Exploiting the Genetic Potential of Cowpea in An Intercropping Complex
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site
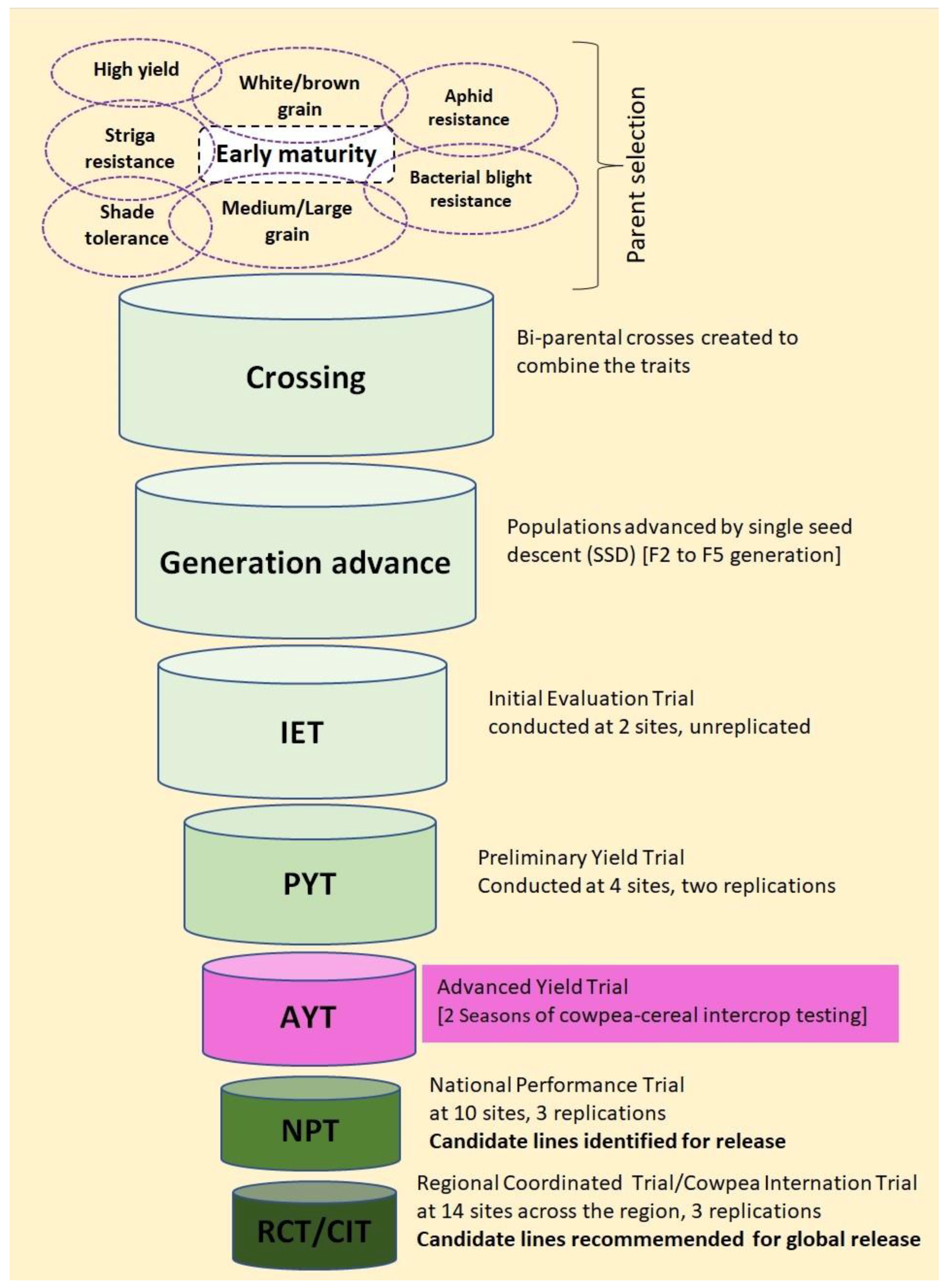
2.2. Genetic Materials
2.3. Design, Layout, and Data Collection
2.4. Statistical Analysis
- represents the average response of the kth genotype in the jth intercropping pattern in the ith replication and lth year, and is the response of kth genotype in the ith replication and lth year.
- i = 1…r replications and is the replication factor.
- k = 1…g genotypes and is the genotype factor.
- j = 1…p cropping patterns and is the cropping pattern factor.
- l = 1…y years and is the year factor.
- Total plots (N) = 2 replications × 35 genotypes × 3 patterns = 210 plots per year.
- is the population mean.
- is a term for replication that is nested within the year.
- is a two-way interaction term for genotype and intercropping pattern.
- is a two-way interaction term for genotype and year.
- is a two-way interaction term for intercropping pattern and year.
- is a three-way interaction term for genotype, intercropping pattern, and year.
- is residual or error variance for the RCBD model.
- and are the main-plot and sub-plot error terms, respectively.
