The Relationship between Cadmium-Related Gene Sequence Variations in Rice and Cadmium Accumulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Measurement of Cd Concentrations in Rice Grains
2.3. Sequence Data Analysis
2.4. Association Analysis
2.5. Analysis of Allele Phenotypic Effects
3. Results
3.1. Phenotypic Variations in Cd Levels in Rice Grains from Natural Population Variants
3.2. Sequence Polymorphism Analyses
3.3. Cd-Associated Gene Sequence Diversity
3.4. Identification of Cd Accumulation Groups of Genotypes Based on 12 Candidate Genes
3.5. Association Analysis of SNPs Related to Cd Accumulation in Rice Grains
3.6. Combination of Favorable Alleles
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bin, H.E.; Zhaojun, Y.; Jianbo, S.; Guibin, J.J. Research progress of heavy metal pollution in China: Sources, analytical methods, status, and toxicity. Chin. Sci. Bull. 2013, 58, 134–140. [Google Scholar]
- Zhang, J.; Zhu, Y.; Yu, L.; Yang, M.; Zou, X.; Yin, C.; Lin, Y. Research Advances in Cadmium Uptake, Transport and Resistance in Rice (Oryza sativa L.). Cells 2022, 11, 569. [Google Scholar] [CrossRef]
- Nawrot, T.S.; Staessen, J.A.; Roels, H.A.; Munters, E.; Cuypers, A.; Richart, T.; Ruttens, A.; Smeets, K.; Clijsters, H.; Vangronsveld, J. Cadmium exposure in the population: From health risks to strategies of prevention. Biometals Int. J. Role Met. Ions Biol. Biochem. Med. 2010, 23, 769–782. [Google Scholar] [CrossRef]
- Nordberg, G.F. Cadmium and health in the 21st century—Historical remarks and trends for the future. Biometals Int. J. Role Met. Ions Biol. Biochem. Med. 2004, 17, 485–489. [Google Scholar] [CrossRef]
- Shifaw, E. Review of Heavy Metals Pollution in China in Agricultural and Urban Soils. J. Health Pollut. 2018, 8, 180607. [Google Scholar] [CrossRef]
- Chen, J.; Huang, X.Y.; Salt, D.E.; Zhao, F.J. Mutation in OsCADT1 enhances cadmium tolerance and enriches selenium in rice grain. New Phytol. 2020, 226, 838–850. [Google Scholar] [CrossRef]
- Miyadate, H.; Adachi, S.; Hiraizumi, A.; Tezuka, K.; Nakazawa, N.; Kawamoto, T.; Katou, K.; Kodama, I.; Sakurai, K.; Takahashi, H.; et al. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 2011, 189, 190–199. [Google Scholar] [CrossRef]
