Insight into the Boron Toxicity Stress-Responsive Genes in Boron-Tolerant Triticum dicoccum Shoots Using RNA Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material, Boron Treatment and Estimation of Physiological Parameters
2.2. RNA Extraction, Formation of cDNA Libraries, Shoot Transcriptome Sequencing of T. dicoccum Genotype
2.3. Assessment of Differentially Expressed Genes
2.4. Functional Assessment and Enrichment Analysis of DEGs
2.5. Involved Transcription Factors (TFs)
2.6. RT-qPCR Based Expression Analysis
3. Results
3.1. Physiological Changes in T. dicoccum, PI94655 and Bolal 2973 under B toxicity Stress
3.2. Transcriptome Sequencing and Genome Mapping of Sequencing Reads
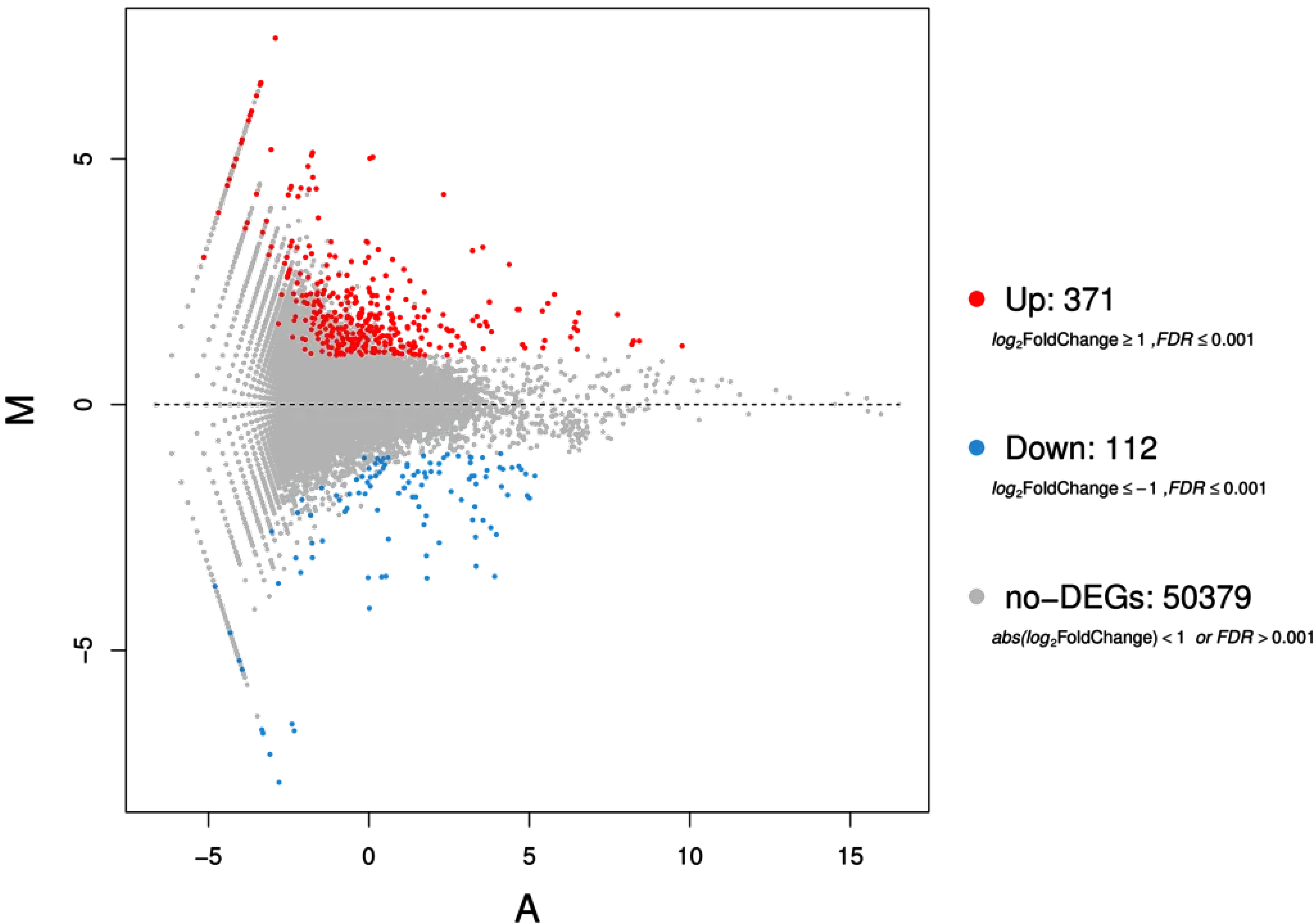
3.3. Differentially Regulated Genes of T. dicoccum Shoots under High B
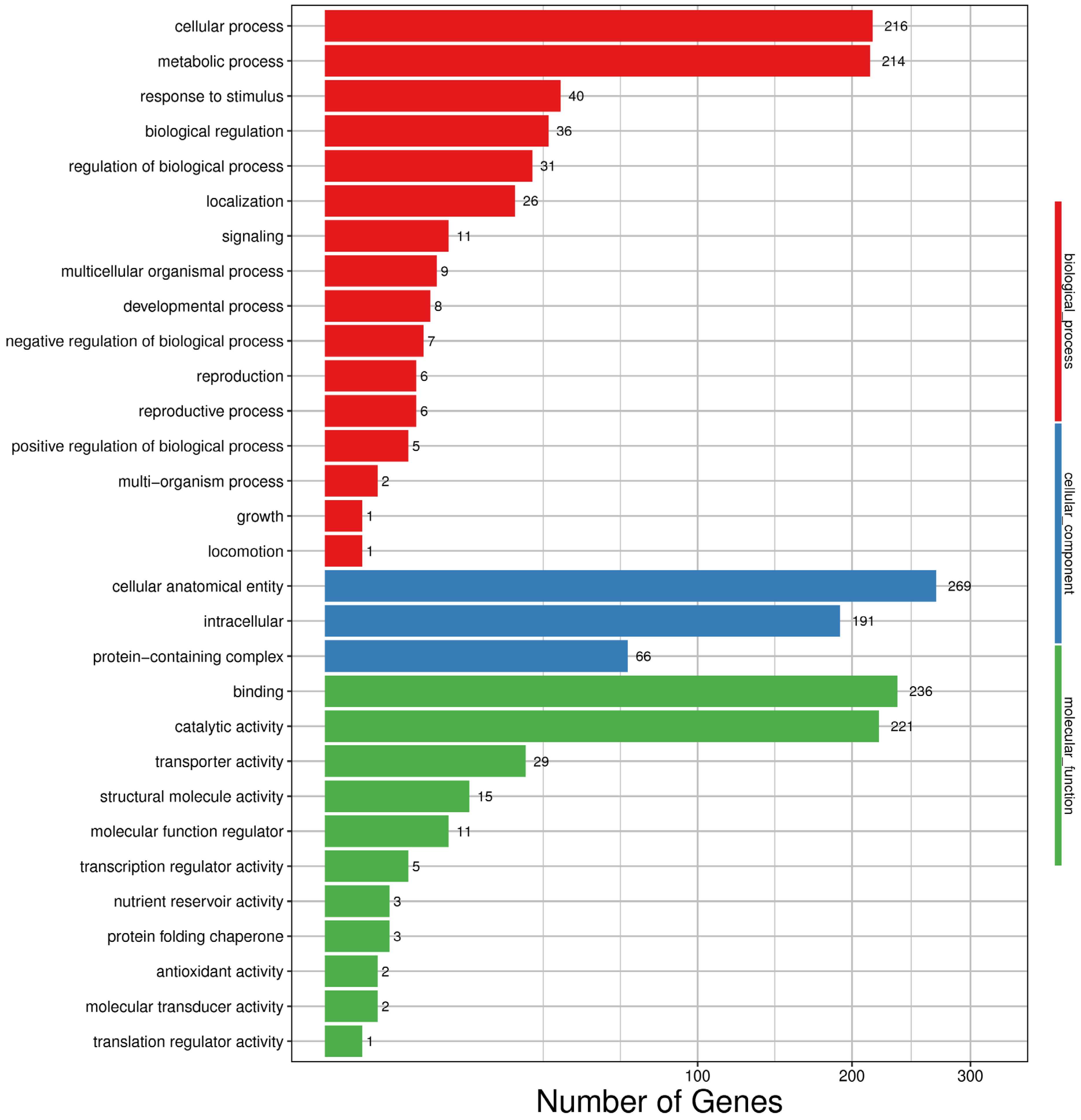
3.4. Gene Ontology (GO) Analysis of Differentially Expressed Genes
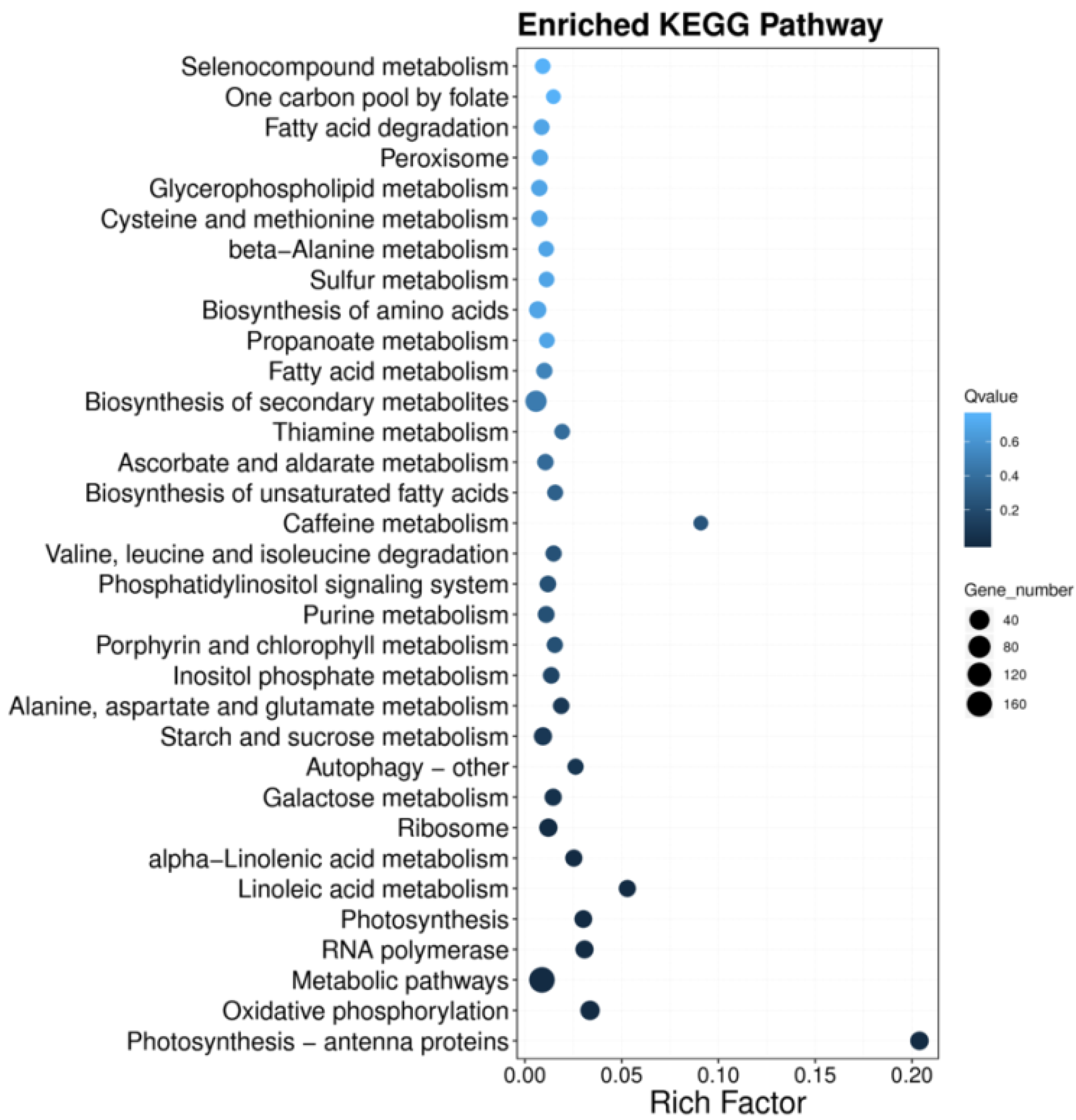
3.5. KEGG Pathway Enrichment (Functional Regulatory Network Analysis) of DEGs of T. dicoccum under High B
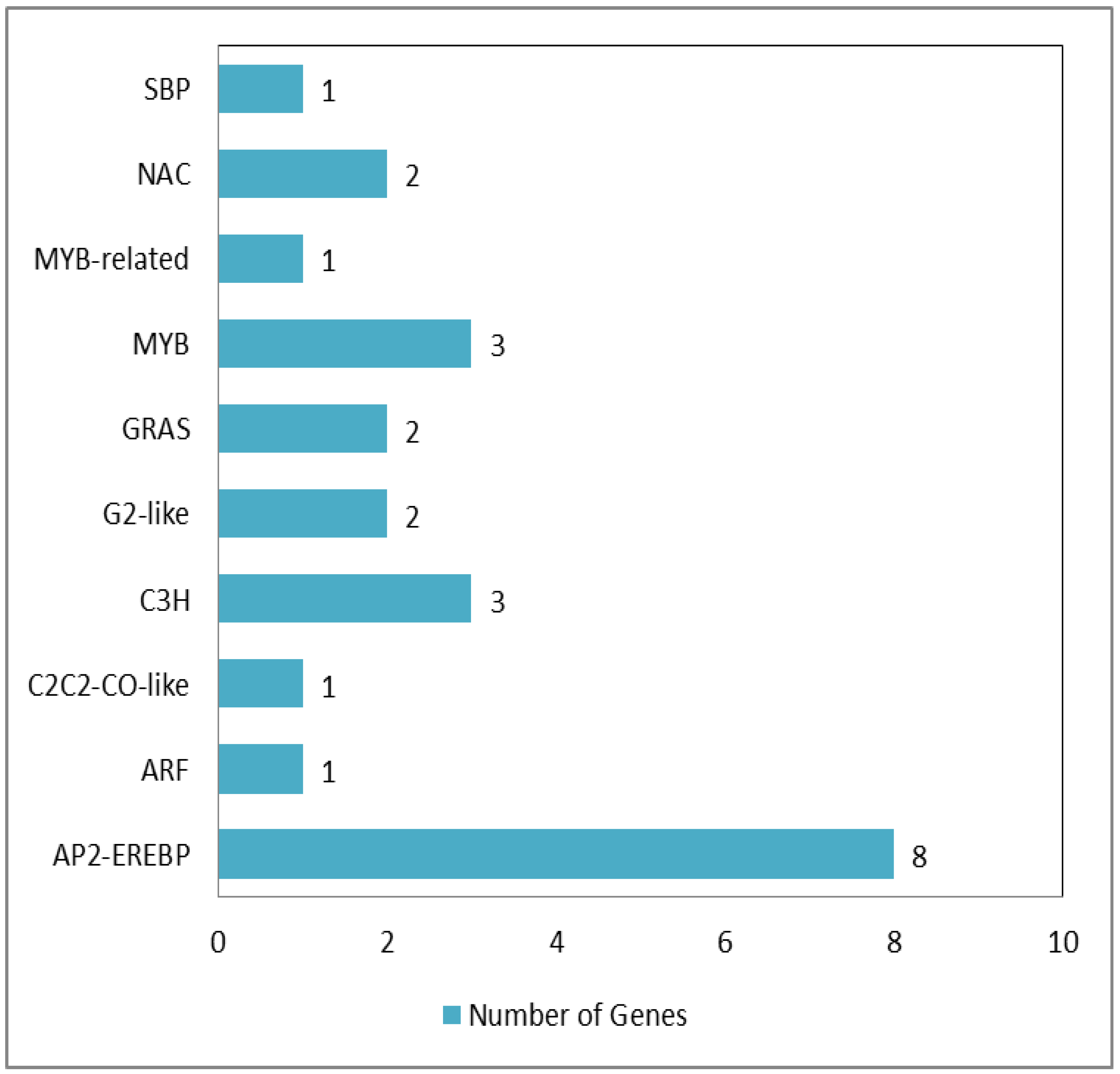
3.6. High-B-Responsive Transcription Factors of PI94655
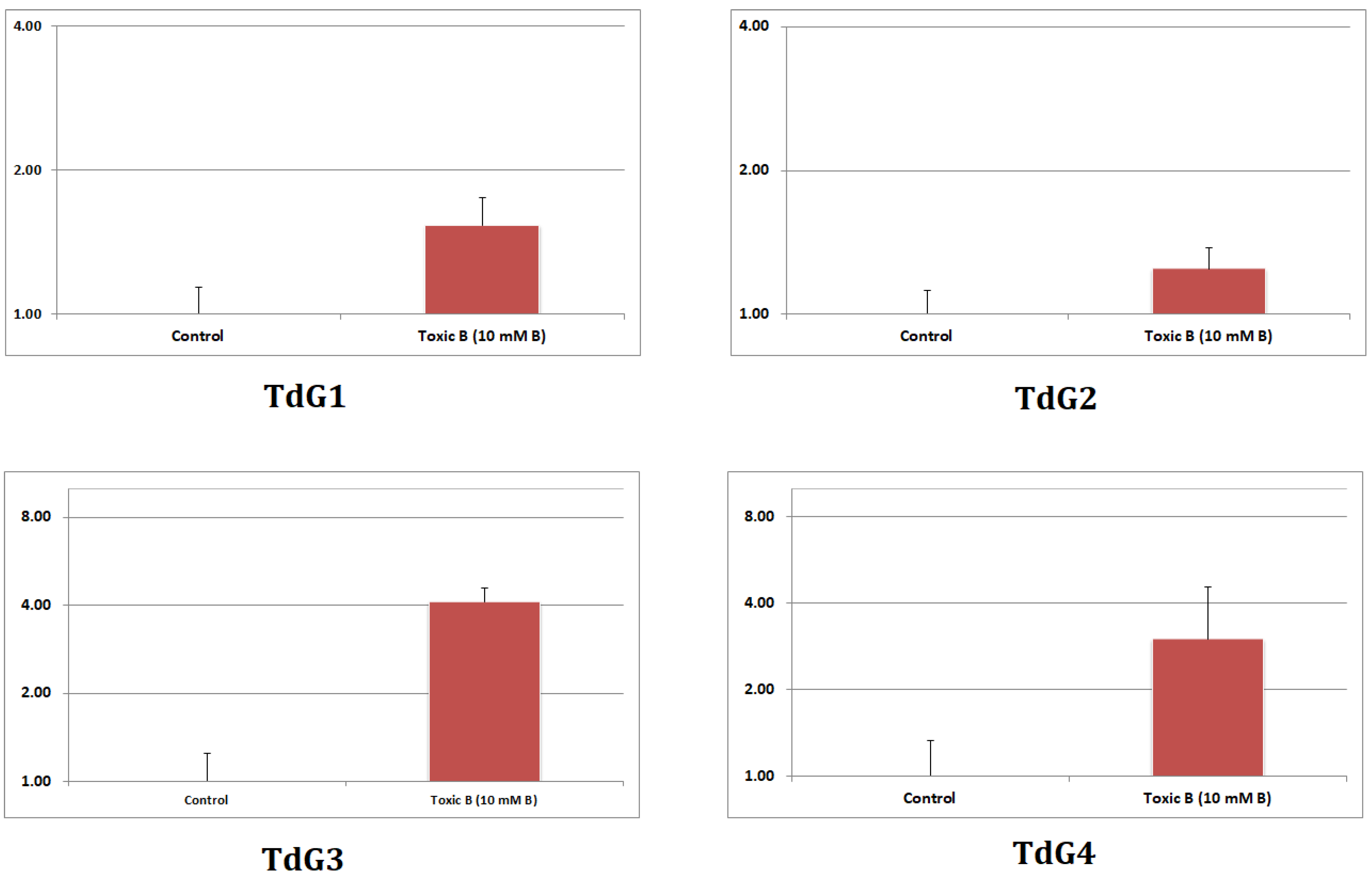
3.7. RT-qPCR Analysis for Confirmation of Sequencing Results
4. Discussion
4.1. Higher B Toxicity Tolerance in T. dicoccum PI94655 as Compared to Bolal 2973
4.2. Gene Ontology Analysis
4.3. Involved Transcription Factors
4.4. Transporters
4.5. Involved KEGG Pathways