3. Results
3.1. Effects of Genotypes, Cropping Patterns, and Years
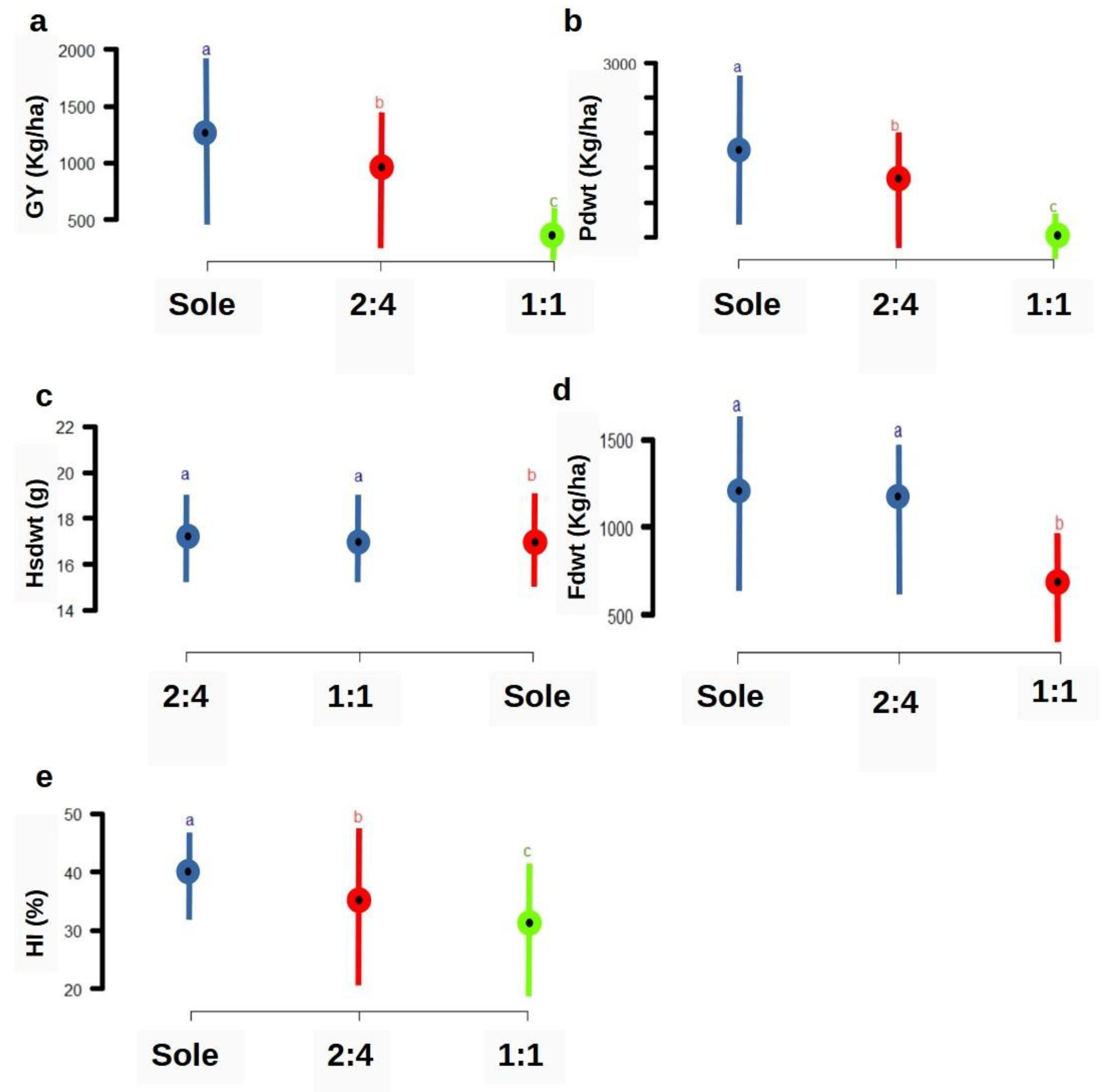
3.2. Mean Comparison of Cropping Patterns
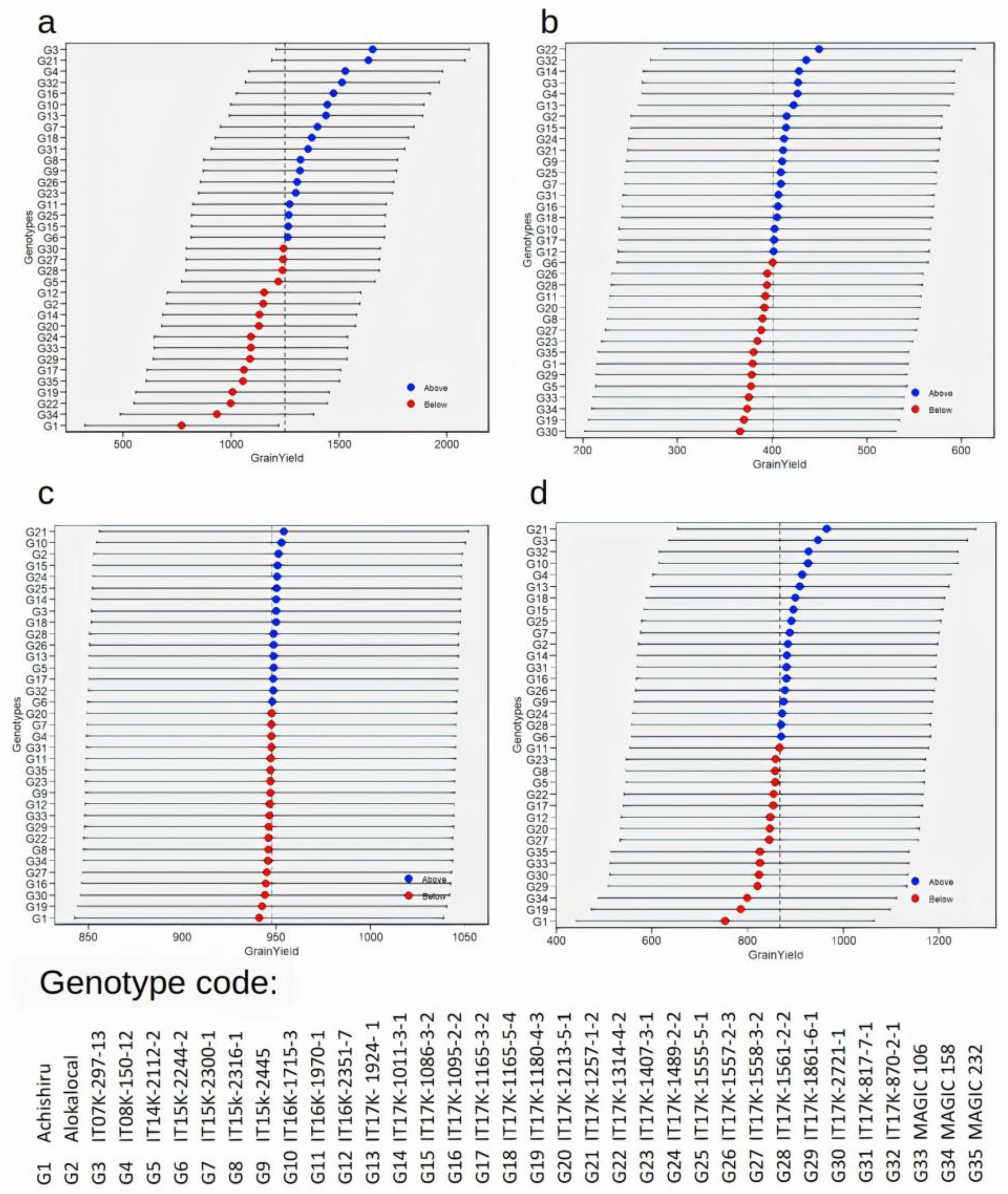
3.3. Performance of Genotypes
3.4. Heritability and Genetic Advance
3.5. Genetic and Phenotypic Correlations
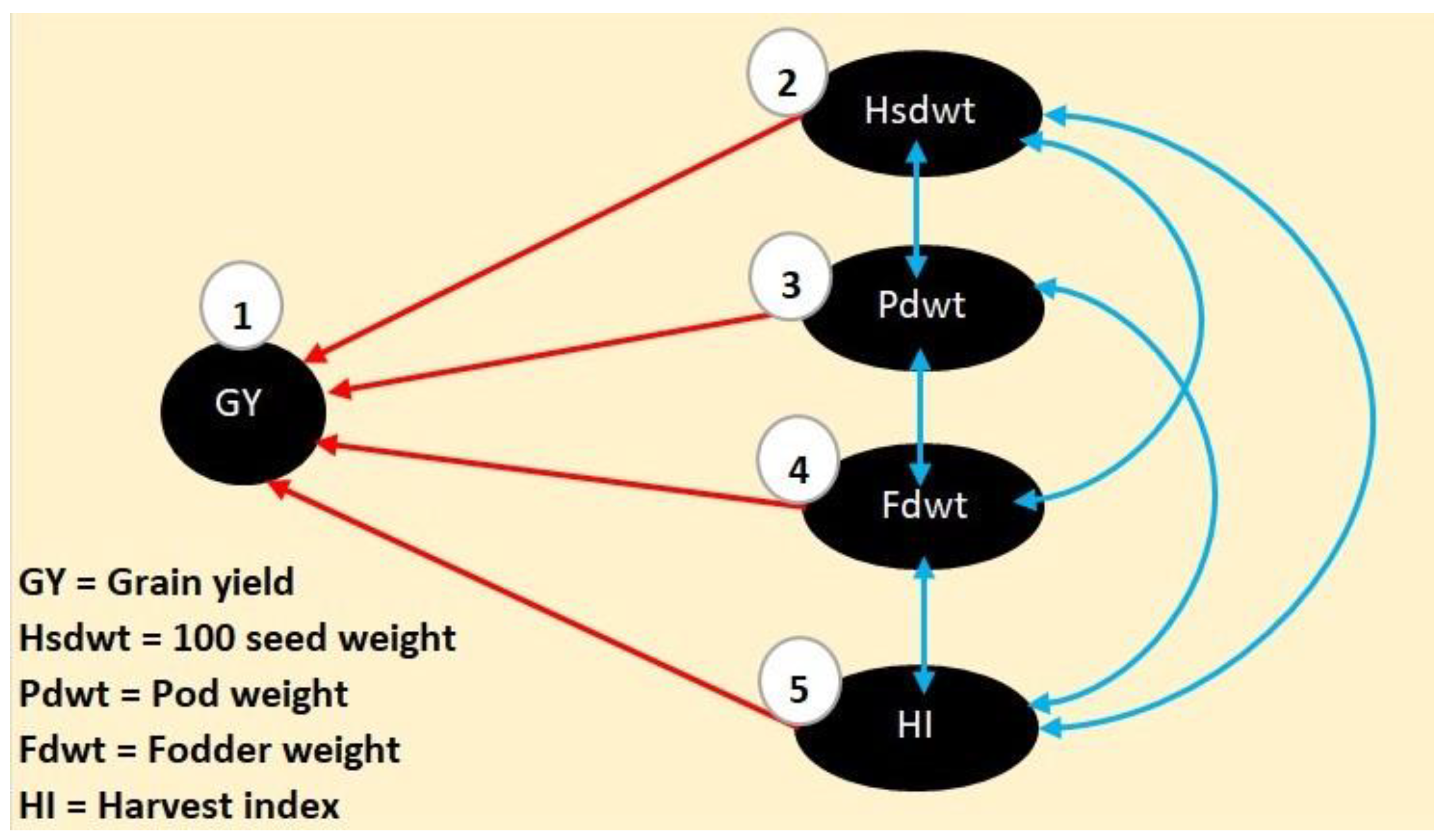
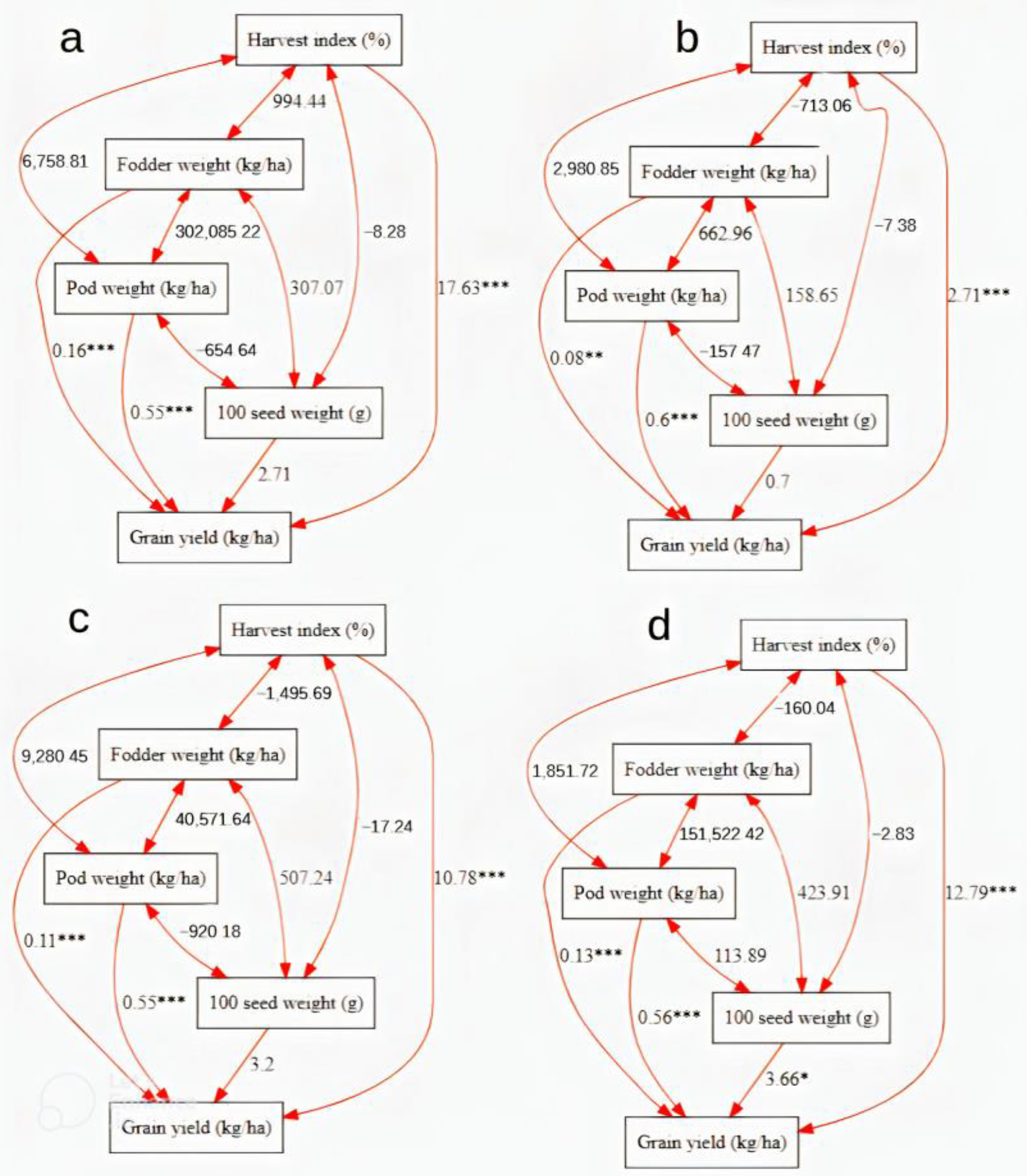
3.6. Path Coefficient Analysis
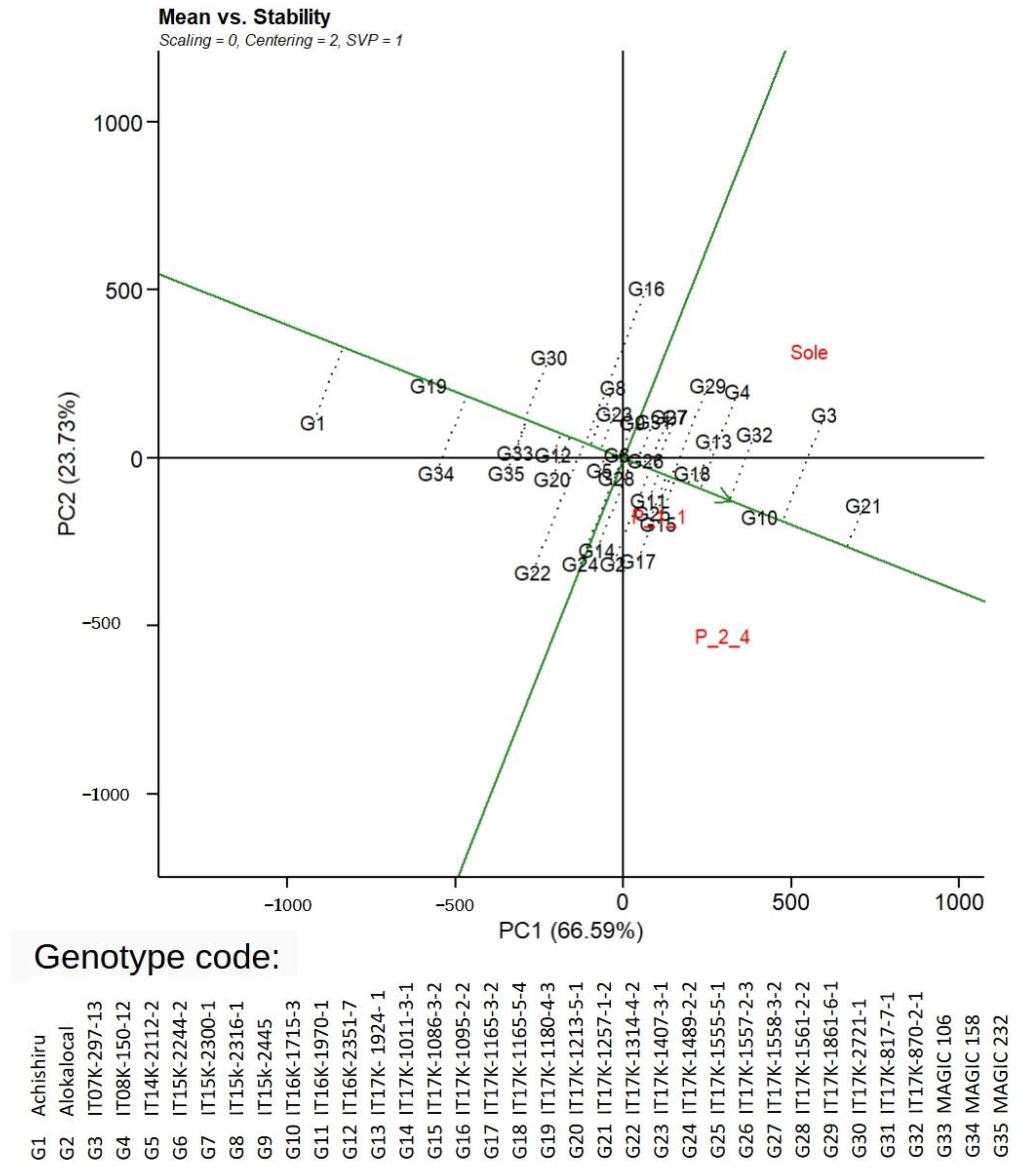
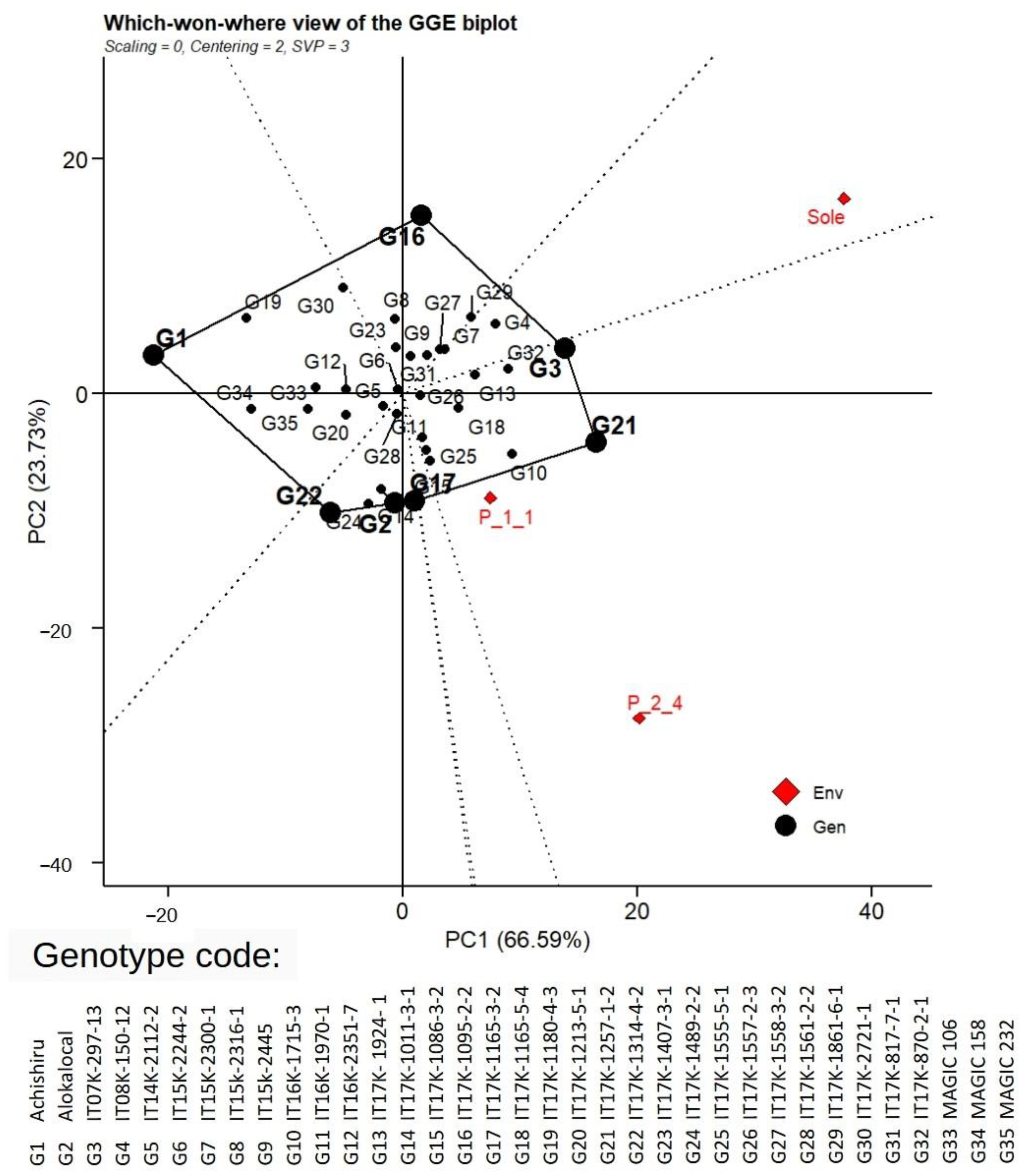
3.7. Adaptation and Stability of Genotypes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Timko, M.P.; Singh, B.B. Cowpea, a Multifunctional Legume. In Genomics of Tropical Crop Plants; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Singh, B.B. Cowpea: The Food Legume of the 21st Century; Crop Science Society of America: Madison, WI, USA, 2014; ISBN 978-0-891-18621-2. [Google Scholar]
- Boukar, O.; Togola, A.; Chamarthi, S.; Belko, N.; Ishikawa, H.; Suzuki, K.; Fatokun, C. Cowpea [Vigna Unguiculata (L.) Walp.] Breeding. In Advances in Plant Breeding Strategies: Legumes; Springer International Publishing: Cham, Switzerland, 2019; Volume 7, pp. 201–243. ISBN 9783030234003. [Google Scholar]
- da Silva, A.C.; da Costa Santos, D.; Junior, D.L.T.; da Silva, P.B.; dos Santos, R.C.; Siviero, A. Cowpea: A Strategic Legume Species for Food Security and Health. In Legume Seed Nutraceutical Research; IntechOpen: London, UK, 2018; ISBN 978-1-78985-398-8. [Google Scholar]
- Jayathilake, C.; Visvanathan, R.; Deen, A.; Bangamuwage, R.; Jayawardana, B.C.; Nammi, S.; Liyanage, R. Cowpea: An Overview on Its Nutritional Facts and Health Benefits. J. Sci. Food Agric. 2018, 98, 4793–4806. [Google Scholar] [CrossRef] [PubMed]
- Shivay, Y.S.; Singh, U.; Prasad, R.; Kaur, R. Agronomic Interventions for Micronutrient Biofortification of Pulses. Indian J. Agron. 2016, 4, 61–72. [Google Scholar]
- Kebede, E.; Bekeko, Z. Expounding the Production and Importance of Cowpea (Vigna Unguiculata (L.) Walp.) in Ethiopia. Cogent Food Agric. 2020, 6, 1769805. [Google Scholar] [CrossRef]
- Mohammed, S.B.; Dzidzienyo, D.K.; Umar, M.L.; Ishiyaku, M.F.; Tongoona, P.B.; Gracen, V. Appraisal of Cowpea Cropping Systems and Farmers’ Perceptions of Production Constraints and Preferences in the Dry Savannah Areas of Nigeria. CABI Agric. Biosci. 2021, 2, 25. [Google Scholar] [CrossRef]
- Maitra, S.; Hossain, A.; Brestic, M.; Skalicky, M.; Ondrisik, P.; Gitari, H.; Brahmachari, K.; Shankar, T.; Bhadra, P.; Palai, J.B.; et al. Intercropping—A Low Input Agricultural Strategy for Food and Environmental Security. Agronomy 2021, 11, 343. [Google Scholar] [CrossRef]
- Kremer, R.J. Soil Health Benefits of the Solar Corridor Crop System. In The Solar Corridor Crop System: Implementation and Impacts; Deichman, C.L., Robert, J.K., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 79–101. ISBN 9780128147931. [Google Scholar]