- Satoh-Nagasawa, N.; Mori, M.; Nakazawa, N.; Kawamoto, T.; Nagato, Y.; Sakurai, K.; Takahashi, H.; Watanabe, A.; Akagi, H. Mutations in rice (Oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol. 2012, 53, 213–224. [Google Scholar] [CrossRef]
- Song, W.Y.; Lee, H.S.; Jin, S.R.; Ko, D.; Martinoia, E.; Lee, Y.; An, G.; Ahn, S.N. Rice PCR1 influences grain weight and Zn accumulation in grains. Plant Cell Environ. 2015, 38, 2327–2339. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, Y.; Zhang, L.; Hu, J.; Zhang, X.; Lu, K.; Dong, H.; Wang, D.; Zhao, F.J.; Huang, C.F.; et al. OsNRAMP5 contributes to manganese translocation and distribution in rice shoots. J. Exp. Bot. 2014, 65, 4849–4861. [Google Scholar] [CrossRef]
- Chang, J.D.; Huang, S.; Yamaji, N.; Zhang, W.; Ma, J.F.; Zhao, F.J. OsNRAMP1 transporter contributes to cadmium and manganese uptake in rice. Plant Cell Environ. 2020, 43, 2476–2491. [Google Scholar] [CrossRef]
- Yang, W.; Chen, L.; Ma, Y.; Hu, R.; Wang, J.; Li, W.; Dong, J.; Yang, T.; Zhou, L.; Chen, J.; et al. OsNRAMP2 facilitates Cd efflux from vacuoles and contributes to the difference in grain Cd accumulation between japonica and indica rice. Crop J. 2022, in press. [Google Scholar] [CrossRef]
- Chu, C.; Huang, R.; Liu, L.; Tang, G.; Xiao, J.; Yoo, H.; Yuan, M. The rice heavy-metal transporter OsNRAMP1 regulates disease resistance by modulating ROS homoeostasis. Plant Cell Environ. 2022, 45, 1109–1126. [Google Scholar] [CrossRef]
- Huang, J.; Jiang, J.; Meng, J.; Shu, Q.; Xu, S.; Liu, S.; Liu, Y.; Li, Y.; Tan, Y. Characterization and Evaluation of OsLCT1 and OsNramp5 Mutants Generated Through CRISPR/Cas9-Mediated Mutagenesis for Breeding Low Cd Rice. Rice Sci. 2019, 26, 88–97. [Google Scholar] [CrossRef]
- Ishikawa, S.; Ishimaru, Y.; Igura, M.; Kuramata, M.; Abe, T.; Senoura, T.; Hase, Y.; Arao, T.; Nishizawa, N.K.; Nakanishi, H. Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low-cadmium rice. Proc. Natl. Acad. Sci. USA 2012, 109, 19166–19171. [Google Scholar] [CrossRef]
- Bari, M.A.; El-Shehawi, A.M.; Elseehy, M.M.; Naheen, N.N.; Rahman, M.M.; Kabir, A.H. Molecular characterization and bioinformatics analysis of transporter genes associated with Cd-induced phytotoxicity in rice (Oryza sativa L.). Plant Physiol. Biochem. PPB 2021, 167, 438–448. [Google Scholar] [CrossRef]
