Abstract
Tartary buckwheat (Fagopyrum tataricum Gaertn.) belongs to the family of Polygonaceae and is used as a multi-functional plant. R2R3-MYB transcription factors play a crucial part in plant growth and are involved in many biological processes where they regulate their internal environment. To date, there is no documented systematic research on the R2R3-MYB gene family in Tartary buckwheat. Here, domain features, chromosomal location, motif prediction, gene structure, cis-acting elements, as well as the expression pattern of R2R3-MYB transcription factors were analyzed comprehensively in Tartary buckwheat using a bioinformatic approach. Additionally, one R2R3-MYB gene was verified by transgenic Arabidopsis. Results indicate that a total of 152 R2R3-MYB genes were identified with special R2R3 domains and were distributed on 8 chromosomes of Tartary buckwheat. They were further classified into 25 sub-categories via phylogenetic analysis in terms of the R2R3-MYB gene family classification principles of Arabidopsis thaliana. This classification was further supported by analysis of exon–intron structure, motif, and cis-elements. Tandem and segmental duplication existed among the R2R3-MYB gene family of Tartary buckwheat, and there were 5, 8, 27, and 36 FtR2R3-MYB homologous genes, respectively, when comparing with Zea mays, Oryza sativa, Arabidopsis thaliana, and Solanum melongena by synteny analysis. The expression pattern of FtR2R3-MYB genes in different tissue and under salt stress and different light condition showed that members had tissue-specific expression levels and that these members may play diverse functions in plant growth and adaptation to varying environments. In addition, one of the FtR2R3-MYB gene families, FtMYB43, a homologue of AtTT2, clustered with R2R3-MYB from other plant species, which were reported to be involved in the regulation of anthocyanin or proanthocyanidin biosynthesis. This gene was located in the nucleus, and had transcriptional activation activity, indicating that FtMYB43 may be a positive transcript factor of anthocyanin or proanthocyadin biosynthesis. Moreover, the function of FtMYB43 was further verified to improve the production of anthocyanin in transgenic Arabidopsis by overexpression, and qRT-PCR assay implied that FtMYB43 may regulate the expression levels of most structural genes of the anthocyanin biosynthesis pathway in Arabidopsis seedlings. These results provide more insights into the structure and function of the R2R3-MYB gene family and may accelerate the breeding of ornamental buckwheat cultivars.
1. Introduction
In most eukaryotes, transcription factors (TFs) are a class of proteins that exhibit polytypism, multifunction, universality, and multidomain structure. They also have distinctive structures called structural domains [1,2]. The MYB structural domain contains four types of MYB proteins, namely, 1R-, R2R3-, 3R-, and 4R-MYB proteins. The TFs family name is based on the number of N-terminal MYB domains with imperfect repeats [1,2]. Each R domain is comprised of approximately 50 amino residues, including conserved Trp (W), which encodes for helices. This conserved Trp cluster and helix-turn-helix (HTH) combines with DNA, resulting in the execution of protein functions [2]. The MYB TFs family is one of the largest families in plants, and the first MYB gene was isolated from Zea mays [2]. Since then, an increasing number of MYB genes were discovered in other species and published (http://planttfdb.gao-lab.org/ (accessed on 10 April 2023)). The largest subfamily of MYB TFs, R2R3-MYB TFs, are involved in regulating a variety of processes [3,4], including plant morphology [5,6,7], secondary metabolism (such as flavonoid [8], especially anthocyanin biosynthesis [9], carotenoid biosynthesis [10], lignin biosynthesis [11]), biotic and abiotic response [12], and other life processes. They either do so individually or in combination with other TFs [11,12]. Therefore, research of R2R3-MYB TFs attracted attention from both domestic and foreign researchers [1]. At present, R2R3-MYB gene families were identified in many plants, including Arabidopsis thaliana (126) [2], Oryza sativa (130) [13], Hypericum perforatum (109) [14], and Camellia sinensis (122) [15] among others [16,17].
Tartary buckwheat (Fagopyrum tataricum Gaertn.) is a kind of important pseudocereal and domesticated crop that belongs to the genus Fagopyrum within the family Polygonaceae. Tartary buckwheat is native to the mountains of southwest China and widely grown around the world, including in Asia, America, and Europe, and it is characterized as a natural functionality plant, which is a combination of medicine, nutrient, and health [18,19,20]. Edible seeds and sprouts of Tartary buckwheat not only contain rich nutrients, including dietary fiber, protein, starch, numerous amino acids, etc., but also contain various bioactive compounds, especially abundant flavonoids, including rutin, anthocyanins, and proanthocyanidins [20,21], and a large number of studies suggested that flavonoids are beneficial for human health, where they function in sterilization, anti-infection, oxidation resistance, and prevention of arteriosclerosis [18,22,23,24,25,26]. Therefore, Tartary buckwheat became a popular daily food and found favor in the consumer’s eyes [27]. Flavonoid biosynthesis requires not only synthetase enzymes, but also transcription factors for modulation [28,29]. Numerous works of literature reported that R2R3-MYB TFs are involved in flavonoid biosynthesis processes in many species, such as Zea mays, Arabidopsis thaliana, Solanum lycopersicum, Malus domestica, and Medicago truncatula [17,30,31,32]. Recent studies suggested that significant accumulations of flavonoid were detected through overexpression of FtPinG0002593000.01, FtPinG0009153900.01, and FtPinG0004608100.01 (GenBank ID: MT333222) of R2R3-MYB members in Tartary buckwheat [3,33,34]. However, systematic and comprehensive bioinformatic analysis of R2R3-MYB TFs of Tartary buckwheat remains elusive, through which more MYBs TFs involved in regulation of flavonoid biosynthesis may be identified.
The Tartary buckwheat genome was sequenced and published [35], which is useful for researching its gene families and gene functions. In this study, 152 R2R3-MYB genes were identified from the Tartary buckwheat genome and bioinformatics analysis including structure, chromosomal distribution, conserved motifs, phylogeny, cis-acting elements, synteny, and expression patterns were conducted. Additionally, one FtR2R3-MYB gene was verified to regulate anthocyanin accumulation. These results provide a research basis and scientific reference for subsequent functional studies of Tartary buckwheat transcription factors.
2. Materials and Methods
2.1. Plant Materials, RNA Extraction, and cDNA Synthesis
Seeds of Tartary buckwheat cultivar ‘Jinqiao No. 2′ from the Research Center of Buckwheat Industry Technology, Guizhou Normal University, were cultivated according to the previous method [35]. Seven-day-old seedlings were collected and snap-frozen in liquid N2, then stored at −80 °C until use.
Total RNAs of samples were extracted using the OminiPlant RNA kit (Kangwei, China), and the concentration and quality of RNAs were determined by spectrophotometer (Yuanxi, China), then cDNA was synthesized according to the manufacturer’s instruction of ReverTra Ace® qPCR RT kit (Toyobo, Japan).
2.2. Identification and Chromosomal Localization Analysis of FtR2R3-MYBs
Initially, the gene ID and classification principle of the Arabidopsis R2R3-MYB subfamily with 126 members were obtained from previous literature [2], and their protein sequences were downloaded from the TAIR database (https://www.arabidopsis.org/ (accessed on 12 April 2023)). At the same time, the Tartary buckwheat genome information was searched and downloaded from the Tartary buckwheat genome database (http://www.mbkbase.org/Pinku1/ (accessed on 12 April 2023)). Furthermore, based on the AtR2R3-MYBs, protein query using BLASTP programs to retrieve local Tartary buckwheat protein data was performed, which obtained the preliminary homologous sequences of Tartary buckwheat. This was followed by the removal of the repeat sequences. The candidates were then submitted to Expasy (https://web.expasy.org/protparam/ (accessed on 14 April 2023)) and SMART (http://smart.embl.de/ (accessed on 14 April 2023)) software for a conserved domain of R2R3-MYB proteins analysis, where protein sequences without these typical characteristics were deleted. These identified sequences were finally analyzed to visualize the chromosomal location using the TBtools program (v1.068) [36].
2.3. Physico-Chemical Analysis of R2R3-MYB Transcription Factors
Physicochemical properties, subcellular localization, and secondary structure of identified FtR2R3-MYB proteins above were analyzed using ExPaSy (https://web.expasy.org/protparam/ (accessed on 15 April 2023)), PSOR (https://www.genscript.com/psort.html#_NO_LINK_PROXY_ (accessed on 15 April 2023)), and SOPMA (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html (accessed on 16 April 2023)) websites, respectively.
2.4. Conserved Domains and Phylogenetic Tree Analysis
Sequence alignment analysis of these identified FtR2R3-MYB proteins was performed by MEGA 7.0 software and the R2 and R3 conserved sequences were extracted. The graphical R2 and R3 logos were obtained by WebLogo (https://weblogo.berkeley.edu/logo.cgi (accessed on 16 April 2023)). The multiple-sequence alignment of these FtR2R3-MYB proteins was performed using ClustalW with default settings, and the neighbor-joining (NJ) phylogenetic tree (boot strap 1000 repeats) was constructed by MEGA 7.0 software.
Thirteen proteins (AtMYB60, AtMYB3, AtMYB4, AtMYB7, AtMYB123, AtMYB75, AtMYB90, AtMYB113, AtMYB114, AtMYB11, AtMYB12, AtMYB3, and AtMYB111) play a regulatory role in anthocyanin accumulation in Arabidopsis [1,2,3,4,5,8,9,37], which were chosen as the query sequences to screen out the MYB proteins with similar function from 152 FtR2R3-MYB. An NJ tree (1000 replicates) was built by the MEGA 7.0 program according to the sequences mentioned above. Furthermore, BLAST, based on the sequence of candidate proteins, was utilized to retrieve R2R3-MYB factors modulating anthocyanins in other species. An NJ tree was finally constructed to further explore function.
2.5. Conserved Motif, Gene Structure, Cis-Elements and Synteny Analysis
To better understand the structural characteristics and promoter regions of FtR2R3-MYB genes as well as their evolutionary relationship, the exon–intron structures, cis-elements, and conserved motifs were predicted; 2000 bp upstream of the start codon of the promoter region of each gene was chosen by TBtools software. All FtR2R3-MYB genes were submitted to the GSDS (http://gsds.gao-lab.org/ (accessed on 17 April 2023)) and PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) websites for analysis of gene structure and cis-elements, respectively. Conserved motifs were predicted by the MEME (http://memesuite.org/tools/meme (accessed on 17 April 2023)) online tool using FtR2R3-MYB protein sequences. Finally, these results were uploaded to the TBtools program for graphical display [36].
Synteny analysis was conducted to explore the conservation homology of R2R3-MYB genes and relative order among Tartary buckwheat and other plants (including O. sativa, A. thaliana, Z. mays, and S. lycopersicum) during their evolutionary processes by TBtools software. All R2R3-MYB protein sequences and annotation information of these four plant species were downloaded from the Rice Genome Database (http://rice.uga.edu/ (accessed on 18 April 2023)), TAIR database, and the plant genome project (http://plantgdb.org (accessed on 18 April 2023)), respectively. While the information on the Tartary buckwheat genome was obtained as mentioned above. The syntenic results were eventually visualized via the Multiple Collinear Scan Kit (MCScanX) of the TBtools program [36].
2.6. Expression Patterns of Ftr2r3-MYB in Different Tissues, under Different Light and Salt Treatments
The expression patterns of these FtR2R3-MYB genes in five different tissues (root, stem, leaf, flower, and seed at three different stages of development) were analyzed by the TBtools program based on RNA-seq data for investigating the biological functions in the growth and development process, and RNA-seq data were obtained from a previous article by Zhao et al. [35]. Meanwhile, the published RNA-seq data of different illumination treatments (blue light, dark, far-red light, and red light) were obtained from NCBI (SRP157461) [38] for exploring the responses of FtR2R3-MYBs to light signals. In addition, the transcriptional data of FtR2R3-MYB genes’ response to salt stress under 0 h and 24 h treatments were downloaded from NCBI (SRP118793) [39]. The TBtools program was utilized to draw heatmaps of these gene expression levels based on the normalized TPM values of these RNA-seq data [36].