- Asseng, S.; Zhu, Y.; Basso, B.; Wilson, T.; Cammarano, D. Simulation Modeling: Applications in Cropping Systems. In Encyclopedia of Agriculture and Food Systems; Neal, K.V.A., Ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 102–112. ISBN 9780080931395. [Google Scholar]
- Mortimore, M.; Singh, B.; Harris, F.; Blade, S. Cowpea in Traditional Cropping Systems. In Advances in Cowpea Research; Singh, B., Mohan, R.D., Dashiel, K., Jackai, L., Eds.; Co-publication of International Institute of Tropical Agriculture (IITA) and Japan International Research Center for Agricultural Sciences (JIRCAS): Ibadan, Nigeria, 1997; pp. 99–113. [Google Scholar]
- Ewansiha, S.U.; Kamara, A.Y.; Onyibe, J.E. Performance of Cowpea Cultivars When Grown as an Intercrop with Maize of Contrasting Maturities. Arch. Agron. Soil Sci. 2013, 60, 597–608. [Google Scholar] [CrossRef]
- Chamkhi, I.; Cheto, S.; Geistlinger, J.; Zeroual, Y.; Kouisni, L.; Bargaz, A.; Ghoulam, C. Legume-Based Intercropping Systems Promote Beneficial Rhizobacterial Community and Crop Yield under Stressing Conditions. Ind. Crops Prod. 2022, 183, 114958. [Google Scholar] [CrossRef]
- Lo Presti, E.; Badagliacca, G.; Romeo, M.; Monti, M. Does Legume Root Exudation Facilitate Itself P Uptake in Intercropped Wheat? J. Soil Sci. Plant Nutr. 2021, 21, 3269–3283. [Google Scholar] [CrossRef]
- Reddy, K.C.; Van Der Ploeg, J.; Maga, I. Genotype Effects in Millet/Cowpea Intercropping in the Semi-Arid Tropics of Niger. Exp. Agric. 1990, 26, 387–396. [Google Scholar] [CrossRef]
- Terao, T.; Watanabe, I.; Matsunaga, R.; Hakoyama, S.; Singh, B.B. Agro-Physiological Constraints in Intercropped Cowpea: An Analysis. In Advances in Cowpea Research; Singh, B.B., Mohan, R.D.R., Dashiel, K.E., Jackai, L.E.N., Eds.; Co-publication of International Institute of Tropical Agriculture (IITA) and Japan International Research Center for Agricultural Sciences (JIRCAS): Ibadan, Nigeria, 1997; pp. 129–140. [Google Scholar]
- Ehlers, J.D. Correlation of Performance of Sole-Crop and Intercrop Cowpeas with and without Protection from Insect Pests. Field Crops Res. 1994, 36, 133–143. [Google Scholar] [CrossRef]
- Ntare, B.R. Evaluation of Cowpea Cultivars for Intercropping with Pearl Millet in the Sahelian Zone of West Africa. Field Crops Res. 1989, 20, 31–40. [Google Scholar] [CrossRef]
- Smith, M.E.; Zobel, R.W. Plant Genetic Interactions in Alternative Cropping Systems: Considerations for Breeding Methods. In Plant Breeding and Sustainable Agriculture: Considerations for Objectives and Methods; Sleper chair, D.A., Barker, T.C., Bramel-Cox, P.J., Eds.; John Wiley & Sons, Ltd.: Madison, WI, USA, 1991; pp. 57–81. ISBN 9780891185970. [Google Scholar]