- Maghrebi, M.; Baldoni, E.; Lucchini, G.; Vigani, G.; Valè, G.; Sacchi, G.A.; Nocito, F.F. Analysis of Cadmium Root Retention for Two Contrasting Rice Accessions Suggests an Important Role for OsHMA2. Plants 2021, 10, 806. [Google Scholar] [CrossRef]
- Liu, E.; Liu, X.; Zeng, S.; Zhao, K.; Zhu, C.; Liu, Y.; Breria, M.C.; Zhang, B.; Hong, D. Time-course association mapping of the grain-filling rate in rice (Oryza sativa L.). PLoS ONE 2015, 10, e0119959. [Google Scholar] [CrossRef]
- Tavarez, M.; Grusak, M.A.; Sankaran, R.P. Effects of Zinc Fertilization on Grain Cadmium Accumulation, Gene Expression, and Essential Mineral Partitioning in Rice. Agronomy 2022, 12, 2182. [Google Scholar] [CrossRef]
- Sasaki, A.; Yamaji, N.; Ma, J.F. Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. J. Exp. Bot. 2014, 65, 6013–6021. [Google Scholar] [CrossRef]
- Lee, S.; Kim, Y.Y.; Lee, Y.; An, G. Rice P1B-type heavy-metal ATPase, OsHMA9, is a metal efflux protein. Plant Physiol. 2007, 145, 831–842. [Google Scholar] [CrossRef]
- Shimo, H.; Ishimaru, Y.; An, G.; Yamakawa, T.; Nakanishi, H.; Nishizawa, N.K. Low cadmium (LCD), a novel gene related to cadmium tolerance and accumulation in rice. J. Exp. Bot. 2011, 62, 5727–5734. [Google Scholar] [CrossRef]
- Tian, S.; Liang, S.; Qiao, K.; Wang, F.; Zhang, Y.; Chai, T. Co-expression of multiple heavy metal transporters changes the translocation, accumulation, and potential oxidative stress of Cd and Zn in rice (Oryza sativa). J. Hazard. Mater. 2019, 380, 120853. [Google Scholar] [CrossRef]
- Uraguchi, S.; Kamiya, T.; Sakamoto, T.; Kasai, K.; Sato, Y.; Nagamura, Y.; Yoshida, A.; Kyozuka, J.; Ishikawa, S.; Fujiwara, T. Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proc. Natl. Acad. Sci. USA 2011, 108, 20959–20964. [Google Scholar] [CrossRef]
- Oda, K.; Otani, M.; Uraguchi, S.; Akihiro, T.; Fujiwara, T. Rice ABCG43 is Cd inducible and confers Cd tolerance on yeast. Biosci. Biotechnol. Biochem. 2011, 75, 1211–1213. [Google Scholar] [CrossRef]
- Yan, H.; Xu, W.; Xie, J.; Gao, Y.; Wu, L.; Sun, L.; Feng, L.; Chen, X.; Zhang, T.; Dai, C.; et al. Variation of a major facilitator superfamily gene contributes to differential cadmium accumulation between rice subspecies. Nat. Commun. 2019, 10, 2562. [Google Scholar] [CrossRef]
- Luo, J.S.; Huang, J.; Zeng, D.L.; Peng, J.S.; Zhang, G.B.; Ma, H.L.; Guan, Y.; Yi, H.Y.; Fu, Y.L.; Han, B.; et al. A defensin-like protein drives cadmium efflux and allocation in rice. Nat. Commun. 2018, 9, 645. [Google Scholar] [CrossRef]