2.7. Arabidopsis Protoplast Transient Expression Assay
The CDS of FtMYB43 were amplified and cloned into the pC1300S-GFP vector by homologous recombination method, and the specific primer sequences are listed in Table S1. The vector Ghd7:RFP was used as a reference, which is located in the nucleus. These two vectors pC1300S-GFP-FtMYB43 and Ghd7:RFP were co-transformed into Arabidopsis protoplast according to the protocol described by Xiong et al. [40], while empty vector pC1300S-GFP and Ghd7:RFP were co-transformed as a control. Fluorescence signals of GFP or RFP were observed using confocal laser scanning microscopy (STELLARIS 8).
2.8. Yeast-Two-Hybrid Analysis
The CDS of FtMYB43 was amplified by specific primers (listed in Table S1) and recombined into a pGBKT7 (BD) vector. Then, the combination of FtMYB43-BD was co-transformed into the AH109 yeast strain with the pGADT7 (AD) vector. While AD co-transformed with BD and pGADT7-T co-transformed with pGBKT7-p53 were used as a negative and positive control, respectively. Additionally, the yeast cells were cultured on a medium lacking Leu and Trp (SD/-L/-T), then transferred several clones on a medium lacking Leu, Trp, His, and Ade (SD/-L/-T/-H/-A) and SD/-L/-T/-H/-A with X-α-gal, respectively.
2.9. Overexpression of FtMYB43 in Arabidopsis
Using the cDNA obtained above as the template, and full-length specific primers of FtMYB43 FtMYB43-F1/FtMYB43-R1 (Containing SpeI and AscI cleavage sites, respectively, Table S1), the FtMYB43 gene fragment was amplified by PCR program by high-fidelity thermostable DNA polymerase (TaKaRa, Beijing, China). The amplified PCR fragment was recombined with overexpression vector pMDC83 using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China). Furthermore, the 35S:FtMYB43 recombinant plasmid was transformed into Arabidopsis by Agrobacterium (GV3101)-mediated floral-dip method [41,42]. The T3 homozygous transgenic lines were used for further analysis.
2.10. Determination of Anthocyanin Content
Total anthocyanins were extracted (three biological replicates) from fresh leaves of WT and transgenic Arabidopsis plants according to Park et al. [43]. The extracted solutions were measured at absorptions of 530 and 657 nm by UV9600 spectrophotometer (Beifen-Ruili, Beijing, China), and total anthocyanin content was calculated according to the formula QAnthocyanin = (A530 − 0.25 × A657)/M (Q represents anthocyanin content, M represents sample weight).
2.11. Real-Time Quantitative PCR (qRT-PCR)
The expression levels of structural genes on the anthocyanin synthesis pathway (CHS, CHI, F3′H, DFR, ANS, and UFGT) in wild-type (WT) and transgenic Arabidopsis thaliana seedlings were detected by qRT-PCR. QRT-PCR was carried on the C1000™ thermal cycler coupled with a CFX96TM detection module (Bio-Rad, Hercules, CA, USA) using the 2× iQ™ SYBR Green Super mix (Bio-Rad, USA). AtActin [44] was used as the internal control. The PCR procedure was: 95 °C for 120 s, 40 cycles of 95 °C for 15 s, and 55 °C for 30 s. The expression levels of anthocyanin pathway genes were obtained by the 2−ΔΔCT method with three technical replicates. The primers of these genes are listed in Table S1.
3. Results
3.1. Identification and Chromosome Location of FtR2R3-MYB Genes
A total of 152 R2R3-MYB genes were identified from the Tartary buckwheat genome based on the number of MYB DNA-binding domains with the name of FtMYB1~FtMYB152 according to their chromosome physical position (Table S2 and Figure 1). These genes were randomly distributed on the eight chromosomes of the Tartary buckwheat, among which, Chr 1 contained the most abundant members, followed by Chr 7 and Chr 2, on which, 25 and 23 FtR2R3-MYB genes were distributed, respectively. Chr 8 contained the least number of these genes. Additionally, there were two pairs of tandem repeat genes on Chr1 (FtMYB10 and FtMYB11, FtMYB12 and FtMYB13), and one pair of tandem repeat genes on Chr 8 (FtMYB149 and FtMYB150) (Figure 1).
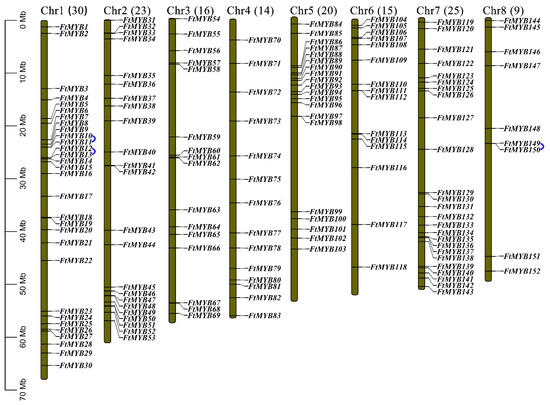
Figure 1.
The distribution mapping of FtR2R3-MYB gene on the eight chromosomes (the blue lines represent tandem repeat of genes).
3.2. Protein Information Analysis of FtR2R3-MYB TFs
These FtR2R3-MYB genes’ structures and their corresponding proteins’ physicochemical characteristics were analyzed and used for classification, with the resulting information being shown in Table S3. The number of amino acids encoded by FtR2R3-MYB genes ranged from 102 (FtMYB89) to 947 amino acids (aa) (FtMYB62), and the range of 200~400 aa was the most abundant, accounting for 80%. The molecular weight (MW) ranged from 11.81 kDa (FtMYB89) to 106.56 kDa (FtMYB62), and the pI values varied from 4.5 (FtMYB40) to 10.66 (FtMYB9). Most proteins (141) were determined as unstable proteins based on an instability index >40. Additionally, the predicted grand average of hydropathicity (GRAVY) values were all less than zero, indicating that all the 152 FtR2R3-MYB proteins were hydrophilic. Moreover, 133 members (accounting for 87.50%) contained random coil, which was the main secondary structure element, followed by an alpha helix. The results of subcellular localization prediction show nearly all these proteins located in the nucleus, indicating that they may function to regulate other genes’ expression as transcription factors.
3.3. Conserved Domains and Phylogenetic Relationship Analysis
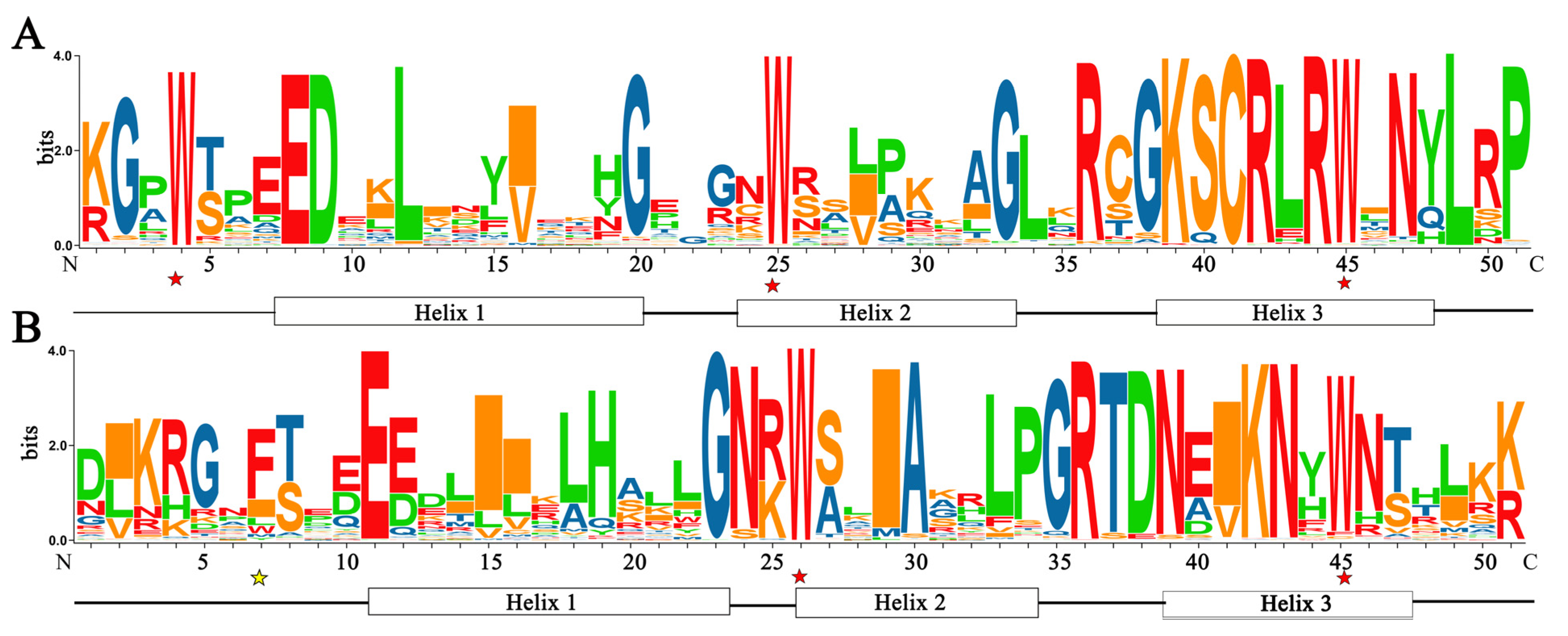
R2R3-MYB transcription factors are typically characterized by two MYB domains (R2 and R3), and each R contains about 50 amino acid residues, which could fold into a helix-turn-helix [45]. Conserved domains analysis revealed that three Trp (W) residues at positions 4, 25, and 45 of the R2 repeat sequence were highly conserved and the second and third Trp residues in the R3 repeat were highly conserved (Figure 2). Generally, the first Trp residue in the R3 repeat was replaced by a Phe (F) or Ile (I) residue [15]. These conserved amino acid residues play an important role in maintaining the structure of HTH and exerting the function of R2R3-MYB TF.
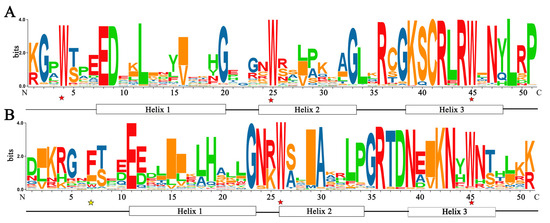
Figure 2.
The domain features of FtR2R3-MYB proteins. (A) R2 MYB repeats. (B) R3 MYB repeats (the abscissa indicates the amino acid position, and the height of the amino acid character indicates the proportion of the position. Completely conserved amino acid sites are marked with red asterisks, slightly conserved amino acid sites are marked with yellow asterisks, and helix1~3 in the boxes represent the positions of the three α-helices; the bit values indicate the specific degree of amino distribution).