- Blade, S.F.; Mather, D.E.; Singh, B.B.; Smith, D.L. Evaluation of Yield Stability of Cowpea under Sole and Intercrop Management in Nigeria. Euphytica 1991, 61, 193–201. [Google Scholar] [CrossRef]
- Padi, F.K. Genotype × Environment Interaction and Yield Stability in a Cowpea-Based Cropping System. Euphytica 2007, 158, 11–25. [Google Scholar] [CrossRef]
- Davis, J.H.C.; Garcia, S. Competitive Ability and Growth Habit of Indeterminate Beans and Maize for Intercropping. Field Crops Res. 1983, 6, 59–75. [Google Scholar] [CrossRef]
- Francis, C.A.; Prager, M.; Tejada, G.; Laing, D.R. Maize Genotype by Cropping Pattern Interactions: Monoculture vs. Intercropping1. Crop Sci. 1983, 23, 302–306. [Google Scholar] [CrossRef]
- Blade, S.F.; Shetty, S.V.R.; Terao, T.; Singh, B.B. Recent Developments in Cowpea Cropping Systems Research. In Advances in Cowpea Research; Singh, B.B., Raj, M., Dashiell, K.E., Jackai, L.E.N., Eds.; Co-publication of International Institute of Tropical Agriculture (IITA) and Japan International Research Center for Agricultural Sciences (JIRCAS): Ibadan, Nigeria, 1997; pp. 114–128. ISBN 978131110X. [Google Scholar]
- Singh, B.B.; Chambliss, O.L.; Sharma, B. Recent Advances in Cowpea Breeding. In Advances in Cowpea Research; International Institute of Tropical Agriculture (IITA)]: Ibadan, Nigeria, 1997; pp. 30–49. [Google Scholar]
- Craufurd, P.Q. Effect of Plant Density on the Yield of Sorghum-Cowpea and Pearl Millet-Cowpea Intercrops in Northern Nigeria. Exp. Agric. 2000, 36, 379–395. [Google Scholar] [CrossRef]
- Ewansiha, S.U.; Udensi, U.E.; Kamara, A.Y.; Meena, V.S. Effect of Tillage on the Growth and Yield of Cowpea Varieties in Sudan Savanna Agroecology of Northern Nigeria. Annu. Res. Rev. Biol. 2015, 5, 275–284. [Google Scholar] [CrossRef][Green Version]
- Bassi, J.A.; Dugje, I.Y.; Aminu, D. Agronomic Performances of Pearl Millet (Pennisetum Glaucum L.) R. Br.) Varieties Intercropped with Legumes in Sahelian Savanna of Nigeria. J. Agric. Crop Res. 2019, 7, 194–203. [Google Scholar] [CrossRef]
- Adeyanju, A.O. Genetics of Harvest and Leaf-Yield Indices in Cowpea. J. Crop Improv. 2009, 23, 266–274. [Google Scholar] [CrossRef]
- Alvarado, G.; Rodríguez, F.M.; Pacheco, A.; Burgueño, J.; Crossa, J.; Vargas, M.; Pérez-Rodríguez, P.; Lopez-Cruz, M.A. META-R: A Software to Analyze Data from Multi-Environment Plant Breeding Trials. Crop J. 2020, 8, 745–756. [Google Scholar] [CrossRef]
- de Mendiburu, F. Agricolae: Statistical Procedures for Agricultural Research. R Package 2021.
- Bates, D.; Mächler, M.; Bolker, B.M.; Walker, S.C. Fitting Linear Mixed-Effects Models Using Lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Allard, R.W. Principles of Plant Breeding, 2nd ed.; John Wiley & Sons, Inc.: New York, NY, USA, 1960; ISBN 978-0-471-02309-8. [Google Scholar]