- Gupta, P.K.; Rustgi, S.; Kulwal, P.L. Linkage disequilibrium and association studies in higher plants: Present status and future prospects. Plant Mol. Biol. 2005, 57, 461–485. [Google Scholar] [CrossRef]
- Zondervan, K.T.; Cardon, L.R. The complex interplay among factors that influence allelic association. Nat. Rev. Genet. 2004, 5, 89–100. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A.; Thuillet, A.C.; Yu, J.; Pressoir, G.; Romero, S.M.; Mitchell, S.E.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S. Maize association population: A high-resolution platform for quantitative trait locus dissection. Plant J. Cell Mol. Biol. 2005, 44, 1054–1064. [Google Scholar] [CrossRef]
- Thornsberry, J.M.; Goodman, M.M.; Doebley, J.; Kresovich, S.; Nielsen, D.; Buckler, E.S.t. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 2001, 28, 286–289. [Google Scholar] [CrossRef]
- Andersen, J.R.; Schrag, T.; Melchinger, A.E.; Zein, I.; Lübberstedt, T. Validation of Dwarf8 polymorphisms associated with flowering time in elite European inbred lines of maize (Zea mays L.). TAG Theor. Appl. Genet. Theor. Angew. Genet. 2005, 111, 206–217. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T. ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Balding, D.J. A tutorial on statistical methods for population association studies. Nat. Rev. Genet. 2006, 7, 781–791. [Google Scholar] [CrossRef]
- Breseghello, F.; Sorrells, M.E. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 2006, 172, 1165–1177. [Google Scholar] [CrossRef]
- Diao, S.Y.; Zhang, L.Z.; Yuan, H. Progress in the toxicity mechanism of cadmium. Dongwu Yixue Jinzhan 2005, 26, 49–51. [Google Scholar]
- Rafati Rahimzadeh, M.; Rafati Rahimzadeh, M.; Kazemi, S.; Moghadamnia, A.A. Cadmium toxicity and treatment: An update. Casp. J. Intern. Med. 2017, 8, 135–145. [Google Scholar] [CrossRef]
- Lestari, P.; Lee, G.; Ham, T.H.; Reflinur; Woo, M.O.; Piao, R.; Jiang, W.; Chu, S.H.; Lee, J.; Koh, H.J. Single nucleotide polymorphisms and haplotype diversity in rice sucrose synthase 3. J. Hered. 2011, 102, 735–746. [Google Scholar] [CrossRef]
