2006, 62, 245–253. [Google Scholar] [CrossRef]
- Kim, H.Y. Statistical notes for clinical researchers: Effect size. Restor. Dent. Endod. 2015, 40, 328–331. [Google Scholar] [CrossRef]
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome datasets are compositional: And this is not optional. Front. Microbiol. 2017, 8, 2224. [Google Scholar] [CrossRef]
- Prerna, D.I.; Govindaraju, K.; Tamilselvan, S.; Kannan, M.; Vasantharaja, R.; Chaturvedi, S.; Shkolnik, D. Influence of nanoscale micro-nutrient α-Fe2O3 on seed germination, seedling growth, translocation, physiological effects and yield of rice (Oryza sativa) and maize (Zea mays). Plant Physiol. Biochem. 2021, 162, 564–580. [Google Scholar] [CrossRef]
- Haydar, M.S.; Ghosh, S.; Mandal, P. Application of iron oxide nanoparticles as micronutrient fertilizer in mulberry propagation. J. Plant Growth Regul. 2022, 41, 1726–1746. [Google Scholar] [CrossRef]
- Le Wee, J.; Law, M.C.; Chan, Y.S.; Choy, S.Y.; Tiong, A.N.T. The potential of Fe-based magnetic nanomaterials for the agriculture sector. ChemistrySelect 2022, 7, e202104603. [Google Scholar] [CrossRef]
- Boutchuen, A.; Zimmerman, D.; Aich, N.; Masud, A.M.; Arabshahi, A.; Palchoudhury, S. Increased plant growth with hematite nanoparticle fertilizer drop and determining nanoparticle uptake in plants using multimodal approach. J. Nanomater. 2019, 2019, 6890572. [Google Scholar] [CrossRef]
- Sutariya, B.P.; Vyas, T.K.; Faldu, P.R.; Patel, K.G.; Vala, A.K. A microcosm study on effect of iron nanoparticles on Paddy (Oryza sativa) growth. J. Inorg. Organomet. Polym. Mater. 2021, 31, 2425–2435. [Google Scholar] [CrossRef]
- Zuverza-Mena, N.; Armendariz, R.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L. Effects of silver nanoparticles on radish sprouts: Root growth reduction and modifications in the nutritional value. Front. Plant Sci. 2016, 7, 90. [Google Scholar] [CrossRef]
- Li, J.; Hu, J.; Ma, C.; Wang, Y.; Wu, C.; Huang, J.; Xing, B. Uptake, translocation and physiological effects of magnetic iron oxide (γ-Fe2O3) nanoparticles in corn (Zea mays L.). Chemosphere 2016, 159, 326–334. [Google Scholar] [CrossRef]
- Martínez-Fernández, D.; Barroso, D.; Komárek, M. Root water transport of Helianthus annuus L. under iron oxide nanoparticle exposure. Environ. Sci. Pollut. Res. 2016, 23, 1732–1741. [Google Scholar] [CrossRef]
- Lei, C.; Sun, Y.; Tsang, D.; Lin, D. Environmental transformations and ecological effects of iron-based nanoparticles. Environ. Pollut. 2018, 232, 10–30. [Google Scholar] [CrossRef]
- Arif, N.; Yadav, V.; Singh, S.; Singh, S.; Ahmad, P.; Mishra, R.K.; Sharma, S.; Tripathi, D.K.; Dubey, N.K.; Chauhan, D.K. Influence of high and low levels of plant-beneficial heavy metal ions on plant growth and development. Front. Environ. Sci. 2016, 4, 69. [Google Scholar] [CrossRef]
- De Souza, A.; Govea-Alcaide, E.; Masunaga, S.H.; Fajardo-Rosabal, L.; Effenberger, F.; Rossi, L.M.; Jardim, R.D.F. Impact of Fe3O4 nanoparticle on nutrient accumulation in common bean plants grown in soil. SN Appl. Sci. 2019, 1, 308. [Google Scholar] [CrossRef]