- Cooper, M.; DeLacy, I.H. Relationships among Analytical Methods Used to Study Genotypic Variation and Genotype-by-Environment Interaction in Plant Breeding Multi-Environment Experiments. Theor. Appl. Genet. 1994, 88, 561–572. [Google Scholar] [CrossRef] [PubMed]
- Sodini, S.M.; Kemper, K.E.; Wray, N.R.; Trzaskowski, M. Comparison of Genotypic and Phenotypic Correlations: Cheverud’s Conjecture in Humans. Genetics 2018, 209, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Rosseel, Y. Lavaan: An R Package for Structural Equation Modeling. J. Stat. Softw. 2012, 48, 1–36. [Google Scholar] [CrossRef]
- Wright, S. Correlation and Causation. J. Agric. Res. 1921, 20, 557–585. [Google Scholar]
- Zimmermann, M.J.O.; Rosielle, A.A.; Waines, J.G.; Foster, K.W. A Heritability and Correlation Study of Grain Yield, Yield Components, and Harvest Index of Common Bean in Sole Crop and Intercrop. Field Crops Res. 1984, 9, 109–118. [Google Scholar] [CrossRef]
- Olivoto, T.; Lúcio, A.D.C. Metan: An R Package for Multi-Environment Trial Analysis. Methods Ecol. Evol. 2020, 11, 783–789. [Google Scholar] [CrossRef]
- Yan, W.; Kang, M.S. GGE Biplot Analysis: A Graphical Tool for Breeders, Geneticists, and Agronomists, 1st ed.; CRC Press: Boca Raton, FL, USA, 2002; ISBN 9780429122729. [Google Scholar]
- Rahmstorf, S.; Coumou, D. Increase of Extreme Events in a Warming World. Proc. Natl. Acad. Sci. USA 2011, 108, 17905. [Google Scholar] [CrossRef]
- Haug, B.; Messmer, M.M.; Enjalbert, J.; Goldringer, I.; Forst, E.; Flutre, T.; Mary-Huard, T.; Hohmann, P. Advances in Breeding for Mixed Cropping—Incomplete Factorials and the Producer/Associate Concept. Front. Plant Sci. 2021, 11, 2176. [Google Scholar] [CrossRef]
- Bedoussac, L.; Journet, E.P.; Hauggaard-Nielsen, H.; Naudin, C.; Corre-Hellou, G.; Jensen, E.S.; Prieur, L.; Justes, E. Ecological Principles Underlying the Increase of Productivity Achieved by Cereal-Grain Legume Intercrops in Organic Farming. A Review. Agron. Sustain. Dev. 2015, 35, 911–935. [Google Scholar] [CrossRef]
- Islam, N.; Zamir, M.S.I.; Din, S.M.U.; Farooq, U.; Arshad, H.; Bilal, A.; Sajjad, M.T.; Islam, N.; Zamir, M.S.I.; Din, S.M.U.; et al. Evaluating the Intercropping of Millet with Cowpea for Forage Yield and Quality. Am. J. Plant Sci. 2018, 9, 1781–1793. [Google Scholar] [CrossRef][Green Version]
- Davis, J.H.C.; Woolley, J.N. Genotypic Requirement for Intercropping. Field Crops Res. 1993, 34, 407–430. [Google Scholar] [CrossRef]
- Woolley, J.N.; Rodríguez, W. Cultivar X Cropping System Interactions in Relay and Row Intercropping of Bush Beans with Different Maize Plant Types. Exp. Agric. 1987, 23, 181–192. [Google Scholar] [CrossRef]
- Kouyaté, Z.; Krasova-Wade, T.; Yattara, I.I.; Neyra, M. Effects of Cropping System and Cowpea Variety on Symbiotic Potential and Yields of Cowpea (Vigna Unguiculata L. Walp) and Pearl Millet (Pennisetum Glaucum L.) in the Sudano-Sahelian Zone of Mali. Int. J. Agron. 2012, 2012, 761391. [Google Scholar] [CrossRef]