- Hao, X.; Wu, C.; Wang, R.; Tian, L.; Song, T.; Tan, H.; Peng, Y.; Zeng, M.; Chen, L.; Liang, M.; et al. Association between sequence variants in cadmium-related genes and the cadmium accumulation trait in thermo-sensitive genic male sterile rice. Breed. Sci. 2019, 69, 455–463. [Google Scholar] [CrossRef]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef]
- Li, W.; Zhu, Z.; Chern, M.; Yin, J.; Yang, C.; Ran, L.; Cheng, M.; He, M.; Wang, K.; Wang, J.; et al. A Natural Allele of a Transcription Factor in Rice Confers Broad-Spectrum Blast Resistance. Cell 2017, 170, 114–126. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Lu, Y.; Zinta, G.; Lang, Z.; Zhu, J.K. UTR-Dependent Control of Gene Expression in Plants. Trends Plant Sci. 2018, 23, 248–259. [Google Scholar] [CrossRef]
- Rose, A.B. The effect of intron location on intron-mediated enhancement of gene expression in Arabidopsis. Plant J. Cell Mol. Biol. 2004, 40, 744–751. [Google Scholar] [CrossRef]
- Rose, A.B. Intron-mediated regulation of gene expression. Curr. Top. Microbiol. Immunol. 2008, 326, 277–290. [Google Scholar] [CrossRef]
- Rose, A.B.; Carter, A.; Korf, I.; Kojima, N. Intron sequences that stimulate gene expression in Arabidopsis. Plant Mol. Biol. 2016, 92, 337–346. [Google Scholar] [CrossRef]
- Aguilar-Hernández, V.; Guzmán, P. Spliceosomal introns in the 5′ untranslated region of plant BTL RING-H2 ubiquitin ligases are evolutionary conserved and required for gene expression. BMC Plant Biol. 2013, 13, 179. [Google Scholar] [CrossRef]
- Khan, N.; You, F.M.; Datla, R.; Ravichandran, S.; Jia, B.; Cloutier, S. Genome-wide identification of ATP binding cassette (ABC) transporter and heavy metal associated (HMA) gene families in flax (Linum usitatissimum L.). BMC Genom. 2020, 21, 722. [Google Scholar] [CrossRef]
- Huang, Q.; Qiu, W.; Yu, M.; Li, S.; Lu, Z.; Zhu, Y.; Kan, X.; Zhuo, R. Genome-Wide Characterization of Sedum plumbizincicola HMA Gene Family Provides Functional Implications in Cadmium Response. Plants 2022, 11, 215. [Google Scholar] [CrossRef]
- Li, H.; Li, C.; Sun, D.; Yang, Z. OsPDR20 is an ABCG metal transporter regulating cadmium accumulation in rice. J. Environ. Sci. 2022, 136, 21–34. [Google Scholar] [CrossRef]
- Guo, J.; Chen, M.; Huang, Y.; Xie, S.; Zhang, X.; Zuo, T.; Hu, C.; Wang, G. Chloride application weakens cadmium immobilization by lime in paddy rice soil. Ecotoxicol. Environ. Saf. 2022, 241, 113761. [Google Scholar] [CrossRef]