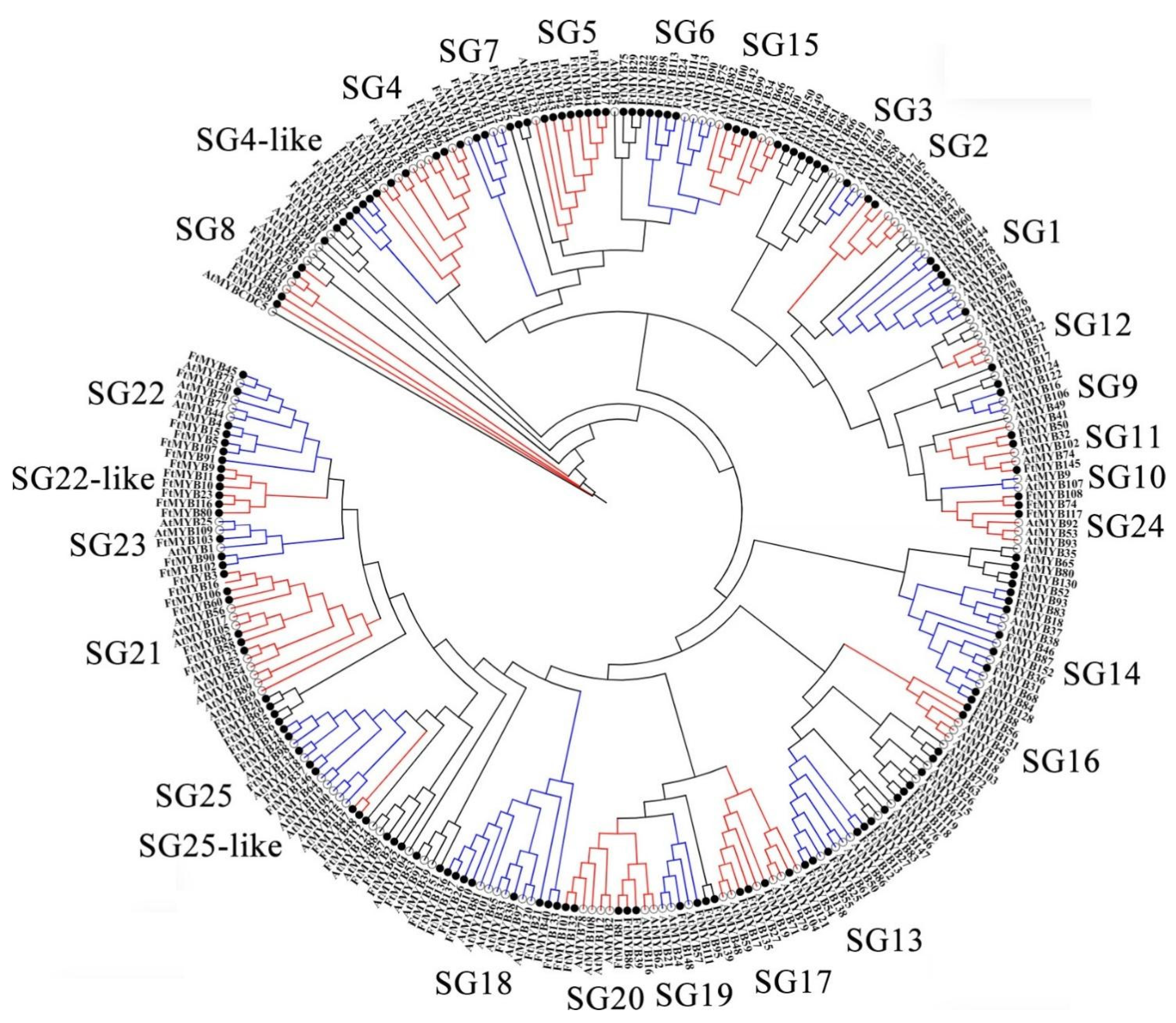
To explore the functions of FtR2R3-MYB TFs, a phylogenetic tree identified 152 FtR2R3-MYBs and well-annotated 126 AtR2R3-MYBs was constructed by the MEGA7 program based on the multiple sequence alignment of these proteins’ sequences. The categorization of FtR2R3-MYB TFs was referred to as the classification of the Arabidopsis R2R3-MYB family described by Stracke et al. [2] and Zhang et al. [35]. All these R2R3-MYBs were classified into 25 subgroups (Figure 3), which were consistent with previous studies [2,35]. Interestingly, no member of FtR2R3-MYBs was among the SG3, SG10, and SG12 subfamilies, and all the 152 FtR2R3-MYBs were divided into 22 subgroups, which implied that the expansion of R2R3-MYB members happened before the divergence of the Tartary buckwheat and Arabidopsis.
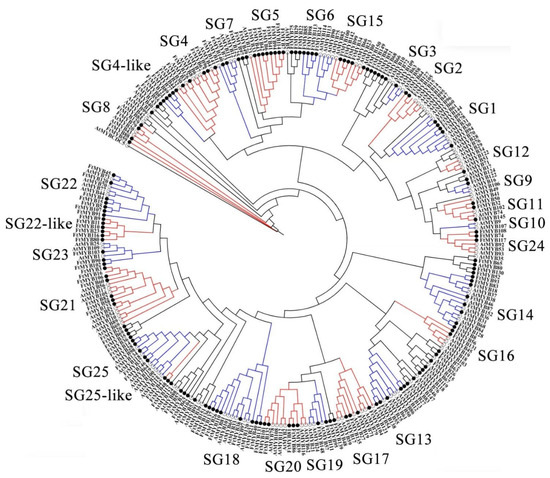
Figure 3.
Phylogenetic tree of R2R3-MYB proteins from Arabidopsis and Tartary buckwheat marked with hollow black circles and solid black circles, respectively.
3.4. Analysis of Motifs, Gene Structure and Cis-Acting Elements of FtR2R3-MYB TFs
To understand the evolutionary relationship and functional differentiation of the FtR2R3-MYB gene family, the gene structural compositions, phylogenetic tree, conserved motif, as well as cis-elements of 152 FtR2R3-MYB genes were investigated (Figure S1). The FtR2R3-MYB proteins were clustered into 32 (A1~A32) subgroups and those proteins that clustered in the same branch had similar conserved motifs, and most of the motif elements were concentrated in the N-terminus (Figure S1A). There were 10 types of motifs identified in the FtR2R3-MYBs family, and each gene contained 3 to 7 types of motifs, while the number of motifs varied from 3 to 7 (Figure S1B). The difference in the type and the number of motifs may be one of the reasons for the diverse functions of R2R3-MYB proteins, which provides a theoretical reference for further research on the functions of R2R3-MYB genes. Structural analysis indicated that up to 98.6% (150) of 152 genes contained no more than 4 exons, except for FtMYB2 and FtMYB57, which contained 4 and 8 introns, respectively (Figure S1C). Among them, 97 FtR2R3-MYBs contained 3 exons, accounting for 63.8% of the total. In addition, 17 members had no introns.
The cis-acting elements of 152 FtR2R3-MYB genes were predicted by the PlantCARE tool for a better understanding of the functions, and 4 types of cis-acting elements were identified, including phytohormone, light, stress-responsive, and plant growth (Figure S1D). Further analysis showed that there were 62 kinds of elements, with 34 for light responses, 7 for stress responses, 11 for phytohormone tolerances, and 10 for plant development-related responses (Table S4a and Figure S2). Among them, elements for light response were the most abundant, such as 449 G-box, 373 box 4, 240 GT1-motifs, and 140 TCT-motifs in all FtR2R3-MYBs’ promoter regions (Table S4b). Meanwhile, response elements to stress and phytohormone mainly contained response to anaerobic (ARE), response to low temperature (LTR), response to ABA (ABRE), and motif response to MeJA (TGACG motif). It is quite interesting that the dehydration responsive element (DRE) was found in the promoters of FtMYB23 and FtMYB66 genes, which leads to the speculation that both might play a regulatory role in multiple stress response processes [46]. In addition, O2-site, CAT-box, and GCN4-motif related to plant growth regulation were identified in the promoters of FtR2R3-MYBs [47]. It should be mentioned that MBSI (an MYB binding site involved in flavonoid biosynthetic genes regulation, with sequence TTTTTACGGTTA) was present in the promoters of 28 members, implying that these may have effects on flavonoid biosynthesis [16].
3.5. Synteny Analysis of R2R3-MYB TFs
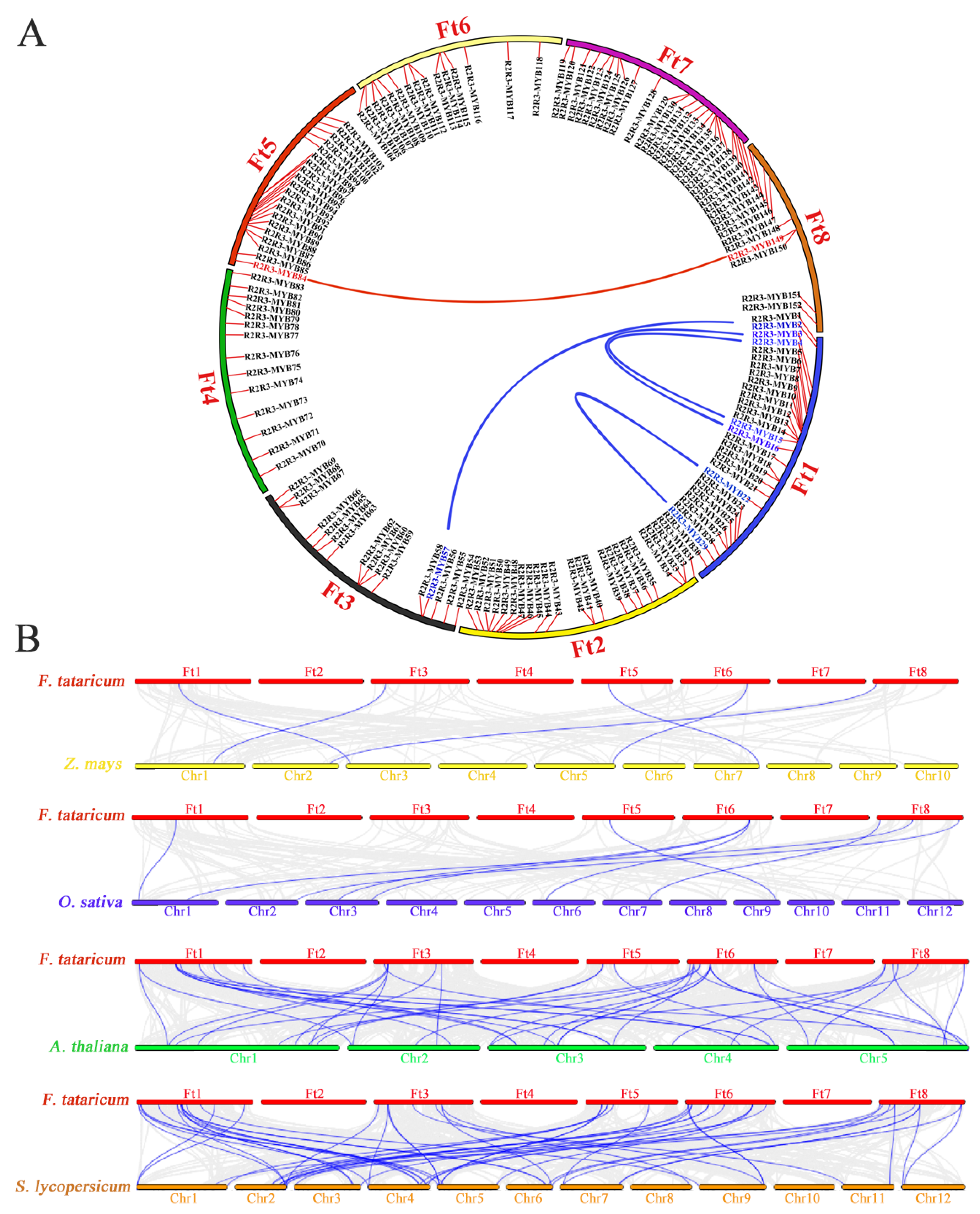
Most plants underwent whole-genome duplication events or polyploidy, and these duplication sequences may augment via natural selection and play an important role in species evolution. Three pairs of tandem duplications were found in the FtR2R3-MYB gene family according to the distribution of chromosomes of FtR2R3-MYB genes including FtMYB10-FtMYB11, FtMYB12-FtMYB13, and FtMYB149-FtMY150 (Figure 1), and further segmental duplication of FtR2R3-MYBs in eight chromosomes indicated that there were another five pairs of segmental duplications, including FtMYB22-FtMYB29, FtMYB3-FtMYB16, FtMYB4-FtMYB15, FtMYB2-FtMYB57, and FtMYB84-FtMYB149, (Figure 4A). Eight pairs of genes were duplicated according to the combined analysis of distribution on chromosomes with segmental duplications of FtR2R3-MYB genes (Figure 1 and Figure 4A), and their values of Ka, Ks, and Ka/Ks ratios are shown in Table S5. Except for FtMYB10-FtMYB11, the other seven pairs of homologous genes with Ka/Ks values in the range 0 to 1 possibly evolved under purifying selection.
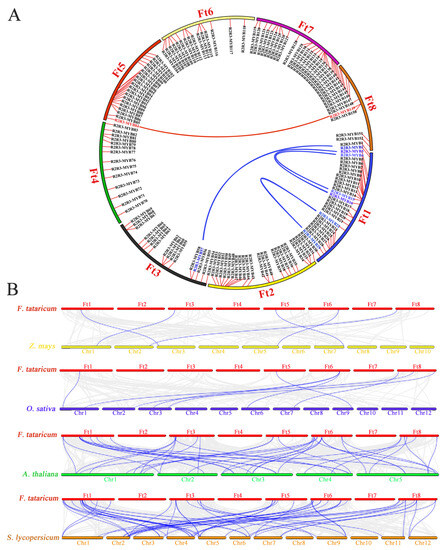
Figure 4.