- Yang, X.; Alidoust, D.; Wang, C. Effects of iron oxide nanoparticles on the mineral composition and growth of soybean (Glycine max L.) plants. Acta Physiol. Plant 2020, 42, 128. [Google Scholar] [CrossRef]
- Tombuloglu, H.; Slimani, Y.; AlShammari, T.M.; Bargouti, M.; Ozdemir, M.; Tombuloglu, G.; Akhtar, S.; Sabit, H.; Hakeem, K.R.; Almessiere, M.; et al. Uptake, translocation, and physiological effects of hematite (α-Fe2O3) nanoparticles in barley (Hordeum vulgare L.). Environ. Pollut. 2020, 266, 115391. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Chen, Y.; Li, H.; Lu, J.; Zhao, H.; Liu, M.; Nechitaylo, G.S.; Glushchenko, N.N. New insights into the cellular responses to iron nanoparticles in Capsicum annuum. Sci. Rep. 2018, 8, 3228. [Google Scholar] [CrossRef]
- Samarajeewa, A.D.; Velicogna, J.R.; Princz, J.I.; Subasinghe, R.M.; Scroggins, R.P.; Beaudette, L.A. Effect of silver nanoparticles on soil microbial growth, activity and community diversity in a sandy loam soil. Environ. Pollut. 2017, 220, 504–513. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, M.; Zhang, W.; Gardea-Torresdey, J.L.; White, J.C.; Ji, R.; Zhao, L. Silver nanoparticles alter soil microbial community compositions and metabolite profiles in unplanted and cucumber-planted soils. Environ. Sci. Technol. 2020, 54, 3334–3342. [Google Scholar] [CrossRef]
- Shah, V.; Collins, D.; Walker, V.K.; Shah, S. The impact of engineered cobalt, iron, nickel and silver nanoparticles on soil bacterial diversity under field conditions. Environ. Res. Lett. 2014, 9, 024001. [Google Scholar] [CrossRef]
- Liu, J.; Cui, X.; Liu, Z.; Guo, Z.; Yu, Z.; Yao, Q.; Sui, Y.; Jin, J.; Liu, X.; Wang, G. The diversity and geographic distribution of cultivable Bacillus-like bacteria across black soils of Northeast China. Front. Microbiol. 2019, 10, 1424. [Google Scholar] [CrossRef]
- Mhete, M.; Eze, P.N.; Rahube, T.O.; Akinyemi, F.O. Soil properties influence bacterial abundance and diversity under different land-use regimes in semi-arid environments. Sci. Afr. 2020, 7, e00246. [Google Scholar] [CrossRef]
- Saxena, A.K.; Kumar, M.; Chakdar, H.; Anuroopa, N.; Bagyaraj, D.J. Bacillus species in soil as a natural resource for plant health and nutrition. J. Appl. Microbiol. 2020, 128, 1583–1594. [Google Scholar] [CrossRef]
- Cao, J.; Feng, Y.; Lin, X.; Wang, J. Arbuscular mycorrhizal fungi alleviate the negative effects of iron oxide nanoparticles on bacterial community in rhizospheric soils. Front. Environ. Sci. 2016, 4, 10. [Google Scholar] [CrossRef]
- Frenk, S.; Ben-Moshe, T.; Dror, I.; Berkowitz, B.; Minz, D. Effect of metal oxide nanoparticles on microbial community structure and function in two different soil types. PLoS ONE 2013, 8, e84441. [Google Scholar] [CrossRef]
- Claudio, C.; Iorio, E.D.; Liu, Q.; Jiang, Z.; Barrón, V. Iron oxide nanoparticles in soils: Environmental and agronomic importance. J. Nanosci. Nanotechnol. 2017, 17, 4449–4460. [Google Scholar] [CrossRef]
- McGee, C.F.; Storey, S.; Clipson, N.; Doyle, E. Soil microbial community responses to contamination with silver, aluminium oxide and silicon dioxide nanoparticles. Ecotoxicology 2017, 26, 449–458. [Google Scholar] [CrossRef]