- Chen, M.; Li, Z.; Huang, J.; Yan, Y.; Wu, T.; Bian, M.; Zhou, J.; Wang, Y.; Lyv, Y.; Hu, G.; et al. Dissecting the meteorological and genetic factors affecting rice grain quality in Northeast China. Genes Genom. 2021, 43, 975–986. [Google Scholar] [CrossRef]
- Moran, B.M.; Payne, C.; Langdon, Q.; Powell, D.L.; Brandvain, Y.; Schumer, M. The genomic consequences of hybridization. eLife 2021, 10, e69016. [Google Scholar] [CrossRef]

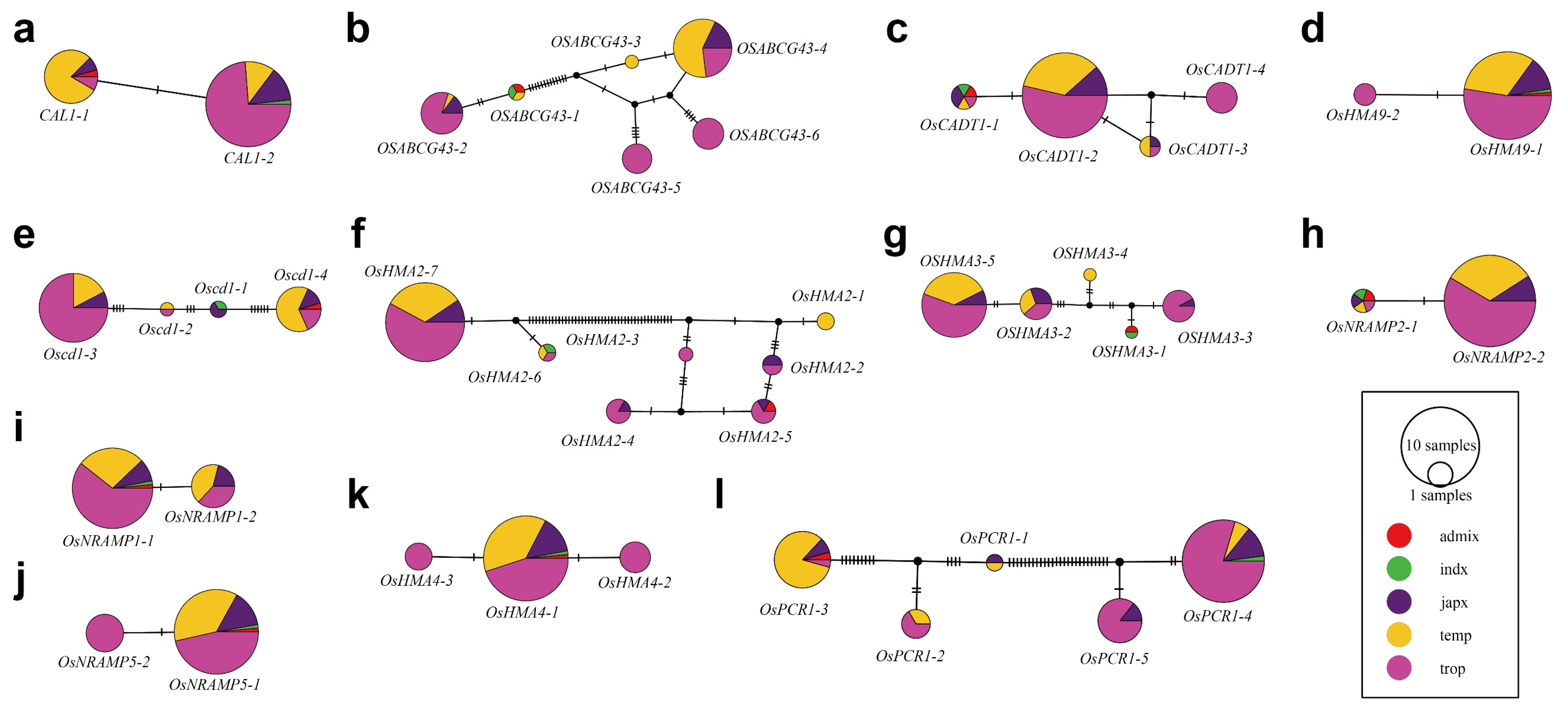
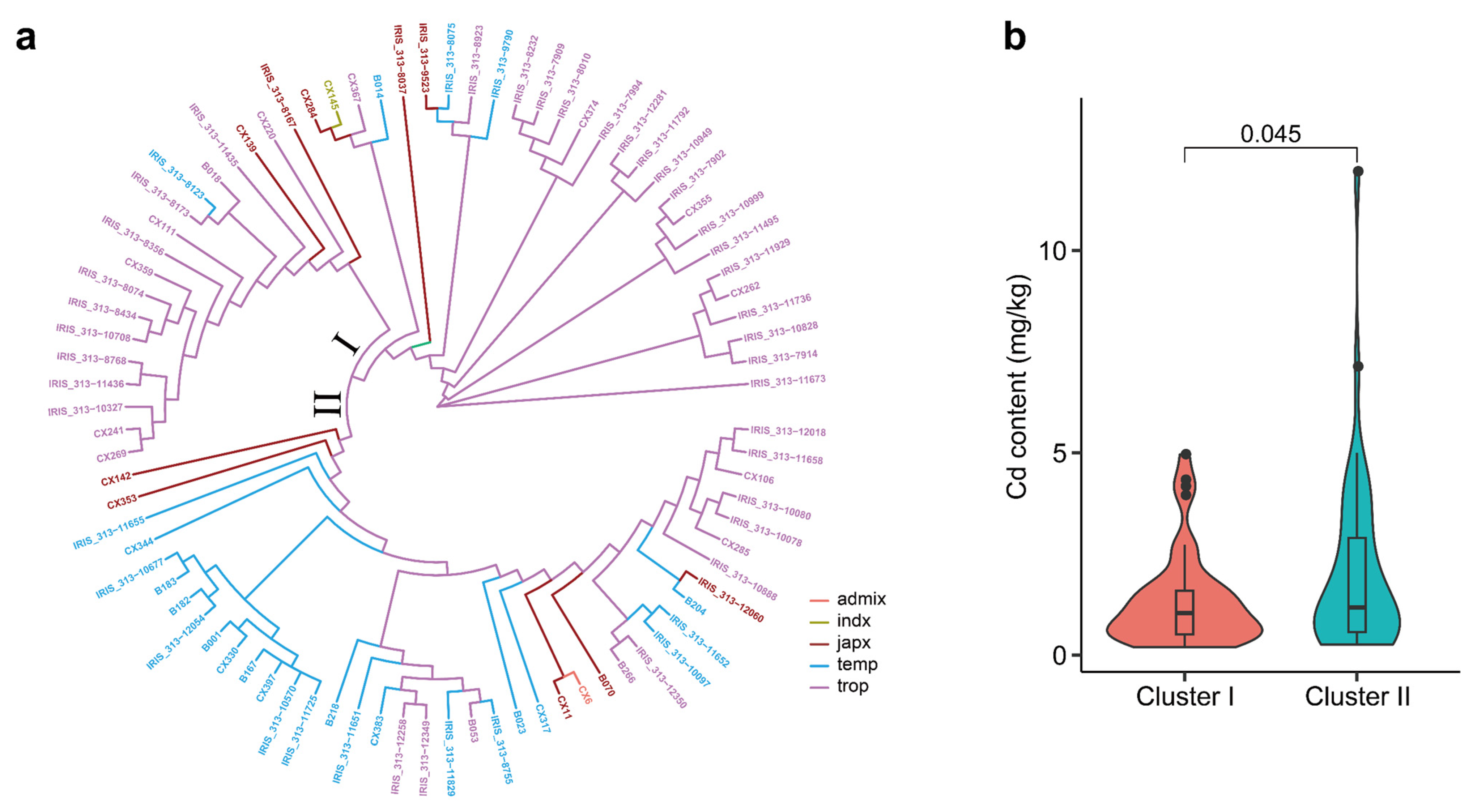
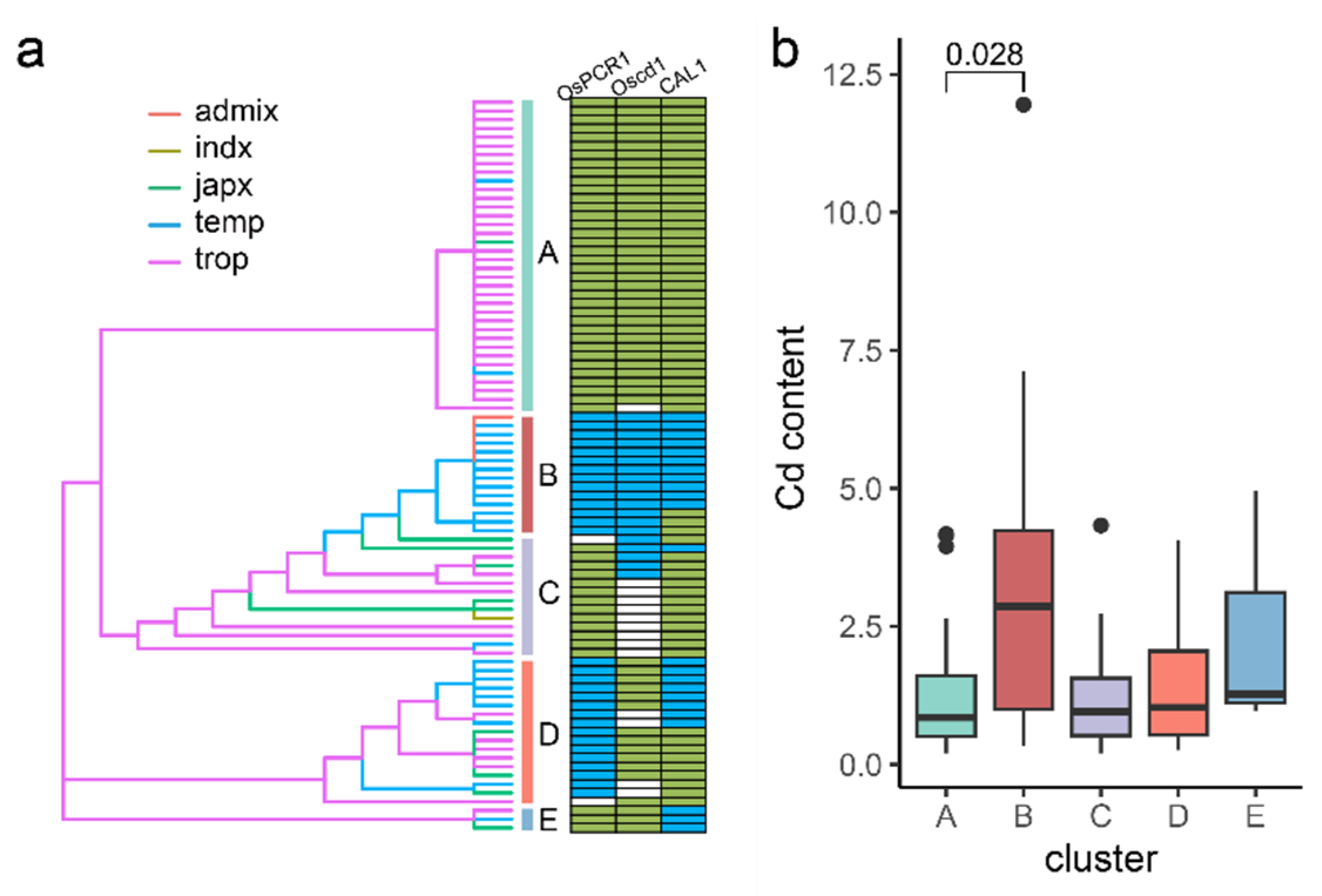
| Gene | Full Length (bp) | No. of Variations | No. of SNP | |||
|---|---|---|---|---|---|---|
| Coding Region | Noncoding Region | 3′-UTR Region | 5′-UTR Region | |||
| CAL1 | 702 | 6 | 2 | 0 | 1 | 3 |
| OsABCG43 | 13,391 | 33 | 7 | 19 | 7 | 0 |
| OsCADT1 | 4206 | 14 | 2 | 10 | 2 | 0 |
| Oscd1 | 3832 | 22 | 1 | 14 | 2 | 5 |
| OsHMA2 | 7276 | 60 | 6 | 17 | 25 | 12 |
| OsHMA3 | 3809 | 19 | 16 | 3 | 0 | 0 |
| OsHMA4 | 5904 | 22 | 11 | 7 | 3 | 1 |
| OsHMA9 | 6505 | 107 | 25 | 68 | 14 | 0 |
| OsNRAMP1 | 4856 | 22 | 5 | 14 | 2 | 1 |
| OsNRAMP2 | 4016 | 10 | 3 | 5 | 0 | 2 |
| OsNRAMP5 | 7263 | 73 | 4 | 58 | 4 | 7 |
| OsPCR1 | 1475 | 48 | 21 | 22 | 2 | 3 |
| Total | 436 | 103 | 237 | 62 | 34 | |
| Gene | Region | No. of Nucleotide Substitutions | Pi | Tajima’s D | Fu and Li’s D* Statistic | Fu and Li’s F* Statistic | Haplotype Diversity |
|---|---|---|---|---|---|---|---|
| CAL1 | Coding | 0 | 0 | 0 | 0 | 0 | 0 |
| Noncoding | 1 | 0.00054 | 1.31708 | 0.50284 | 0.81086 | 0.41 | |
| All | 1 | 0.00054 | 1.31708 | 0.50284 | 0.81086 | 0.41 | |
| OsABCG43 | Coding | 7 | 0.00038 | 1.81635 | 1.20535 | 1.50873 | 0.683 |
| Noncoding | 17 | 0.00098 | 2.63933 | 1.47212 | 1.91827 | 0.696 | |
| All | 24 | 0.00135 | 2.65336 | 1.70392 | 2.12496 | 0.699 | |
| OsCADT1 | Coding | 0 | 0 | 0 | 0 | 0 | 0 |
| Noncoding | 3 | 0.00009 | −0.43289 | 0.69853 | 0.442 | 0.309 | |
| All | 3 | 0.00009 | −0.43289 | 0.69853 | 0.442 | 0.309 | |
| Oscd1 | Coding | 1 | 0.00007 | 1.49554 | 0.50284 | 0.88397 | 0.447 |
| Noncoding | 10 | 0.00066 | 3.06736 | 1.13298 | 1.80449 | 0.547 | |
| All | 11 | 0.00073 | 3.16855 | 1.20535 | 1.92797 | 0.562 | |
| OsHMA2 | Coding | 5 | 0.00047 | 1.37746 | 0.69853 | 0.92065 | 0.361 |
| Noncoding | 46 | 0.00401 | 1.80795 | 1.05055 | 1.35628 | 0.361 | |