4.6. Annotated Genes Confirmed with RT-qPCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Khan, M.K.; Pandey, A.; Hamurcu, M.; Avsaroglu, Z.Z.; Ozbek, M.; Omay, A.H.; Elbasan, F.; Omay, M.R.; Gokmen, F.; Topal, A.; et al. Variability in Physiological Traits Reveals Boron Toxicity Tolerance in Aegilops Species. Front. Plant Sci. 2021, 12, 736614. [Google Scholar] [CrossRef] [PubMed]
- Brdar-Jokanovic, M. Boron Toxicity and Deficiency in Agricultural Plants. Int. J. Mol. Sci. 2020, 21, 1424. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Khan, M.K.; Hamurcu, M.; Brestic, M.; Topal, A.; Gezgin, S. Insight into the Root Transcriptome of a Boron-Tolerant Triticum zhukovskyi Genotype Grown under Boron Toxicity. Agronomy 2022, 12, 2421. [Google Scholar] [CrossRef]
- Camacho-Cristóbal, J.J.; Navarro-Gochicoa, M.T.; Rexach, J.; González-Fontes, A.; Herrera-Rodríguez, M.B. Chapter 6—Plant Response to Boron Deficiency and Boron Use Efficiency in Crop Plants. In Plant Micronutrient Use Efficiency; Hossain, M.A., Kamiya, T., Burritt, D.J., Phan Tran, L.-S., Fujiwara, T., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 109–121. [Google Scholar]
- Khan, M.K.; Pandey, A.; Hamurcu, M.; Germ, M.; Yilmaz, F.G.; Ozbek, M.; Avsaroglu, Z.Z.; Topal, A.; Gezgin, S. Nutrient Homeostasis of Aegilops Accessions Differing in B Tolerance Level under Boron Toxic Growth Conditions. Biology 2022, 11, 1094. [Google Scholar] [CrossRef] [PubMed]
- Reid, R.J.; Hayes, J.E.; Post, A.; Stangoulis, J.C.R.; Graham, R.D. A critical analysis of the causes of boron toxicity in plants. Plant Cell Environ. 2004, 27, 1405–1414. [Google Scholar] [CrossRef]
- Pandey, A.; Khan, M.K.; Hamurcu, M.; Yilmaz, F.G.; Gezgin, S. Boron Toxicity: An Insight on Its Influence on Wheat Growth. In Metal Toxicology Handbook; CRC Press: Boca Raton, FL, USA, 2020; pp. 417–432. [Google Scholar]