Synteny of the R2R3-MYB proteins. (A) Segmental duplication of R2R3-MYB in Tartary buckwheat. Red short lines indicate the location of each FtR2R3-MYB gene on the chromosomes of Tartary buckwheat, and long blue and red lines indicate Synteny among FtR2R3-MYB genes. (B) Collinear of R2R3-MYB in four species. Gray lines represent genome-wide collinear genes between Tartary buckwheat and other plant species, and blue lines represent collinear of R2R3-MYB genes between tartary buckwheat and other plant species.
The syntenic relationships of R2R3-MYB genes among Tartary buckwheat and other 4 plant species suggested that there were 5 genes in Tartary buckwheat that had collinearity with 5 genes in Z. mays, 6 members in Tartary buckwheat that were linked to 8 members in O. sativa, 27 genes in Tartary buckwheat that collinearity with 34 genes in A. thaliana, as well as 36 genes in Tartary buckwheat had synteny with 44 genes in S. lycopersicum (Figure 4B). Remarkably, as one of the dicots, Tartary buckwheat had more homologous pairs with the other two dicots compared with other two monocotyledons, which indicated that Tartary buckwheat was relatively conserved in the evolutionary processes of dicots.
3.6. Expression Patterns of FtR2R3-MYBs in Different Tissues, and under Salt and Light Treatments
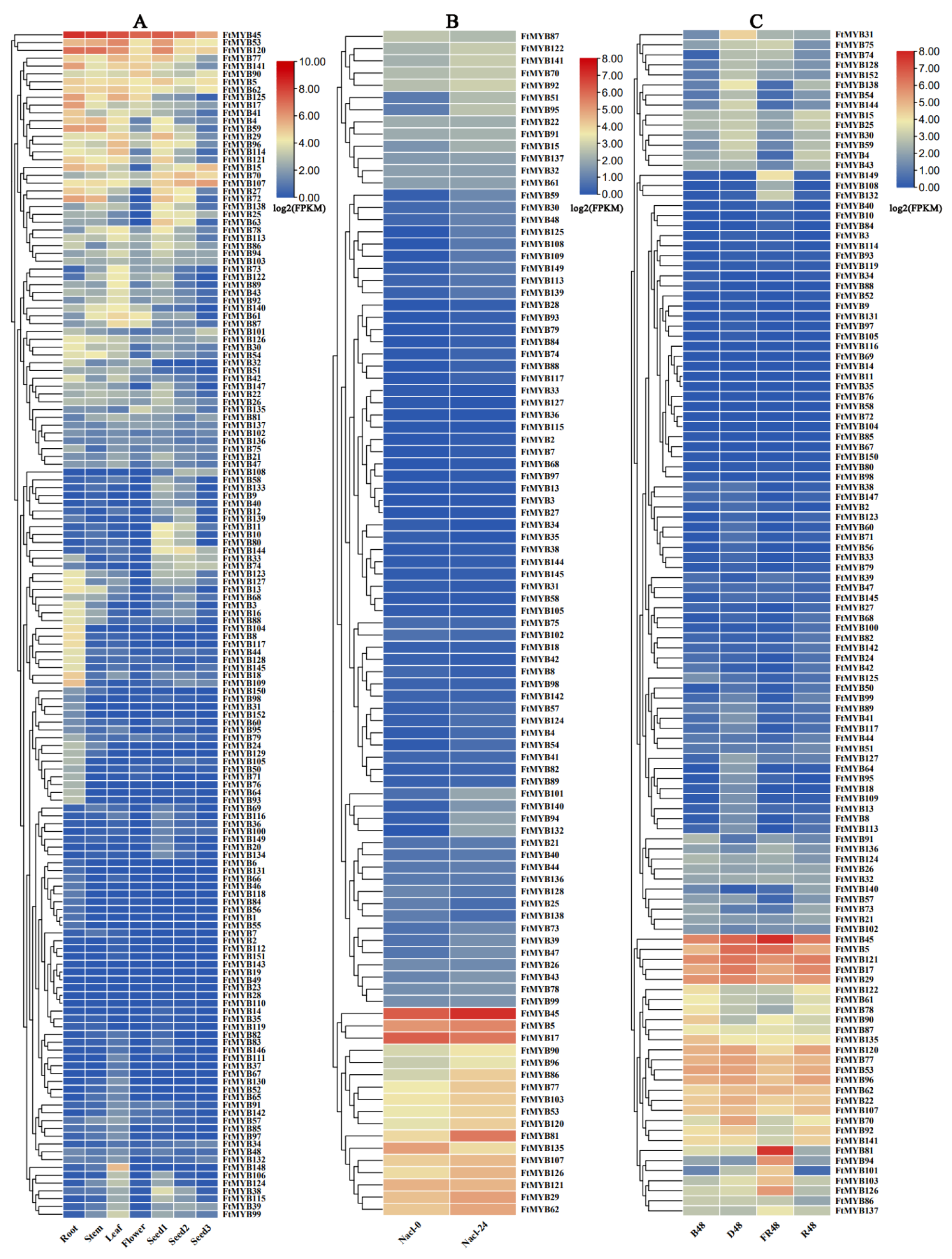
Investigation of the tissue-specific expression of these FtR2R3-MYBs showed that the FtMYB45 gene exhibited a higher expression pattern in each tissue. Originally from the same group as FtMYB45, FtMYB53 and FtMYB120 genes showed a higher expression level, especially in root, stem, leaf, and seed. FtMYB15/17/18/41/59/72/109/125/141, FtMYB4/40, and FtMYB53/62/77/90 exhibited relatively higher expression in root, stem, and flower, respectively. FtMYB15 and FtMYB121, both belonging to the same branch, were highly expressed in seed and increased with seed development. However, FtMYB19 was unexpressed in all tissues, leading to an assumption that it was not detected or could be a pseudogene. FtMYB1/104/131/143, FtMYB111/151, and FtMYB49 were only expressed in roots, leaves, and flowers, respectively (Figure 5A). These results suggest that members in the same branch had similar expression patterns, and most of them shared common cis-elements, such as how FtMYB53, FtMYB90, and FtMYB135 share CGTCAC-motif, G-box, ARE, TCA-element, and TC-rich repeat in their promoters, which may due to their similar function in regulating the growth of Tartary buckwheat.
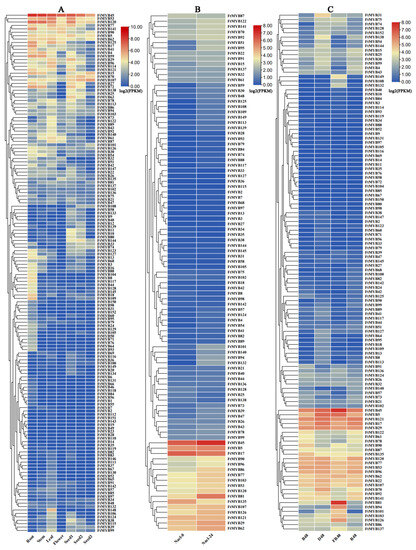
Figure 5.
The expression patterns of R2R3-MYB genes of Tartary buckwheat in terms of RNA-seq data. (A) The expression profiling in different tissues (roots, stems, leaves, flowers and seed with prefilling, filing, and mature stage); (B) the expression profiling under salt stress (Nacl-0: 0 h salinity treatment; Nacl-24: 24 h salinity treatment); and (C) the expression profiling under light treatment for 48 h (D48: dark; FR48: far-red light; B48: blue light; and R48: and red light). Detailed expression values (FPKM) are listed in Table S6.
The expression levels analysis of FtR2R3-MYB genes under salt treatment showed that except for 55 genes that had no detectable expression levels, most genes exhibited a low expression level and had no significant differences under salt treatment for 0 h and 24 h, suggesting that these genes may have no response to salt stress. However, FtMYB6, FtMYB15, FtMYB45, FtMYB51, and FtMYB81 had an increased expression level during salt stress treatment (Figure 5B), which implied that they may likely play a key role in the regulation of Tartary buckwheat response to salt stress.
Cis-acting elements analysis of the FtR2R3-MYB genes revealed that promoters of each gene contained light-responsive elements. The expression patterns of FtR2R3-MYBs in Tartary buckwheat seedlings treated with different light for 48 h (blue light (B48), dark (D48), far-red light (FR48), and red light (R48)) were analyzed. The results show that, except for 34 genes, the rest had detectable expression levels. Among them, FtMYB53, FtMYB90, and FtMYB135 were highly expressed in B48 seedlings compared with other light conditions. FtMYB17, FtMYB85, FtMYB70, and FtMYB121 were expressed highly in D48 seedlings, while FtMYB107 and FtMYB120 had a relatively higher accumulation in R48 seedlings. FtMYB45, FtMYB81, and FtMYB126 had higher accumulation in FR48 seedlings compared with other illumination treatments (Figure 5C). These results suggest that different FtR2R3-MYB members respond to different light treatments.
3.7. Potential FtR2R3-MYB Proteins Involved in Anthocyanin Biosynthesis
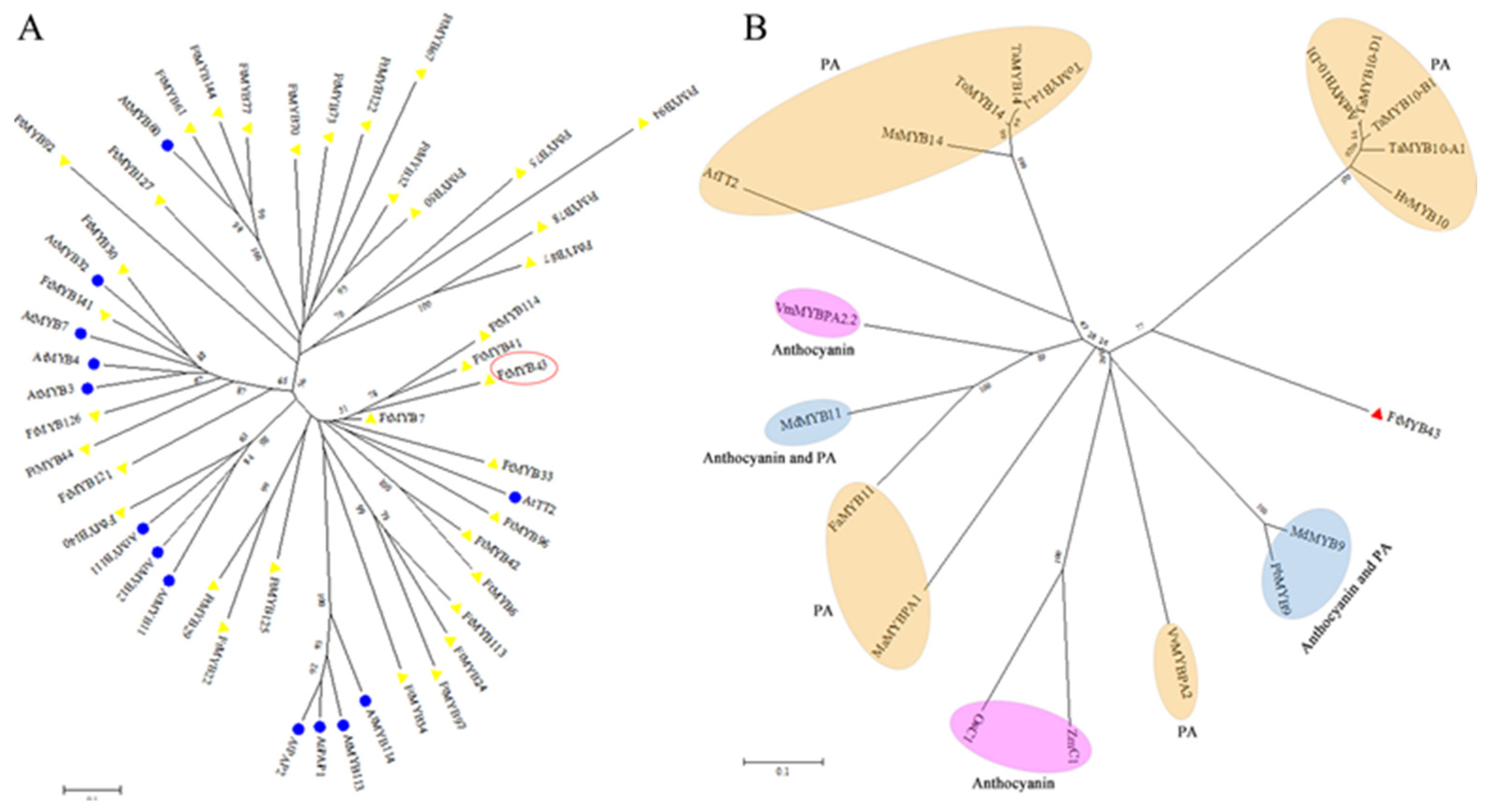
Previous reports demonstrated that some R2R3-MYB TFs had a regulatory effect on anthocyanin production, including the AtMYB3, AtMYB4, AtMYB7, AtMYB60, AtMYB11, AtMYB12, AtMYB111, AtMYB32, AtPAP1 (AtMYB75), AtPAP1 (AtMYB90), AtMYB113, and AtMYB114 from Arabidopsis, whereas AtTT2 (AtMYB123) was confirmed as having a negative regulatory mechanism of anthocyanin while promoting proancyanidins (PA) accumulation [1,2,3,4,5,8,9,37]. Here, 28 FtR2R3-MYB proteins were obtained based on the phylogenic analysis between Arabidopsis proteins mentioned above and Tartary buckwheat (Figure 6A), which perhaps participated in the modulation of anthocyanin synthesis. Among them, 11 FtR2R3-MYB candidate proteins (FtMYB22, FtMYB29, FtMYB30, FtMYB44, FtMYB92, FtMYB121, FtMYB125, FtMYB126, FtMYB127, FtMYB140, and FtMYB141) clustered together with those proteins that have positive effects on anthocyanin synthesis [9,37,42,48] (Figure 6A). Therefore, it was speculated that the 11 members possessed a similar function. Other members (FtMYB7, FtMYB33, FtMYB41, FtMYB43, FtMYB96, and FtMYB114) in the same subclass with AtTT2 may be related to anthocyanin and PA pigmentation (Figure 6B). One TT2 TF type, FtMYB43, clustered with AtTT2 in SG5, indicating its potential function in pigmentation. In addition, an unrooted phylogenetic tree was constructed by utilizing sequences of FtMYB43 and other species of R2R3-MYB proteins, which were reported to regulate anthocyanins and Pas [9,28,37,44,48,49,50,51,52,53]. FtMYB43 had a close relationship with one branch that related to PA biosynthesis and another branch that regulates both anthocyanin and PA biosynthesis. Therefore, FtMYB43 was thought to possess similar functions and its function was further verified as described below.