| All | 51 | 0.00448 | 1.85177 | 1.20535 | 1.53938 | 0.361 | |
| OsHMA3 | Coding | 7 | 0.00056 | 1.7227 | 0.69853 | 1.09012 | 0.539 |
| Noncoding | 1 | 0.00008 | 0.773901 | 0.50444 | 0.62169 | 0.315 | |
| All | 8 | 0.00063 | 1.77265 | 0.69853 | 1.09012 | 0.539 | |
| OsHMA4 | Coding | 2 | 0.00005 | −0.07085 | 0.69853 | 0.46949 | 0.327 |
| Noncoding | 0 | 0 | 0 | 0 | 0 | 0 | |
| All | 2 | 0.00005 | −0.07085 | 0.69853 | 0.46949 | 0.327 | |
| OsHMA9 | Coding | 0 | 0.00000 | 0 | 0 | 0 | 0 |
| Noncoding | 1 | 0.00002 | −0.44067 | 0.50284 | 0.22155 | 0.112 | |
| All | 1 | 0.00002 | −0.44067 | 0.50284 | 0.22155 | 0.112 | |
| OsNRAMP1 | Coding | 0 | 0 | 0 | 0 | 0 | 0 |
| Noncoding | 1 | 0.00007 | 0.912655 | 0.50284 | 0.69455 | 0.351 | |
| All | 1 | 0.00007 | 0.912655 | 0.50284 | 0.69455 | 0.351 | |
| OsNRAMP2 | Coding | 1 | 0.00001 | −0.68637 | 0.50526 | 0.1407 | 0.071 |
| Noncoding | 2 | 0.00003 | −0.8314 | 0.70469 | 0.2003 | 0.074 | |
| All | 3 | 0.00004 | −1.01128 | 0.85097 | 0.16714 | 0.052 | |
| OsNRAMP5 | Coding | 0 | 0 | 0 | 0 | 0 | 0 |
| Noncoding | 1 | 0.00004 | 0.603241 | 0.50284 | 0.55054 | 0.278 | |
| All | 1 | 0.00004 | 0.603241 | 0.50284 | 0.55054 | 0.278 | |
| OsPCR1 | Coding | 13 | 0.00095 | 4.08769 | 0.84089 | 1.5273 | 0.469 |
| Noncoding | 21 | 0.00144 | 4.19166 | 0.84089 | 1.16236 | 0.511 | |
| All | 34 | 0.00239 | 4.55707 | 1.13298 | 1.77874 | 0.511 |
| Gene | Pos | p-Value | R2 | Gene | Pos | p-Value | R2 |
|---|---|---|---|---|---|---|---|
| OsCADT1 | 37964661 | 0.000134 | 0.20677 | OsPCR1 | 825399 | 0.02427 | 0.08877 |
| 37965214 | 0.000223 | 0.18538 | 825477 | 0.01321 | 0.10132 | ||
| CAL1 | 25190520 | 0.000533 | 0.16794 | 825557 | 0.00782 | 0.08513 | |
| Oscd1 | 842611 | 0.02663 | 0.08463 | 825596 | 0.01527 | 0.09926 | |
| 842747 | 0.01065 | 0.07601 | 825772 | 0.01365 | 0.1006 | ||
| 843460 | 0.01065 | 0.07601 | 825773 | 0.01395 | 0.10011 | ||
| 844102 | 0.01024 | 0.07863 | 825910 | 0.00489 | 0.0926 | ||
| 844179 | 0.02797 | 0.08452 | 825926 | 0.0092 | 0.07894 | ||
| 844273 | 0.01173 | 0.10277 | 825938 | 0.01349 | 0.07391 | ||
| 844280 | 0.00971 | 0.07877 | 825980 | 0.02528 | 0.0868 | ||
| 844328 | 0.01363 | 0.09946 | 825981 | 0.03445 | 0.08076 | ||
| 844332 | 0.01065 | 0.07601 | 825989 | 0.02528 | 0.0868 | ||
| 845563 | 0.02315 | 0.08775 | 825994 | 0.03445 | 0.08076 | ||