- Landi, M.; Degl’Innocenti, E.; Pardossi, A.; Guidi, L. Antioxidant and Photosynthetic Responses in Plants under Boron Toxicity: A Review. Am. J. Agric. Biol. Sci. 2012, 7, 255–270. [Google Scholar] [CrossRef]
- Hamurcu, M.; Khan, K.; Pandey, A.; Avşaroğlu, Z.Z.; Elbasan, F.; Gezgin, S. Boron stress exposes differential antioxidant responses in maize cultivars (Zea mays L.). J. Elem. 2020, 24, 1291–1304. [Google Scholar]
- Sakamoto, T.; Inui, Y.T.; Uraguchi, S.; Yoshizumi, T.; Matsunaga, S.; Mastui, M.; Umeda, M.; Fukui, K.; Fujiwara, T. Condensin II alleviates DNA damage and is essential for tolerance of boron overload stress in Arabidopsis. Plant Cell 2011, 23, 3533–3546. [Google Scholar] [CrossRef]
- Catav, S.S.; Genc, T.O.; Kesik Oktay, M.; Kucukakyuz, K. Effect of Boron Toxicity on Oxidative Stress and Genotoxicity in Wheat (Triticum aestivum L.). Bull. Environ. Contam. Toxicol. 2018, 100, 502–508. [Google Scholar] [CrossRef]
- Tanaka, M.; Takano, J.; Chiba, Y.; Lombardo, F.; Ogasawara, Y.; Onouchi, H.; Naito, S.; Fujiwara, T. Boron-dependent degradation of NIP5;1 mRNA for acclimation to excess boron conditions in Arabidopsis. Plant Cell 2011, 23, 3547–3559. [Google Scholar] [CrossRef]
- Reid, R. Identification of boron transporter genes likely to be responsible for tolerance to boron toxicity in wheat and barley. Plant Cell Physiol. 2007, 48, 1673–1678. [Google Scholar] [CrossRef] [PubMed]
- Miwa, K.; Aibara, I.; Fujiwara, T. Arabidopsis thaliana BOR4 is upregulated under high boron conditions and confers tolerance to high boron. Soil Sci. Plant Nutr. 2014, 60, 349–355. [Google Scholar] [CrossRef]