- Nelson, W.C.D.; Hoffmann, M.P.; Vadez, V.; Roetter, R.P.; Whitbread, A.M. Testing Pearl Millet and Cowpea Intercropping Systems under High Temperatures. Field Crop. Res. 2018, 217, 150–166. [Google Scholar] [CrossRef]
- Haider, M.; Sheikh, B.; Rahman, M. Effect of Intercropping on Growth and Yield in Wheat-Lentil and Wheat-Chickpea Intercropping System at Different Planting Configurations. J. Innov. Dev. Strategy 2011, 5, 125–137. [Google Scholar]
- Habib, A.; Mohammad, S.; Jahan, B.; Bashir, A.; Hamayoon, K. Yield and Yield Components of Wheat and Gram Planted in Monoculture and in Combination at Different Row Directions and Crop Geometry. Sarhad J. Agric. 2000, 16, 237–246. [Google Scholar]
- Mandal, B.K.; Saha, S.; Jana, T.K. Yield Performance and Complementarity of Rice (Oryza Sativa) with Greengram (Phaseolus Radiatus), Blackgram (Phaseolus Mungo) and Pigeonpea (Cajanus Cajan) under Different Rice-Legume Associations. Indian J. Agron. 2000, 45, 41–47. [Google Scholar]
- Falconer, D.S. The Problem of Environment and Selection. Am. Nat. 1952, 86, 293–298. [Google Scholar] [CrossRef]
- Moore, V.M.; Schlautman, B.; Fei, S.Z.; Roberts, L.M.; Wolfe, M.; Ryan, M.R.; Wells, S.; Lorenz, A.J. Plant Breeding for Intercropping in Temperate Field Crop Systems: A Review. Front Plant Sci. 2022, 13, 862. [Google Scholar] [CrossRef]
- Brummer, E.C.; Baker, R.F. Breeding for Cropping Systems. In Plant Breeding: The Arnel R. Hallauer International Symposium; Lamkey, K.R., Michael, L., Eds.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2008; pp. 97–106. ISBN 9780813828244. [Google Scholar]
- Bänziger, M.; Cooper, M. Breeding for Low Input Conditions and Consequences for Participatory Plant Breeding: Examples from Tropical Maize and Wheat. Euphytica 2001, 122, 503–519. [Google Scholar] [CrossRef]
- Holland, J.B.; Brummer, E.C. Cultivar Effects on Oat–Berseem Clover Intercrops. Agron. J. 1999, 91, 321–329. [Google Scholar] [CrossRef]
- Lynch, M.; Walsh, B. Genetics and Analysis of Quantitative Traits, 1st ed.; Sinauer Associates: Sunderland, MA, USA, 1998; ISBN 9780878934812. [Google Scholar]
- Yan, W. Crop Variety Trials: Data Management and Analysis, 1st ed.; John Willey & Sons, Inc.: New Delhi, India, 2014; ISBN 978-1-118-68864-9. [Google Scholar]
- Bidinger, F.R.; Weltzien, E.; Mahalakshmi, R.V.; Singh, S.D.; Rao, K.P. Evaluation of Landrace Topcross Hybrids of Pearl Millet for Arid Zone Environments. Euphytica 1994, 76, 215–226. [Google Scholar] [CrossRef]
- Singh, S.; Molina, A.; Gepts, P. Potential of Wild Common Bean for Seed Yield Improvement of Cultivars in the Tropics. Can. J. Plant Sci. 1995, 75, 807–813. [Google Scholar] [CrossRef]

| Source of Variation | Df | MS | Den.df | ||||