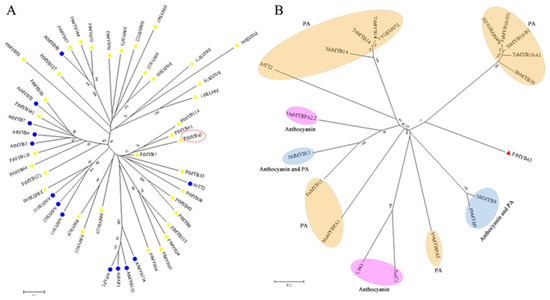
Figure 6.
Phylogenetic tree of R2R3-MYB homologues that regulate anthocyanin biosynthesis. (A) Phylogenetic tree of FtMYBs and AtMYBs. (B) Phylogenetic tree of FtMYBs and MYBs from other species. MdMYB9 (Malus domestica, DQ267900), MdMYB9 (Malus domestica, DQ074463), ZmC1 (Zea may, P10290), OsC1 (Oryza sativa, BAD04030), PbMYB9 (Pyrus bretschneideri, KT601123), TaMYB10-A1, B1, and D1 (Triticum aestivum, AB599721, AB599722, and AB191460), HvMYB10 (Hordeum vulgare, BAL42245.1), VmMYBPA2.2 (Vaccinium myrtillus, QWW89545.1), MsMYB14 (Medicago sativa, AFJ53055.1), AetMYB10-D1 (Aegilops tauschii, AKE33252.1), FaMYB11 (Fragaria x ananassa, AFL02461.1), VvMybPA2 (Vitis vinifera, NP_001267953.1), TaMYB14 (Trifolium arvense, AFJ53053.1), ToMYB14-1 and 14 (Trifolium occidentale, AFJ53051.1 and AFJ53052.1), and MaMYBPA1 (Musa acuminata, UTU02182.1). The protein in red box and marked with red triangle was homologous to AtTT2.
3.8. Characterization of Candidate Gene FtMYB43 That May Promote Anthocyanin Biosynthesis
Previous studies showed that AtTT2 regulated PA biosynthesis [37], while MdMYB9, MdMYB11, and PbMYB9 as its homologous gene regulated both anthocyanin and PA synthesis [9,44]. Here, one member of FtR2R3-MYBs FtMYB43 was the homologous gene of AtTT2 and located between the R2R3-MYB genes from other species that were reported to have functions in the regulation of PA and anthocyanin in the phylogenetic tree (Figure 6); therefore, FtMYB43 might as well have the similar function.
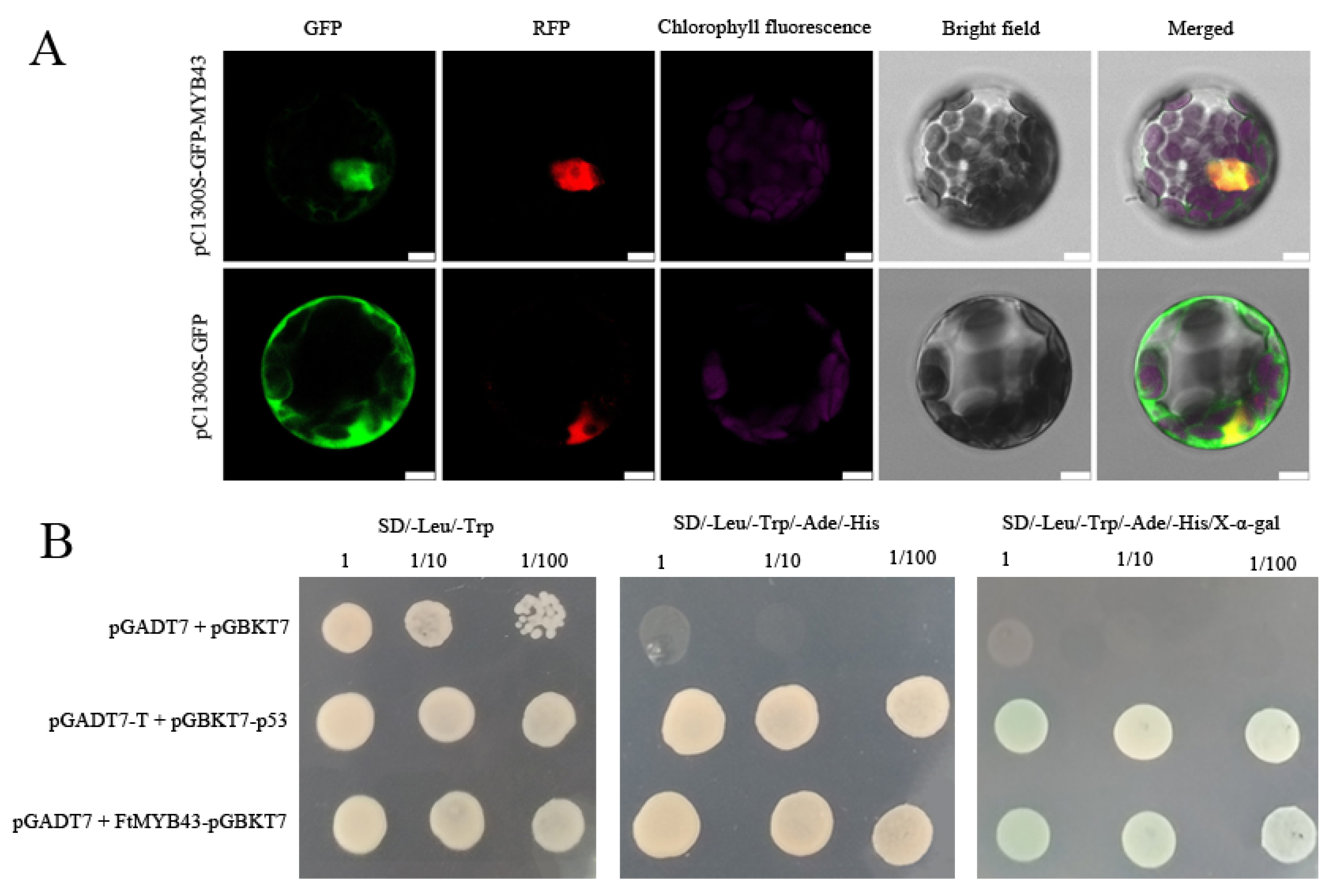
Subcellular location analysis showed that FtMYB43-GFP merged with RFP (the reference for nucleus location) (Figure 7A), while the GFP from the empty vector was detected throughout the cell, indicating that FtMYB43 locates in the nucleus and acts as a transcript factor. Yeast-two-hybrid assay showed that FtMYB43 owns transcriptional activation activity (Figure 7B). These results suggest that FtMYB43 is a transcription factor that plays a positive regulatory role.
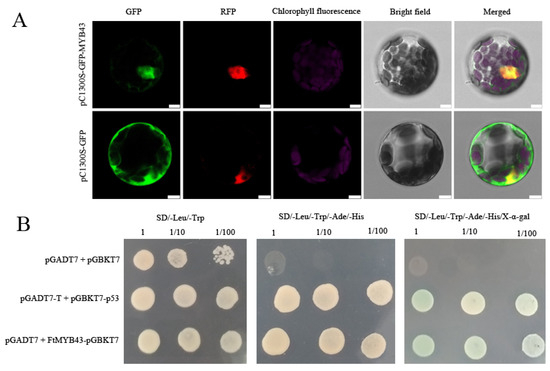
Figure 7.
(A) Subcellular localization of FtMYB43 protein (scale bars = 7.5 μm). (B) Yeast-two-hybrid assay, pGADT7 + pGBKT7, and pGADT7-T + pGBKT7-p53 were used as negative and positive control, respectively.