- Khan, M.K.; Pandey, A.; Hamurcu, M.; Gezgin, S.; Athar, T.; Rajput, V.D.; Gupta, O.P.; Minkina, T. Insight into the Prospects for Nanotechnology in Wheat Biofortification. Biology 2021, 10, 1123. [Google Scholar] [CrossRef] [PubMed]
- Paull, J.G.; Cartwright, B.; Rathjen, A.J. Responses of wheat and barley genotypes to toxic concentrations of soil boron. Euphytica 1988, 39, 137–144. [Google Scholar] [CrossRef]
- Cartwright, B.; Zarcinas, B.; Mayfield, A. Toxic concentrations of boron in a red-brown earth at Gladstone, South Australia. Soil Res. 1984, 22, 261–272. [Google Scholar] [CrossRef]
- Yau, S.-k.; Nachit, M.M.; Ryan, J. Variation in boron-toxicity tolerance in a durum wheat core collection. In Boron in Soils and Plants, Proceedings of the International Symposium on Boron in Soils and Plants held at Chiang Mai, Thailand, 7–11 September 1997; Bell, R.W., Rerkasem, B., Eds.; Springer: Dordrecht, The Netherlands, 1997; pp. 117–120. [Google Scholar]
- Paull, J.; Nable, R.; Rathjen, A. Physiological and genetic control of the tolerance of wheat to high concentrations of boron and implications for plant breeding. Plant Soil 1992, 146, 251–260. [Google Scholar] [CrossRef]
- Paull, J.G. Genetic Studies on the Tolerance of Wheat to High Concentrations of Boron. Ph.D. Thesis, University of Adelaide, Adelaide, Australia, 1990. [Google Scholar]