|---|---|---|---|---|---|---|---|
| Pdwt a | GY b | Fdwt c | Hsdwt d | HI e | |||
| Year (Y) | 1 | 187,976,331 *** | 94,495,477 ** | 2,576,623 ** | 341.89 * | 36143 ** | Y/Rep |
| Y/Rep | 2 | 92,604 | 188,380 ns | 19,411 ns | 10.90 | 159 | Pooled error (a) |
| Pattern (P) | 2 | 48,892,462 *** | 24,942,335 *** | 12437044 ** | 6.29 | 2717 * | Pooled error (a) |
| P × Y | 2 | 15,375,111 ** | 8,003,522 ** | 2,911,634 * | 23.72 | 852 | Pooled error (a) |
| Pooled error (a) | 4 | 461,656 | 272,015 | 258,384 | 4.38 | 278 | |
| Genotype (G) | 34 | 718,325 *** | 345,532 *** | 1,676,127 *** | 81.96 *** | 301 *** | Pooled error (b) |
| G × Y | 34 | 383,060 *** | 169,757 *** | 422,733 *** | 3.52 *** | 117 *** | Pooled error (b) |
| G × P | 68 | 245,121 ** | 110,555 ** | 281,291 ** | 1.74 * | 42 | Pooled error (b) |
| G × P × Y | 67 | 199,054 | 82,951 | 203,459 | 1.54 | 49 | Pooled error (b) |
| Pooled error (b) | 191 | 157,884 | 65,430 | 172,784 | 1.18 | 37 | |
| Trait | Pattern | H | σ2G | σ2GP | σ2GY | σ2GPY | σ2e | µ | LSD | CV (%) | GA | GAM (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GY | Sole | 0.66 | 58,504.85 *** | 17,940.35 | 83,116.71 | 1222.51 | 419.07 | 23.58 | 345.62 | 28.27 | ||
| 1:1 | 0.18 | 2626.50 | 6295.46 | 36,664.47 | 392.51 | 134.59 | 48.78 | 37.71 | 9.61 | |||
| 2:4 | 0.01 | 573.07 | 55,105.39 | 72,908.14 | 920.34 | 628.35 | 29.34 | 4.20 | 0.46 | |||
| Combined | 0.47 | 14,513.00 *** | 6163.00 | 15,249.00 | 8489.00 | 65,430.00 | 867.98 | 59.46 | 29.47 | 144.52 | 16.65 | |
| Podwt | Sole | 0.63 | 108,953.86 *** | 46,326.12 | 159,507.30 | 1707.60 | 593.96 | 23.39 | 461.07 | 27.00 | ||
| 1:1 | 0.22 | 5985.10 | 5663.44 | 71,222.39 | 555.37 | 197.31 | 48.05 | 64.38 | 11.59 | |||
| 2:4 | 0.00 | 29.66 | 121,183.25 | 235,012.00 | 1302.57 | 1004.75 | 36.94 | 0.15 | 0.01 | |||
| Combined | 0.44 | 27,785.00 *** | 9994.00 | 31,345.00 | 23,078.00 | 157,884.00 | 1217.59 | 92.37 | 32.63 | 193.07 | 15.86 | |
| Hsdwt | Sole | 0.94 | 6.85 *** | 0.58 | 0.74 | 16.86 | 2.23 | 5.10 | 4.44 | 26.35 | ||
| 1:1 | 0.89 | 6.44 *** | 0.69 | 1.76 | 17.17 | 2.56 | 7.73 | 4.21 | 24.50 | |||
| 2:4 | 0.94 | 7.14 *** | 0.30 | 1.08 | 17.30 | 1.89 | 6.01 | 4.55 | 26.28 | |||
| Combined | 0.96 | 6.80 *** | 0.06 | 0.36 | 0.08 | 1.20 | 17.10 | 0.25 | 6.35 | 4.48 | 26.17 | |
| Fdwt | Sole | 0.58 | 105,716.36 ** | 84,650.55 | 135,843.93 | 1207.86 | 619.16 | 30.51 | 434.89 | 36.01 | ||
| 1:1 | 0.65 | 50,873.04 *** | 0.00 | 109,030.39 | 671.41 | 389.94 | 49.18 | 319.41 | 47.57 | |||
| 2:4 | 0.68 | 213,137.43 *** | 70,804.09 | 260,766.84 | 1165.40 | 919.58 | 43.82 | 668.13 | 57.33 | |||
| Combined | 0.71 | 101,500.00 *** | 19,490.00 | 36,220.00 | 17,840.00 | 172,784.00 | 1024.11 | 96.62 | 40.59 | 470.27 | 45.92 | |
| HI | Sole | 0.27 | 5.93 | 17.13 | 30.47 | 39.57 | 6.04 | 13.95 | 2.21 | 5.59 | ||