3.9. Overexpression of FtMYB43 in Arabidopsis
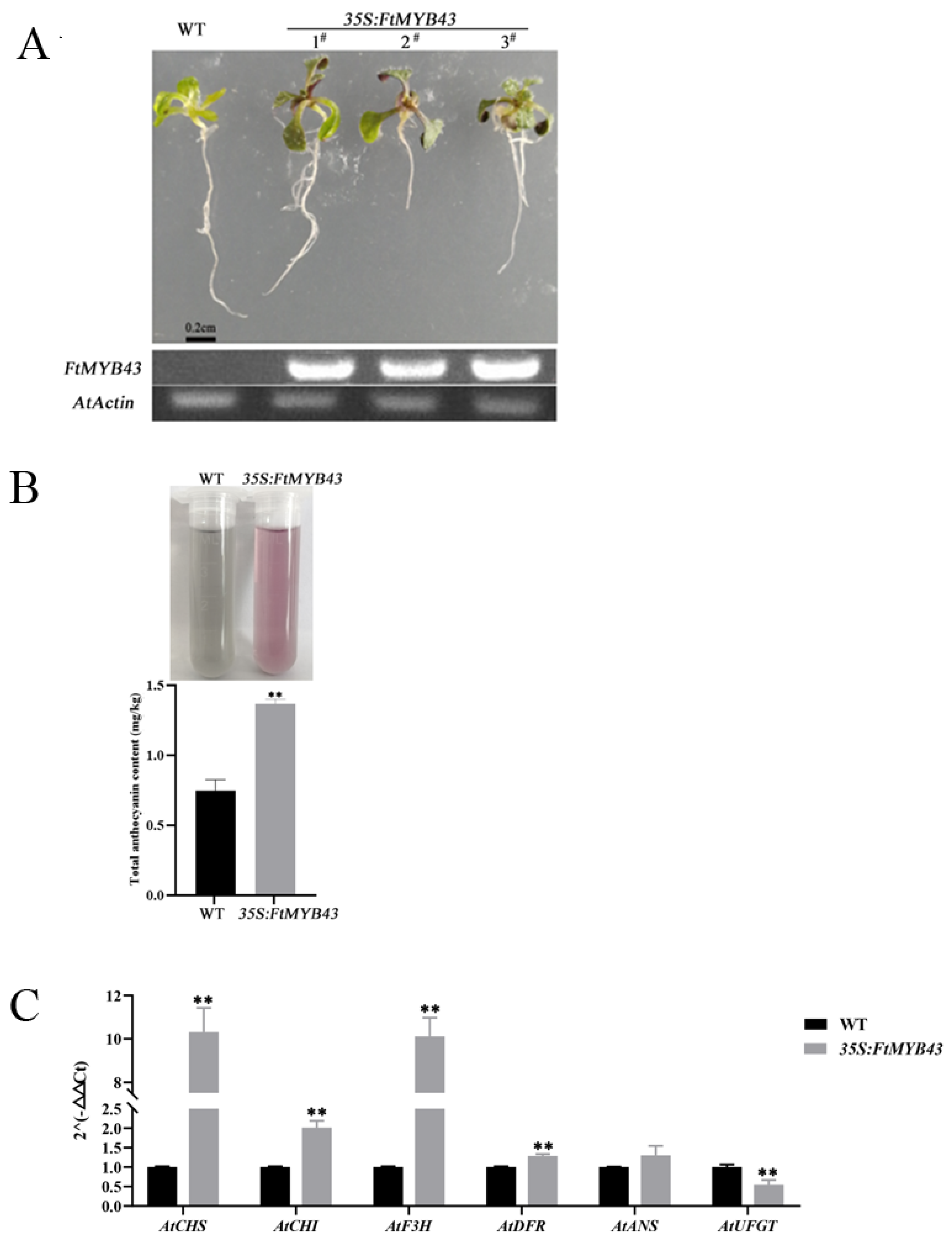
The function of FtMYB43 was verified by overexpression in Arabidopsis and the results show that red pigments could be observed in the leaves of transgenic Arabidopsis seedlings, while no obvious pigment accumulation was observed in the wild-type (WT) seedlings (Figure 8A). After extraction, the extracted solution exhibited red color, indicative of the high anthocyanin content in the transgenic Arabidopsis seedlings, while the extracted solution of the wild type exhibited green color with lower anthocyanin content (Figure 8B). The transcription levels of anthocyanidin-related genes (including CHI, CHS, F3H, DFR, ANS, and UFGT) of transgenic and WT Arabidopsis plants were analyzed through qRT-PCR. The results show that except for UFGT, the other structural genes in the anthocyanin biosynthesis pathway had higher expression levels in transgenic Arabidopsis (Figure 8C). Notably, the expression of CHI, CHS, and F3H increased significantly (Figure 8C), which was consistent with anthocyanidin accumulation (Figure 8B). These results suggest that FtMYB43 played an important role as a positive regulator in anthocyanin biosynthesis.
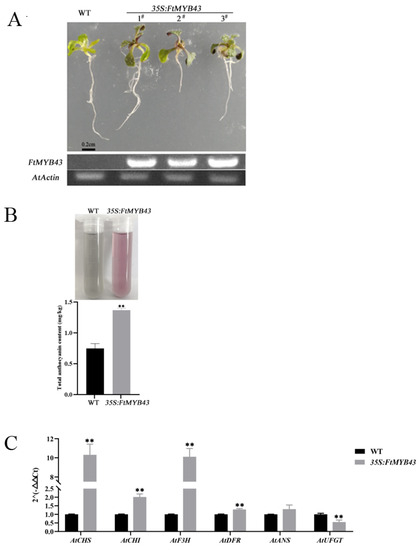
Figure 8.
Functional characterization of FtMYB43. (A) Seedlings of WT control and 35S:FtMYB43 transgenic Arabidopsis. # 1,#2 and #3 represent different tansgenic lines of Arabidopsis. (B) Pigment extract solution and total anthocyanin contents of WT control and 35S:FtMYB43 transgenic plants. (C) Relative expression levels of structural genes in the anthocyanin biosynthesis pathway in seedlings of WT and transgenic Arabidopsis by qRT-PCR analysis (means were calculated from three repeats and error bars represent mean ± SD, ** p < 0.01).
4. Discussion
4.1. Identification and Characteristics of R2R3-MYB Genes in Tartary Buckwheat
The R2R3-MYB gene family is widely distributed among all eukaryotes. It plays an important role in the regulation of secondary metabolic pathways, different signaling pathways, various developmental and morphological processes, and responses to biotic and abiotic stress, among others [1,4]. This gene family was identified in many green plants, such as Arabidopsis thaliana [2], Oryza sativa [13], Hypericum perforatum [14], and Camellia sinensis [15], among others [16,17]. However, previous studies on the MYB TFs of Tartary buckwheat focused on the cloning and functional research of individual genes [53,54,55], and therefore, system characterization of the R2R3-MYB gene family is lacking. Here, a total of 152 FtR2R3-MYB genes were identified from the whole genome sequence of Tartary buckwheat, which possesses more R2R3-MYB members compared with A. thaliana (126) [2], O. sativa (130) [13], H. perforatum (109) [14], and C. sinensis (122) [15]. This could probably be attributed to natural selection and the duplications and recombination of the genome during the evolution [54], implying that there was an expansion in the FtR2R3-MYB gene family as well, which may create a diversity of proteins. It was reported that tandem gene duplications might enhance the environmental fitness of plants by purifying selection [54,55].
4.2. Phylogenesis and Evolution of FtR2R3-MYB Genes
The sequence alignment analysis of R2R3-MYB proteins from Tartary buckwheat and Arabidopsis generated an NJ tree for assessment of their evolutionary relationships. All proteins could be classified broadly into 25 subgroups (Figure 3), which was consistent with previous reports [2,35]. However, the SG3, SG10, and SG12 subgroups did not contain any member of Tartary buckwheat (Figure 3), which may be due to specificity in species. Most of the R2R3-MYB proteins, which had high homology with Arabidopsis, were clustered in the same branch, indicating that they might have similar functions. For instance, AtMYB members (AtMYB75, AtMYB90, AtMYB113, and AtMYB114) clustered in SG6, and are involved in anthocyanins biosynthesis by positively modulating F3H, DFR, and UFGT [5,56]. This suggested that their orthologs genes (FtMYB24, FtMYB85, FtMYB98, as well as FtMYB113) may also be responsible for the deposition of anthocyanins in Tartary buckwheat. A previous study showed that FtMYB15, homologous to AtMYB3, participates in the regulation of anthocyanin biosynthesis [57].
FtR2R3-MYB genes from the same sub-categories share common motifs and had obvious differences from other sub-categories. FtR2R3-MYB members in the same clade shared similar exon–intron gene structure and common motifs, showing conservation exists within these members. While, FtR2R3-MYB genes from different sub-categories had different gene structures, which indicated evolution among them. This conclusion further supported the reliability of FtR2R3-MYB TFs classification.
4.3. Expression Profiles of FtR2R3-MYB Gene in Different Tissues, and under Salt and Light Stress
Analysis of gene expression levels in various tissues could provide crucial information on gene function. In the present study, the expression patterns of FtR2R3-MYB genes in different tissues and responses to light and salt stress were analyzed according to the published RNA-seq data. The results displayed appreciable differences in the spatial expression profiles of the FtR2R3-MYB genes. One member of the FtR2R3-MYB gene family FtMYB17, the homologous gene of AtMYB59, had an increased expression in roots and may perform an analogous function to regulate root growth and cell cycle with AtMYB59 [58].
Plants can grow well in adverse environmental conditions through different adaptive strategies. In the present study, the cis-acting elements of abiotic stress response were distributed in nearly all the FtR2R3-MYBs, such as dehydration, low temperature, anaerobic, and light induction (Table S4a,b), which demonstrated that these members participate in diversified abiotic stress responsiveness. A sum of 69 (42%), 96 (63%), and 2 (1%) FtR2R3-MYB genes, respectively, contained MBS, LTR, and DFR elements, suggesting that these genes could function in drought and cold tolerance. A total of 412 ABRE, 268 CGTCA-motif, and 268 TGACG-motif were identified in FtR2R3-MYB genes, which may be the main cis-elements for response to ABA and MeJA. Previous literature suggested that AtMYB2, AtMYB14, AtMYB15, AtMYB44, AtMYB70, AtMYB77, AtMYB73, and AtMYB96 play crucial roles in multiple stresses, such as low temperature, water loss, and salt stress [56]. The mechanisms of response to various stresses may be roughly analogous in other species, and overexpression of OsMYB2, OsMYB2P-1, OsMYB3R-2, and OsMYB48-1 resulted in increased salt, low temperature, and drought tolerance in rice [59]. Similarly, as the homologous genes with Arabidopsis clustered in the same branch, FtMYB9 and FtMYB30 were also validated to participate in response to drought, salt, as well as ABA treatments by transgenic Arabidopsis [2]. Moreover, FtMYB116, as a light-induced transcription factor, directly combined with the promoter region of F3′H and improved the accumulation of rutin [38].
4.4. FtMYB43 Is a Transcript Factor and Participates in Positively Regulating Anthocyanin Biosynthesis
Some gene functions can be revealed through genome-wide analysis of gene family, and this method is well documented and utilized in lotus [60], wheat [61,62,63,64], and cotton [65], among others. In the present study, one FtR2R3-MYB member, FtMYB43, was identified by genome-wide analysis of this gene family, and might also play a similar role with its homologous gene in Arabidopsis (AtTT2) in the regulation of anthocyanin biosynthesis.
FtMYB43-GFP merged with RFPnucleus in the nucleusnucleus when they were co-expressed transiently in Arabidopsis protoplast, while the empty GFP protein could be detected throughout the cell (Figure 7A). This nucleus localization, therefore, validates the transcription factor role of FtMYB43.
Additionally, we hypothesized that FtMYB43 could regulate the expression of the anthocyanin-related genes on the basis that it was the homologous gene of TT2 type, and that PbMYB9 and MdMYB9 were reported to regulate anthocyanin biosynthesis [9,44]. To confirm this assumption, the FtMYB43 was cloned and used to construct overexpression vector pMDC83-FtMTB43 and genetically transformed into Arabidopsis thaliana. The results show that the heterologous expression of FtMYB43 could significantly increase anthocyanin accumulation and effectively regulate the expression of anthocyanin-related genes in transgenic Arabidopsis plants. We found that FtMYB43 and previously reported genes such as FtMYB1 [66] and FtMYB116 [38] were the same transcription factor gene by sequence alignment. Bai et al. reported that overexpression of FtMYB1 in N. tabacum could significantly promote PA biosynthesis [66]. Zhang et al. found that FtMYB116 was able to promote the synthesis of rutin [38]. The above results are coincident with the study on MdMYB9, MdMYB11, and PbMYB9, which were AtTT2-homologous genes and they improved both PA and anthocyanin accumulation [9,44]. Taken together, FtMYB43 could positively regulate the biosynthesis of three types of flavonoids, namely anthocyanins, PA, and flavones. These results may provide valuable information for the understanding of the mechanism of anthocyanin biosynthesis modulation in Tartary buckwheat, which is of great academic and practical significance in molecular breeding and developing new breeds.