- Moody, D.B.; Rathjen, A.J.; Cartwright, B. Yield evaluation of a gene for boron tolerance using backcross-derived lines. In Proceedings of the Genetic Aspects of Plant Mineral Nutrition: The Fourth International Symposium on Genetic Aspects of Plant Mineral Nutrition, Canberra, Australia, 30 September–4 October 1991; Randall, P.J., Delhaize, E., Richards, R.A., Munns, R., Eds.; Springer: Dordrecht, The Netherlands, 1993; pp. 363–366. [Google Scholar]
- Yau, S.K.; Ryan, J. Boron Toxicity Tolerance in Crops: A Viable Alternative to Soil Amelioration. Crop Sci. 2008, 48, 854. [Google Scholar] [CrossRef]
- Nable, R.O.; Bañuelos, G.S.; Paull, J.G. Boron toxicity. Plant Soil 1997, 193, 181–198. [Google Scholar] [CrossRef]
- Nable, R.O. Resistance to boron toxicity amongst several barley and wheat cultivars: A preliminary examination of the resistance mechanism. Plant Soil 1988, 112, 45–52. [Google Scholar] [CrossRef]
- Furlani, Â.M.C.; Carvalho, C.P.; Freitas, J.G.d.; Verdial, M.F. Wheat cultivar tolerance to boron deficiency and toxicity in nutrient solution. Sci. Agric. 2003, 60, 359–370. [Google Scholar] [CrossRef]
- Torun, A.A.; Yazici, A.; Erdem, H.; Çakmak, İ. Genotypic variation in tolerance to boron toxicity in 70 durum wheat genotypes. Turk. J. Agric. For. 2006, 30, 49–58. [Google Scholar]
- Yau, S.; Nachit, M.; Ryan, J.; Hamblin, J. Phenotypic variation in boron-toxicity tolerance at seedling stage in durum wheat (Triticum durum). Euphytica 1995, 83, 185–191. [Google Scholar] [CrossRef]
- Hua, T.; Zhang, R.; Sun, H.; Liu, C. Alleviation of boron toxicity in plants: Mechanisms and approaches. Crit. Rev. Environ. Sci. Technol. 2020, 51, 2975–3015. [Google Scholar] [CrossRef]
- Reid, R. Can we really increase yields by making crop plants tolerant to boron toxicity? Plant Sci. 2010, 178, 9–11. [Google Scholar] [CrossRef]