| 1:1 | 0.71 | 30.75 *** | 5.20 | 40.69 | 30.65 | 9.11 | 20.82 | 8.18 | 26.70 | |||
| 2:4 | 0.36 | 13.79 | 28.35 | 40.74 | 34.97 | 14.54 | 18.25 | 3.91 | 11.18 | |||
| Combined | 0.66 | 18.31 *** | 0.00 | 12.27 | 1.25 | 37.00 | 35.50 | 1.41 | 17.04 | 6.10 | 17.19 |
| Trait | Sole vs. 1:1 | Sole vs. 2:4 | 1:1 vs. 2:4 | |
|---|---|---|---|---|
| GY | rg | 1.00 ** | 1.00 ** | 1.00 ** |
| rp | 0.37 * | 0.40 * | 0.42 ** | |
| Podwt | rg | 0.90 | 1.00 ** | 1.00 ** |
| rp | 0.29 | 0.47 ** | 0.34 * | |
| Hsdwt | rg | 0.99 | 1.00 ** | 1.00 ** |
| rp | 0.88 *** | 0.94 *** | 0.91 *** | |
| Fdwt | rg | 1.00 ** | 1.00 ** | 1.00 ** |
| rp | 0.62 *** | 0.70 *** | 0.66 *** | |
| HI | rg | 1.00 ** | 1.00 ** | 1.00 ** |
| rp | 0.69 *** | 0.69 *** | 0.65 *** |
| Sole | 1:1 | 2:4 | ||||
|---|---|---|---|---|---|---|
| Trait | rg | rp | rg | rp | rg | rp |
| GY vs. Podwt | 1.00 *** | 0.95 *** | 0.93 *** | 0.98 *** | 0.96 *** | 0.86 *** |
| GY vs. Fdwt | 0.72 *** | 0.63 *** | 0.18 | 0.36 * | 0.79 *** | 0.54 *** |
| GY vs. Hsdwt | 0.57 *** | 0.33 | 0.28 | 0.04 | 0.61 *** | 0.09 |
| GY vs. HI | 0.32 | 0.15 | 0.5 ** | 0.28 | −0.19 | −0.09 |
| Hsdwt vs. Podwt | 0.5 ** | 0.26 | 0.42 ** | 0.01 | 0.42 ** | −0.02 |
| Hsdwt vs. Fdwt | 0.67 *** | 0.62 *** | 0.42 ** | 0.33 * | 0.53 *** | 0.42 ** |
| Hsdwt vs. HI | −0.21 | −0.41 ** | −0.25 | −0.17 | −0.18 | −0.44 ** |
| Podwt vs. Fdwt | 0.78 *** | 0.65 *** | 0.33 * | 0.39 * | 0.84 *** | 0.54 *** |
| Podwt vs. HI | 0.23 | 0.03 | 0.41 * | 0.19 | −0.45 ** | −0.27 |
| Fdwt vs. HI | −0.49 ** | −0.59 *** | −0.84 *** | −0.70 *** | −0.81 *** | −0.77 *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ongom, P.O.; Fatokun, C.; Togola, A.; Mohammed, S.B.; Ishaya, D.J.; Bala, G.; Popoola, B.; Mansur, A.; Tukur, S.; Ibikunle, M.; et al. Exploiting the Genetic Potential of Cowpea in An Intercropping Complex. Agronomy 2023, 13, 1594. https://doi.org/10.3390/agronomy13061594
Ongom PO, Fatokun C, Togola A, Mohammed SB, Ishaya DJ, Bala G, Popoola B, Mansur A, Tukur S, Ibikunle M, et al. Exploiting the Genetic Potential of Cowpea in An Intercropping Complex. Agronomy. 2023; 13(6):1594. https://doi.org/10.3390/agronomy13061594
Chicago/Turabian StyleOngom, Patrick Obia, Christian Fatokun, Abou Togola, Saba B. Mohammed, Daniel Jockson Ishaya, Garba Bala, Bosede Popoola, Ahmad Mansur, Sagir Tukur, Mumini Ibikunle, and et al. 2023. "Exploiting the Genetic Potential of Cowpea in An Intercropping Complex" Agronomy 13, no. 6: 1594. https://doi.org/10.3390/agronomy13061594
APA StyleOngom, P. O., Fatokun, C., Togola, A., Mohammed, S. B., Ishaya, D. J., Bala, G., Popoola, B., Mansur, A., Tukur, S., Ibikunle, M., Abdulkazeem, B., & Boukar, O. (2023). Exploiting the Genetic Potential of Cowpea in An Intercropping Complex. Agronomy, 13(6), 1594. https://doi.org/10.3390/agronomy13061594