- Dizaj, S.M.; Lotfipour, F.; Barzegar-Jalali, M.; Zarrintan, M.H.; Adibkia, K. Antimicrobial activity of the metals and metal oxide nanoparticles. Mater. Sci. Eng. C 2014, 44, 278–284. [Google Scholar] [CrossRef]
- Gold, K.; Slay, B.; Knackstedt, M.; Gaharwar, A.K. Antimicrobial activity of metal and metal-oxide based nanoparticles. Adv. Ther. 2018, 1, 1700033. [Google Scholar] [CrossRef]
- Koskey, G.; Mburu, S.W.; Kimiti, J.M.; Ombori, O.; Maingi, J.M.; Njeru, E.M. Genetic characterization and diversity of Rhizobium isolated from root nodules of mid-altitude climbing bean (Phaseolus vulgaris L.) varieties. Front. Microbiol. 2018, 9, 968. [Google Scholar] [CrossRef]
- Meier, M.J.; Dodge, A.E.; Samarajeewa, A.D.; Beaudette, L.A. Soil exposed to silver nanoparticles reveals significant changes in community structure and altered microbial transcriptional profiles. Environ. Pollut. 2020, 258, 113816. [Google Scholar] [CrossRef]
- Wu, J.; Zhai, Y.; Liu, G.; Bosker, T.; Vijver, M.G.; Peijnenburg, W.J. Dissolution dynamics and accumulation of Ag nanoparticles in a microcosm consisting of a soil-lettuce-rhizosphere bacterial community. ACS Sustain. Chem. Eng. 2021, 9, 16172–16181. [Google Scholar] [CrossRef]
- Liu, G.; Zhang, M.; Jin, Y.; Fan, X.; Xu, J.; Zhu, Y.; Fu, Z.; Pan, X.; Qian, H. The effects of low concentrations of silver nanoparticles on wheat growth, seed quality, and soil microbial communities. Water Air Soil Pollut. 2017, 228, 348. [Google Scholar] [CrossRef]
- Montes de Oca-Vásquez, G.; Solano-Campos, F.; Vega-Baudrit, J.R.; López-Mondéjar, R.; Odriozola, I.; Vera, A.; Moreno, J.L.; Bastida, F. Environmentally relevant concentrations of silver nanoparticles diminish soil microbial biomass but do not alter enzyme activities or microbial diversity. J. Hazard. Mater. 2020, 391, 122224. [Google Scholar] [CrossRef] [PubMed]
- Ling, N.; Wang, T.; Kuzyakov, Y. Rhizosphere bacteriome structure and functions. Nat. Commun. 2022, 13, 836. [Google Scholar] [CrossRef] [PubMed]
- Kögel-Knabner, I. The macromolecular organic composition of plant and microbial residues as inputs to soil organic matter: Fourteen years on. Soil Biol. Biochem. 2017, 105, A3–A8. [Google Scholar] [CrossRef]
- Shi, S.; Richardson, A.E.; O’Callaghan, M.; DeAngelis, K.M.; Jones, E.E.; Stewart, A.; Firestone, M.K.; Condron, L.M. Effects of selected root exudate components on soil bacterial communities. FEMS Microbiol. Ecol. 2011, 77, 600–610. [Google Scholar] [CrossRef]
- Dutta, J.; Bora, U. Rhizosphere microbiome and plant probiotics. In New and Future Developments in Microbial Biotechnology and Bioengineering; Singh, J.S., Ed.; Elsevier: New York, NY, USA, 2019; pp. 273–281. [Google Scholar] [CrossRef]
- Zhou, Y.; Yao, Q.; Zhu, H. Soil organic carbon attenuates the influence of plants on root-associated bacterial community. Front. Microbiol. 2020, 11, 594890. [Google Scholar] [CrossRef]
- Xie, C.; Guo, Z.; Zhang, P.; Yang, J.; Zhang, J.; Ma, Y.; He, X.; Lynch, I.; Zhang, Z. Effect of CeO2 nanoparticles on plant growth and soil microcosm in a soil-plant interactive system. Environ. Pollut. 2022, 300, 118938. [Google Scholar] [CrossRef]