- Zaharieva, M.; Ayana, N.G.; Hakimi, A.A.; Misra, S.C.; Monneveux, P. Cultivated emmer wheat (Triticum dicoccon Schrank), an old crop with promising future: A review. Genet. Resour. Crop Evol. 2010, 57, 937–962. [Google Scholar] [CrossRef]
- Ozkan, H.; Brandolini, A.; Schäfer-Pregl, R.; Salamini, F. AFLP analysis of a collection of tetraploid wheats indicates the origin of emmer and hard wheat domestication in southeast Turkey. Mol. Biol. Evol. 2002, 19, 1797–1801. [Google Scholar] [CrossRef]
- Luo, M.C.; Yang, Z.L.; You, F.M.; Kawahara, T.; Waines, J.G.; Dvorak, J. The structure of wild and domesticated emmer wheat populations, gene flow between them, and the site of emmer domestication. Theor. Appl. Genet. 2007, 114, 947–959. [Google Scholar] [CrossRef]
- Kayıhan, C.; Öz, M.T.; Eyidoğan, F.; Yücel, M.; Öktem, H.A. Physiological, Biochemical, and Transcriptomic Responses to Boron Toxicity in Leaf and Root Tissues of Contrasting Wheat Cultivars. Plant Mol. Biol. Report. 2017, 35, 97–109. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, J.; Jiang, W.; Liu, J.; Yang, S.; Gai, J.; Li, Y. Identification and Analysis of NaHCO3 Stress Responsive Genes in Wild Soybean (Glycine soja) Roots by RNA-seq. Front. Plant Sci. 2016, 7, 1842. [Google Scholar] [CrossRef]
- Mahboobi, H.; Yucel, M.; Öktem, H.A. Cell wall uronic acid concentrations of resistant and sensitive cultivars of wheat and barley under boron toxicity. J. Plant Nutr. 2001, 24, 1965–1973. [Google Scholar] [CrossRef]