| OsHMA2 | 29477961 | 0.02189 | 0.089 | 826042 | 0.00375 | 0.09796 | |
| 29478764 | 0.01517 | 0.09941 | 826066 | 0.0092 | 0.07894 | ||
| 29478784 | 0.00209 | 0.14462 | 826091 | 0.00453 | 0.09524 | ||
| 29480672 | 0.000521 | 0.14561 | 826100 | 0.00717 | 0.0859 | ||
| 29480848 | 0.00285 | 0.13472 | 826103 | 0.00717 | 0.0859 | ||
| OsPCR1 | 824973 | 0.03713 | 0.0898 | 826118 | 0.0092 | 0.07894 | |
| 824981 | 0.00938 | 0.0866 | 826121 | 0.0092 | 0.07894 | ||
| 824990 | 0.00919 | 0.08181 | 826172 | 0.0153 | 0.07218 | ||
| 824995 | 0.00919 | 0.08181 | 826262 | 0.02212 | 0.06533 | ||
| 825147 | 0.0092 | 0.07894 |
| Gene | Region | Position (bp) | Allele | Allele Effect |
|---|---|---|---|---|
| OsPCR1 | Exon1 | 824990 | G | 0.00 |
| C | −1.07 | |||
| Exon3 | 825477 | T | 0.00 | |
| A | −1.16 | |||
| Exon4 | 826066 | C | 0.00 | |
| A | −1.04 | |||
| 3′-UTR | 824981 | T | 0.00 | |
| G | −0.63 | |||
| 5′-UTR | 826262 | A | 0.00 | |
| G | −0.38 | |||
| CAL1 | 5′-UTR | 25190520 | C | 0.00 |
| G | −1.67 | |||
| Oscd1 | 5′-UTR | 842611 | A | 0.00 |
| G | −1.15 | |||
| 842747 | T | 0.00 | ||
| C | −0.83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Xu, F.; Cai, T.; Zhao, W.; Lin, J.; Huang, J.; Wang, L.; Bian, J.; Fu, J.; Ouyang, L.; et al. The Relationship between Cadmium-Related Gene Sequence Variations in Rice and Cadmium Accumulation. Agronomy 2023, 13, 800. https://doi.org/10.3390/agronomy13030800
Li W, Xu F, Cai T, Zhao W, Lin J, Huang J, Wang L, Bian J, Fu J, Ouyang L, et al. The Relationship between Cadmium-Related Gene Sequence Variations in Rice and Cadmium Accumulation. Agronomy. 2023; 13(3):800. https://doi.org/10.3390/agronomy13030800
Chicago/Turabian StyleLi, Weixing, Feng Xu, Tingting Cai, Wanling Zhao, Jianting Lin, Jiayu Huang, Liguo Wang, Jianmin Bian, Junru Fu, Linjuan Ouyang, and et al. 2023. "The Relationship between Cadmium-Related Gene Sequence Variations in Rice and Cadmium Accumulation" Agronomy 13, no. 3: 800. https://doi.org/10.3390/agronomy13030800
APA StyleLi, W., Xu, F., Cai, T., Zhao, W., Lin, J., Huang, J., Wang, L., Bian, J., Fu, J., Ouyang, L., Cai, Y., He, H., Sun, X., & Zhu, C. (2023). The Relationship between Cadmium-Related Gene Sequence Variations in Rice and Cadmium Accumulation. Agronomy, 13(3), 800. https://doi.org/10.3390/agronomy13030800