- Ge, Y.; Priester, J.H.; Van De Werfhorst, L.C.; Walker, S.L.; Nisbet, R.M.; An, Y.J.; Schimel, J.P.; Gardea-Torresdey, J.L.; Holden, P.A. Soybean plants modify metal oxide nanoparticle effects on soil bacterial communities. Environ. Sci. Technol. 2014, 48, 13489–13496. [Google Scholar] [CrossRef]
- Vives-Peris, V.; de Ollas, C.; Gómez-Cadenas, A.; Pérez-Clemente, R.M. Root exudates: From plant to rhizosphere and beyond. Plant Cell Rep. 2020, 39, 3–17. [Google Scholar] [CrossRef]
- Peng, C.; Tong, H.; Yuan, P.; Sun, L.; Jiang, L.; Shi, J. Aggregation, sedimentation, and dissolution of copper oxide nanoparticles: Influence of low-molecular-weight organic acids from root exudates. Nanomaterials 2019, 9, 841. [Google Scholar] [CrossRef]
- Yang, J.; Duan, H.; Wang, X.; Zhang, H.; Zhang, Z. Effects of rice root exudates on aggregation, dissolution and bioaccumulation of differently charged Ag nanoparticles. RSC Adv. 2022, 12, 9435–9444. [Google Scholar] [CrossRef]
- Zhai, Y.; Chen, L.; Liu, G.; Song, L.; Arenas-Lago, D.; Kong, L.; Peijnenburg, W.; Vijver, M.G. Compositional and functional responses of bacterial community to titanium dioxide nanoparticles varied with soil heterogeneity and exposure duration. Sci. Total Environ. 2021, 773, 144895. [Google Scholar] [CrossRef]
- Jiao, S.; Chen, W.; Wei, G. Resilience and assemblage of soil microbiome in response to chemical contamination combined with plant growth. Appl. Environ. Microbiol. 2019, 85, e02523-18. [Google Scholar] [CrossRef]
- Tian, H.; Kah, M.; Kariman, K. Are nanoparticles a threat to mycorrhizal and rhizobial symbioses? A Critical Review. Front. Microbiol. 2019, 10, 1660. [Google Scholar] [CrossRef]
- Louca, S.; Polz, M.F.; Mazel, F.; Albright, M.B.N.; Huber, J.A.; O’Connor, M.I.; Ackermann, M.; Hahn, A.S.; Srivastava, D.S.; Crowe, S.A.; et al. Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2018, 2, 936–943. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zarco-González, K.E.; Valle-García, J.D.; Navarro-Noya, Y.E.; Fernández-Luqueño, F.; Dendooven, L. Silver and Hematite Nanoparticles Had a Limited Effect on the Bacterial Community Structure in Soil Cultivated with Phaseolus vulgaris L. Agronomy 2023, 13, 2341. https://doi.org/10.3390/agronomy13092341
Zarco-González KE, Valle-García JD, Navarro-Noya YE, Fernández-Luqueño F, Dendooven L. Silver and Hematite Nanoparticles Had a Limited Effect on the Bacterial Community Structure in Soil Cultivated with Phaseolus vulgaris L. Agronomy. 2023; 13(9):2341. https://doi.org/10.3390/agronomy13092341
Chicago/Turabian StyleZarco-González, Karla E., Jessica D. Valle-García, Yendi E. Navarro-Noya, Fabián Fernández-Luqueño, and Luc Dendooven. 2023. "Silver and Hematite Nanoparticles Had a Limited Effect on the Bacterial Community Structure in Soil Cultivated with Phaseolus vulgaris L." Agronomy 13, no. 9: 2341. https://doi.org/10.3390/agronomy13092341
APA StyleZarco-González, K. E., Valle-García, J. D., Navarro-Noya, Y. E., Fernández-Luqueño, F., & Dendooven, L. (2023). Silver and Hematite Nanoparticles Had a Limited Effect on the Bacterial Community Structure in Soil Cultivated with Phaseolus vulgaris L. Agronomy, 13(9), 2341. https://doi.org/10.3390/agronomy13092341