- Pallotta, M.; Schnurbusch, T.; Hayes, J.; Hay, A.; Baumann, U.; Paull, J.; Langridge, P.; Sutton, T. Molecular basis of adaptation to high soil boron in wheat landraces and elite cultivars. Nature 2014, 514, 88–91. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zaim, S.R.; Aberasturi, D.; Berghout, J.; Li, H.; Kenost, C.; Zhang, H.H.; Lussier, Y.A. iDEG: A singlesubject method for assessing gene differential expression from two transcriptomes of an individual. bioRxiv 2018. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Schnurbusch, T.; Langridge, P.; Sutton, T. The Bo1-specific PCR marker AWW5L7 is predictive of boron tolerance status in a range of exotic durum and bread wheats. Genome 2008, 51, 963–971. [Google Scholar] [CrossRef]
- Metwally, A.; El-Shazoly, R.; Hamada, A.M. Effect of boron on growth criteria of some wheat cultivars. J. Biol. Earth Sci. 2012, 2, B1–B9. [Google Scholar]
- Coskun, Y.; Olgunsoy, P.; Karatas, N.; Bulut, F.; Yarar, F. Mannitol application alleviates boron toxicity in wheat seedlings. Commun. Soil Sci. Plant Anal. 2014, 45, 944–952. [Google Scholar] [CrossRef]
- Amirbakhtiar, N.; Ismaili, A.; Ghaffari, M.R.; Mirdar Mansuri, R.; Sanjari, S.; Shobbar, Z.S. Transcriptome analysis of bread wheat leaves in response to salt stress. PLoS ONE 2021, 16, e0254189. [Google Scholar] [CrossRef]
- Cooper, R. Re-discovering ancient wheat varieties as functional foods. J. Tradit. Complement. Med. 2015, 5, 138–143. [Google Scholar] [CrossRef]
- Khan, M.K.; Arifuzzaman, M.; Pandey, A.; Turin, M.T.S.; Hamurcu, M.; Athar, T.; Masuda, M.S.; Gokmen Yilmaz, F.; Topal, A.; Gezgin, S. Chapter 21—Advancement in mitigating the effects of boron stress in wheat. In Abiotic Stresses in Wheat; Khan, M.K., Pandey, A., Hamurcu, M., Gupta, O.P., Gezgin, S., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 329–338. [Google Scholar]