5. Conclusions
In conclusion, a genome-wide search identified a total of 152 R2R3-MYB TF genes in the Tartary buckwheat genome, and their physicochemical property, phylogeny, gene structure, synteny, cis-acting elements, conserved domain, and motifs as well as gene expression profiles were systematically analyzed by bioinformatics methods. The functions of the FtR2R3-MYB gene family members were relatively conserved and numerous genes selected purification pressure during evolution. Most FtR2R3-MYB genes may participate in plant growth and respond to salt and light treatments or other abiotic stresses according to their expression patterns and cis-elements prediction. In addition, one member of FtR2R3-MYB gene family FtMYB43, congenerous to AtMYB123 (AtTT2) and belonging to the SG5 subgroup, was verified to be a positive TF by subcellular localization and transcriptional activity assay. FtMYB43 was further identified to positively regulate anthocyanin biosynthesis in transgenic Arabidopsis. This study might offer important information for research on the function of other gene families and provide excellent genetic resources for breeding of Tartary buckwheat.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy13082117/s1, Table S2. Renaming and domain analysis of FtR2R3-MYB genes. Additional File S1: Table S1. The primers sequences of genes used for qRT-PCR of Arabidopsis and vector construction of Tartary buckwheat. Table S3. The bioinformation of 152 FtR2R3-MYB genes. Table S4a. Putative cis-acting elements identified in the promoter regions of FtR2R3-MYB genes. Table S4b. Putative cis-acting elements with their number and function identified in the promoter regions of FtR2R3-MYB genes. Table S5. Ka/Ks analysis for the duplicated FtR2R3-MYB paralogs. Table S6. Expression profiles of FtR2R3-MYB genes. Figure S1. Structure of FtR2R3-MYBs gene family. (A) Phylogenetic tree of the FtR2R3-MYBs proteins. (The MYB proteins were clustered into 32 (A1~A32) subfamilies according to their motifs and intron/exon characteristics); (B) Motif identification of FtR2R3-MBs proteins. (The color corresponds to the number, representing the same kind of motif); (C) Exon-intron structure of FtR2R3-MYBs genes. (The different colors in structures of FtR2R3-MYB genes represent the following: yellow, CDS; brown, UTR; black, intron; the phases of the introns were shown by the figure 0, 1 and 2); (D) Cis-acting elements. (Different colors of box represent different categories of cis-elements). Figure S2. Analysis of the cis-elements of the FtR2R3-MYB genes. (A) Heat map of the number of the cis-elements. (Circle sizes and colour line, respectively, represent the number and type of cis-elements. The blue, orange, gray, and yellow colour bars, which, respectively, correspond to the light, stress, phytohormone responsive and plant growth cis-elements). (B) A histogram of different colors is representation of the total of cis-elements per category.
Author Contributions
J.D., Q.C. and R.N.D. designed the research; L.W., J.Z. and T.W. analyzed the data; L.W. and J.D. wrote the manuscript; L.Z. (Lan Zhang) and C.Y. carried out the qRT-PCR analysis and measured anthocyanin content; J.H. and T.S. constructed vectors and transgenic experiment; L.Z. (Liwei Zhu), Z.M. and F.C. planted Arabidopsis and observed phenotype. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Joint Fund of the National Natural Science Foundation of China and the Karst Science Research Center of Guizhou Provincial (Grant No. U1812401), Guizhou Provincial Basic Research Program (Natural Science) (QianKeHeJiChu-ZK [2023] YiBan 278), the National Science Foundation of China (32102422), the Training Program of Guizhou Normal University (QianKeHePingTaiRenCai (11904/0520070)), and China Agriculture Research System (CARS-07-A5).
Data Availability Statement
The gene expression level used in this study are publicly available at the National Center for Biotechnology Information (NCBI) https://www.ncbi.nlm.nih.gov/ (accessed on 20 April 2023) with accession numbers- SRP157461, SRP118793, SRR5433730, SRR5433731, SRR5433732 and SRR5433734.
Acknowledgments
We thank the company of Towin Biotechnology (Wuhan, China) for Arabidopsis protoplast transient expression assay.
Conflicts of Interest
The authors declare no conflict interest.
References
- Thakur, S.; Vasudev, P.G. MYB Transcription Factors and their Role in Medicinal Plants. Mol. Biol. Rep. 2022, 49, 10995–11008. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB Gene Family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold EWeisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, X.; He, K.; Liu, M.; Li, J.; Gao, Z.; Lin, Z.; Zhang, Y.; Wang, X.; Qiu, X.; et al. The MYB Transcription Factor Superfamily of Arabidopsis: Expression Analysis and Phylogenetic Comparison with the Rice MYB Family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar]
- Baumann, K.; Perez-Rodriguez, M.; Bradley, D.; Venail, J.; Bailey, P.; Jin, H.; Koes, R.; Roberts, K.; Martin, C. Control of Cell and Petal Morphogenesis by R2R3 MYB Transcription Factors. Development 2007, 134, 1691–1701. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB Transcription Factor Genes as Regulators for Plant Responses: An Overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef]
- Oshima, Y.; Mitsuda, N. The MIXTA-Like Transcription Factor MYB16 is a Major Regulator of Cuticle Formation in Vegetative Organs. Plant Signal. Behav. 2013, 8, e26826. [Google Scholar] [CrossRef]
- Rosany, C.R.; Beatriz, V.T.; Blanca, S.S. MiR858-Mediated Regulation of Flavonoid-Specific MYB Transcription Factor Genes Controls Resistance to Pathogen Infection in Arabidopsis. Plant Cell Physiol. 2018, 1, 190–204. [Google Scholar]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhang, X.; Zhao, M.; Chiang, V.L.; Sederoff, R.R.; Zhao, X. MYB-Mediated Regulation of Anthocyanin Biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef]
- He, Y.; Li, M.; Wang, Y.; Shen, S. The R2R3-MYB transcription factor MYB44 modulates carotenoid biosynthesis in Ulva prolifera. Algal Res. 2022, 62, 102578. [Google Scholar] [CrossRef]
- Jia, N.; Liu, J.; Tan, P.; Sun, Y.; Lv, Y.; Liu, J.; Sun, J.; Huang, Y.; Lu, J.; Jin, N.; et al. Citrus Sinensis MYB Transcription Factor CsMYB85 Induce Fruit Juice Sac Lignification through Interaction with other CsMYB Transcription Factors. Front. Plant Sci. 2019, 25, 213. [Google Scholar] [CrossRef]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription Factors Associated with Abiotic and Biotic Stress Tolerance and Their Potential for Crops Improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef]
- Jin, J.P.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a Central Hub for Transcription Factors and Regulatory Interactions in Plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zhang, Q.; Sun, Y.; Yang, L.; Wang, Z. Genome-Wide Identification and Characterization of R2R3-MYB Family in Hypericum perforatum under Diverse Abiotic Stresses. Int. J. Biol. Macromol. 2020, 145, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, P.; Gu, M.; Lin, X.; Hou, B.; Zheng, Y.; Sun, Y.; Jin, S.; Ye, N. R2R3-MYB Transcription Factor Family in Tea Plant (Camellia sinensis): Genome-Wide Characterization, Phylogeny, Chromosome Location, Structure and Expression Patterns. Genomics 2021, 11, 1565–1578. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; He, W.; Guo, X.; Pan, J. Genome-Wide Identification, Classification and Expression Analysis of the MYB Transcription Factor Family in Petunia. Int. J. Mol. Sci. 2021, 22, 4838. [Google Scholar] [CrossRef] [PubMed]
- Naik, J.; Rajput, R.; Pucker, B.; Stracke, R.; Pandey, A. The R2R3-MYB Transcription Factor MtMYB134 Orchestrates Flavonol Biosynthesis in Medicago truncatula. Plant Mol. Biol. 2021, 106, 157–172. [Google Scholar] [CrossRef]
- Zhou, X.L.; Chen, Z.D.; Zhou, Y.M.; Shi, R.H.; Li, Z.J. The Effect of Tartary Buckwheat Flavonoids in inhibiting the Proliferation of MGC80-3 Cells during Seed Germination. Molecules 2019, 24, 3092. [Google Scholar] [CrossRef]
- Cao, P.; Wu, Y.; Li, Y.; Xiang, L.; Cheng, B.; Hu, Y.; Jiang, X.; Wang, Z.; Wu, S.; Si, L.; et al. The Important Role of Glycerophospholipid Metabolism in the Protective Effects of Polyphenol-Enriched Tartary Buckwheat Extract against Alcoholic Liver Disease. Food Funct. 2022, 13, 10415–10425. [Google Scholar] [CrossRef]
- Li, H.; Lv, Q.; Liu, A.; Wang, J.; Sun, X.; Deng, J.; Chen, Q.; Wu, Q. Comparative Metabolomics Study of Tartary (Fagopyrum tataricum (L.) Gaertn) and Common (Fagopyrum esculentum Moench) Buckwheat Seeds. Food Chem. 2021, 371, 131125. [Google Scholar] [CrossRef]
- Chen, Y.; Zhu, Y.; Qin, L.K. The Cause of Germination Increases the Phenolic Compound Contents of Tartary Buckwheat (Fagopyrum tataricum). J. Future Foods 2022, 2, 372–379. [Google Scholar] [CrossRef]
- Kubatka, P.; Mazurakova, A.; Samec, M.; Koklesova, L.; Zhai, K.; Al-Ishaq, R.; Kajo, K.; Biringer, K.; Vybohova, D.; Brockmueller, A.; et al. Flavonoids Against Non-Physiologic Inflammation Attributed to Cancer Initiation, Development, and Progression-3PM Pathways. EPMA J. 2021, 12, 559–587. [Google Scholar] [CrossRef]
- George, V.C.; Dellaire, G.; Rupasinghe, H.P.V. Plant Flavonoids in Cancer Chemoprevention: Role in Genome Stability. J. Nutr. Biochem. 2017, 45, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Joshi, D.C.; Zhang, K.; Wang, C.; Chandora, R.; Khurshid, M.; Li, J.; He, M.; Georgiev, M.I.; Zhou, M. Strategic Enhancement of Genetic Gain for nutraceutical development in buckwheat: A Genomics-Driven Perspective. Biotechnol. Adv. 2020, 39, 107479. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Hou, Z.; Liu, D.; Yang, X. Tartary buckwheat Flavonoids Protect Hepatic Cells against High Glucose-Induced Oxidative Stress and Insulin Resistance via MAPK Signaling Pathways. Food Funct. 2016, 7, 1523–1536. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.G.; Jang, I.S.; Yang, K.E.; Yoon, S.J.; Baek, S.; Lee, J.Y.; Suzuki, T.; Chung, K.Y.; Woo, S.H.; Choi, J.S. Effect of Rutin from Tartary Buckwheat Sprout on serum Glucose-Lowering in Animal Model of Type 2 Diabetes. Acta Pharm. 2016, 66, 297–302. [Google Scholar] [CrossRef]
- Tatsuro Suzuki, T.; Morishita, T.; Takigawa, S.; Noda, T.; Ishiguro, K. Development of Rutin-Rich Noodles using Trace-Rutinosidase Variety of Tartary buckwheat (Fagopyrum tataricum. Gaertn.) ‘Manten-Kirari’. Food Sci. Technol. Res. 2019, 25, 915–920. [Google Scholar] [CrossRef]
- Schaart, J.G.; Dubos, C.; Romero De La Fuente, I.; van Houwelingen, A.M.M.L.; de Vos, R.C.H.; Jonker, H.H.; Xu, W.; Routaboul, J.M.; Lepiniec, L.; Bovy, A.G. Identification and Characterization of MYB-bHLH-WD40 Regulatory Complexes Controlling Proanthocyanidin Biosynthesis in Strawberry (Fragaria × ananassa) Fruits. New Phytol. 2013, 197, 454–467. [Google Scholar] [CrossRef]
- Liu, H.; Chen, H.X.; Chen, X.X.; Lu, J.; Chen, D.L.; Luo, C.; Cheng, X.; Huang, C.L. Transcriptomic and Metabolomic Analyses Reveal that MYB transcription Factors Regulate Anthocyanin Synthesis and Accumulation in the Disc Florets of the Anemone form of Chrysanthemum Morifolium. Sci. Hortic. 2023, 307, 110847. [Google Scholar] [CrossRef]
- Subban, P.; Prakash, S.; Bootbool Mann, A.; Kutsher, Y.; Evenor, D.; Levin, I.; Reuveni, M. Functional Analysis of MYB Alleles from Solanum chilense and Solanum lycopersicum in Controlling Anthocyanin Levels in Heterologous Tobacco Plants. Physiol. Plant. 2021, 172, 1630–1640. [Google Scholar] [CrossRef]
- Wang, N.; Xu, H.; Jiang, S.; Zhang, Z.; Lu, N.; Qiu, H.; Qu, C.; Wang, Y.; Wu, S.; Chen, X. MYB12 and MYB22 Play Essential Roles in Proanthocyanidin and Flavonol Synthesis in Red-Fleshed Apple (Malus sieversii f. niedzwetzkyana). Plant J. 2017, 90, 276–292. [Google Scholar] [CrossRef] [PubMed]
- Lafferty, D.J.; Espley, R.V.; Deng, C.H.; Dare, A.P.; Günther, C.S.; Jaakola, L.; Karppinen, K.; Boase, M.R.; Wang, L.; Luo, H.; et al. The Coordinated Action of MYB Activators and Repressors Controls Proanthocyanidin and Anthocyanin Biosynthesis in Vaccinium. Front. Plant Sci. 2022, 13, 910155. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Du, W.; Hao, Y.; Han, Y.; Li, H.; Liu, L.; Zhang, K.; Zhou, M.; Sun, Z. Elucidation of the Regulatory Network of Flavonoid Biosynthesis by Profiling the Metabolome and Transcriptome in Tartary Buckwheat. J. Agric. Food Chem. 2021, 69, 7218–7229. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lv, Q.; Ma, C.; Qu, J.; Cai, F.; Deng, J.; Huang, J.; Ran, P.; Shi, T.; Chen, Q. Metabolite Profiling and transcriptome Analyses Provide Insights into the Flavonoid Biosynthesis in the Developing Seed of Tartary Buckwheat (Fagopyrum tataricum). J. Agric. Food Chem. 2019, 67, 11262–11276. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Ma, B.; Gao, Q.; Du, H.; Han, Y.; Li, Y.; Cao, Y.; Qi, M.; Zhu, Y.; et al. The Tartary Buckwheat Genome Provides Insights into Rutin Biosynthesis and Abiotic Stress Tolerance. Mol. Plant. 2017, 10, 1224–1237. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Nesi, N.; Jond, C.; Debeaujon, I.; Caboche, M.; Lepiniec, L. The Arabidopsis TT2 Gene Encodes an R2R3 MYB Domain Protein that Acts as a Key Determinant for Proanthocyanidin Accumulation in Developing Seed. Plant Cell. 2001, 13, 2099–2114. [Google Scholar] [CrossRef]
- Zhang, D.; Jiang, C.; Huang, C.; Wen, D.; Lu, J.; Chen, S.; Zhang, T.; Shi, Y.; Xue, J.; Ma, W.; et al. The light-Induced Transcription Factor FtMYB116 Promotes Accumulation of Rutin in Fagopyrum tataricum. Plant Cell Environ. 2019, 42, 1340–1351. [Google Scholar] [CrossRef]
- Wu, Q.; Bai, X.; Zhao, W.; Xiang, D.; Wan, Y.; Yan, J.; Zou, L.; Zhao, G. De Novo Assembly and Analysis of Tartary Buckwheat (Fagopyrum tataricum Garetn.) Transcriptome Discloses Key Regulators Involved in Salt-Stress Response. Genes 2017, 8, 255. [Google Scholar] [CrossRef]
- Xiong, L.; Li, C.; Li, H.; Lyu, X.; Zhao, T.; Liu, J.; Zuo, Z.; Liu, B. A transient Expression System in Soybean Mesophyll Protoplasts Reveals the Formation of Cytoplasmic GmCRY1 Photobody-Like Structures. Sci. China Life Sci. 2019, 2, 1070–1077. [Google Scholar] [CrossRef]
- Zale, J.M.; Agarwal, S.; Loar, S.; Steber, C.M. Evidence for stable transformation of wheat by floral dip in Agrobacterium tumefaciens. Plant Cell Rep. 2009, 28, 903–913. [Google Scholar] [CrossRef]
- Labra, M.; Vannini, C.; Grassi, F.; Bracale, M.; Balsemin, M.; Basso, B.; Sala, F. Genomic Stability in Arabidopsis thaliana Transgenic Plants Obtained by Floral Dip. Theor. Appl. Genet. 2004, 109, 1512–1518. [Google Scholar] [CrossRef]
- Park, N.I.; Li, X.; Suzuki, T.; Kim, S.J.; Woo, S.H.; Park, C.H.; Park, S.U. Differential Expression of Anthocyanin Biosynthetic Genes and Anthocyanin Accumulation in Tartary Buckwheat Cultivars ‘Hokkai t8’ and ‘Hokkai t10’. J. Agric. Food Chem. 2011, 59, 2356–2361. [Google Scholar] [CrossRef]
- An, X.H.; Tian, Y.; Chen, K.Q.; Liu, X.J.; Liu, D.D.; Xie, X.B.; Cheng, C.G.; Cong, P.H.; Hao, Y.J. MdMYB9 and MdMYB11 are Involved In the regulation of the JA-Induced Biosynthesis of Anthocyanin and Proanthocyanidin in Apples. Plant Cell Physiol. 2015, 56, 650–662. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Liang, L.X.; Li, L.B.; Wang, T. Genome-Wide Analysis of R2R3-MYB Transcription Factors in Phalaenopsis equestris. For. Res. 2018, 31, 104–113. [Google Scholar]
- Zhang, M.; Liu, W.; Bi, Y.P. Dehydration-Responsive Element-Binding (DREB) Transcription Factor in Plants and its Role during Abiotic Stresses. Yi Chuan 2009, 31, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Suzuki, A.; Washida, H.; Takaiwa, F. The GCN4 Motif in a Rice Glutelin Gene is Essential for endosperm-Specific Gene Expression and is Activated by Opaque-2 in Transgenic Rice Plants. Plant J. 1998, 14, 673–683. [Google Scholar] [CrossRef]
- Himi, E.; Maekawa, M.; Miura, H.; Noda, K. Development of PCR Markers for Tamyb10 Related to R-1, Red grain Color Gene in Wheat. Theor. Appl. Genet. 2011, 122, 1561–1576. [Google Scholar] [CrossRef]
- Zhong, Y.; Chen, Y.; Zheng, D.; Pang, J.; Liu, Y.; Luo, S.; Meng, S.; Qian, L.; Wei, D.; Dai, S.; et al. Chromosomal-Level Genome Assembly of the Orchid Tree Bauhinia variegata (Leguminosae; Cercidoideae) Supports the Allotetraploid Origin Hypothesis of Bauhinia. DNA Res. 2022, 29, dsac012. [Google Scholar] [CrossRef]
- Terrier, N.; Torregrosa, L.; Ageorges, A.; Vialet, S.; Verriès, C.; Cheynier, V.; Romieu, C. Ectopic Expression of VvMybPA2 Promotes Proanthocyanidin Biosynthesis in Grapevine and Suggests Additional Targets in the Pathway. Plant Physiol. 2009, 149, 1028–1041. [Google Scholar] [CrossRef]
- Hancock, K.R.; Collette, V.; Fraser, K.; Greig, M.; Xue, H.; Richardson, K.; Jones, C.; Rasmussen, S. Expression of the R2R3-MYB Transcription Factor TaMYB14 from Trifolium Arvense Activates Proanthocyanidin Biosynthesis in the Legumes Trifolium repens and Medicago sativa. Plant Physiol. 2012, 159, 1204–1220. [Google Scholar] [CrossRef]
- Rajput, R.; Naik, J.; Stracke, R.; Pandey, A. Interplay between R2R3 MYB-Type Activators and Repressors Regulates Proanthocyanidin Biosynthesis in Banana (Musa acuminata). New Phytol. 2022, 236, 1108–1127. [Google Scholar] [CrossRef]
- Dong, Z.D.; Chen, J.; Li, T.; Chen, F.; Cui, D.Q. Molecular Survey of Tamyb10-1 Genes and their Association with Grain Colour and Germinability in Chinese Wheat and Aegilops tauschii. J. Genet. 2015, 94, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Pu, X.; Gao, R.; Demurtas, O.C.; Fleck, S.J.; Richter, M.; He, C.; Ji, A.; Sun, W.; Kong, J.; et al. Tandem Gene Duplications Drive Divergent Evolution of Caffeine and Crocin Biosynthetic Pathways in Plants. BMC Biol. 2020, 18, 63. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lin-Wang, K.; Liu, Z.; Allan, A.C.; Qin, S.; Zhang, J.; Liu, Y. Genome-Wide Analysis and Expression Profiles of the StR2R3-MYB transcription Factor Superfamily in Potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 2020, 148, 817–832. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Han, G.; Sun, C.; Sui, N. Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal. Behav. 2019, 14, 1613131. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.P.; Zhao, H.X.; Yao, P.F.; Li, Q.Q.; Huang, Y.J.; Li, C.L.; Chen, H.; Wu, Q. An R2R3-MYB Transcription Factor FtMYB15 Involved in the Synthesis of Anthocyanin and Proanthocyanidins from Tartary Buckwheat. J. Plant Growth Regul. 2018, 37, 76–84. [Google Scholar] [CrossRef]
- Mu, R.L.; Cao, Y.R.; Liu, Y.F.; Lei, G.; Zou, H.F.; Liao, Y.; Wang, H.W.; Zhang, W.K.; Ma, B.; Du, J.Z.; et al. An R2R3-Type Transcription Factor Gene AtMYB59 Regulates Root Growth and Cell Cycle Progression in Arabidopsis. Cell Res. 2009, 19, 1291–1304. [Google Scholar] [CrossRef]
- Gao, F.; Zhao, H.X.; Yao, H.P.; Li, C.L.; Chen, H.; Wang, A.H.; Park, S.U.; Wu, Q. Identification, Isolation and Expression Analysis of Eight Stress-Related R2R3-MYB Genes in Tartary Buckwheat (Fagopyrum tataricum). Plant Cell Rep. 2016, 35, 1385–1396. [Google Scholar] [CrossRef]
- Lin, Z.; Cao, D.; Damaris, R.N.; Yang, P. Genome-Wide Identification of MADS-box Gene Family in Sacred Lotus (Nelumbo nucifera) Identifies a SEPALLATA Homolog Gene Involved in Floral Development. BMC Plant Biol. 2020, 20, 497. [Google Scholar] [CrossRef]
- Kumar, M.; Kherawat, B.S.; Dey, P.; Saha, D.; Singh, A.; Bhatia, S.K.; Ghodake, G.S.; Kadam, A.A.; Kim, H.U.; Manorama Chung, S.M. Genome-Wide Identification and Characterization of PIN-FORMED (PIN) Gene Family Reveals Role in Developmental and Various Stress Conditions in Triticum aestivum L. Int. J. Mol. Sci. 2021, 22, 7396. [Google Scholar] [CrossRef] [PubMed]
- Kesawat, M.S.; Kherawat, B.S.; Singh, A.; Dey, P.; Kabi, M.; Debnath, D.; Saha, D.; Khandual, A.; Rout, S.; Manorama; et al. Genome-Wide Identification and Characterization of the Brassinazole-resistant (BZR) Gene Family and Its Expression in the Various Developmental Stage and Stress Conditions in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2021, 22, 8743. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Chen, W.; Zhu, X.; Zhou, X.; Jin, Y.; Zhan, C.; Liu, G.; Liu, X.; Ma, D.; Qiao, Y. Genome-Wide Identification and Characterization of Wheat 14-3-3 Genes Unravels the Role of TaGRF6-A in Salt Stress Tolerance by Binding MYB Transcription Factor. Int. J. Mol. Sci. 2021, 22, 1904. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Kherawat, B.S.; Singh, A.; Dey, P.; Routray, S.; Mohapatra, C.; Saha, D.; Ram, C.; Siddique, K.H.M.; Kumar, A.; et al. Genome-Wide Analysis and Characterization of the Proline-Rich Extensin-like Receptor Kinases (PERKs) Gene Family Reveals Their Role in Different Developmental Stages and Stress Conditions in Wheat (Triticum aestivum L.). Plants 2022, 11, 496. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Y.; Xu, N.; Rui, C.; Fan, Y.; Wang, J.; Han, M.; Wang, Q.; Sun, L.; Chen, X.; et al. Genome-Wide Expression Analysis of Phospholipase A1 (PLA1) Gene Family Suggests Phospholipase A1-32 Gene Responding to Abiotic Stresses in Cotton. Int. J. Biol. Macromol. 2021, 192, 1058–1074. [Google Scholar] [CrossRef]
- Bai, Y.C.; Li, C.L.; Zhang, J.W.; Li, S.J.; Luo, X.P.; Yao, H.P.; Chen, H.; Zhao, H.X.; Park, S.U.; Wu, Q. Characterization of Two Tartary Buckwheat R2R3-MYB Transcription Factors and their Regulation of Proanthocyanidin Biosynthesis. Physiol. Plant. 2014, 152, 431–440. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).