- Nejad, S.G.; Savaghebi, G.; Farahbakhsh, M.; Amiri, R.M.; Rezaei, H. Tolerance of some wheat varieties to boron toxicity. Cereal Res. Commun. 2015, 43, 384–393. [Google Scholar] [CrossRef]
- Schnurbusch, T.; Collins, N.C.; Eastwood, R.F.; Sutton, T.; Jefferies, S.P.; Langridge, P. Fine mapping and targeted SNP survey using rice-wheat gene colinearity in the region of the Bo1 boron toxicity tolerance locus of bread wheat. Theor. Appl. Genet. 2007, 115, 451–461. [Google Scholar] [CrossRef]
- Kalayci, M.; Alkan, A.; Cakmak, I.; Bayramoğlu, O.; Yilmaz, A.; Aydin, M.; Ozbek, V.; Ekiz, H.; Ozberisoy, F. Studies on differential response of wheat cultivars to boron toxicity. Euphytica 1998, 100, 123–129. [Google Scholar] [CrossRef]
- Ozturk, S.E.; Goktay, M.; Has, C.; Babaoglu, M.; Allmer, J.; Doganlar, S.; Frary, A. Boron Hyperaccumulation Mechanisms in Puccinellia distans as Revealed by Transcriptomic Analysis. bioRxiv 2017. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.; Shao, H.; Tang, X. Recent Advances in Utilizing Transcription Factors to Improve Plant Abiotic Stress Tolerance by Transgenic Technology. Front. Plant Sci. 2016, 7, 67. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.-J.; Vogel, M.O.; Viehhauser, A. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 2010, 245, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef] [PubMed]
- Tombuloglu, H.; Kekec, G.; Sakcali, M.S.; Unver, T. Transcriptome-wide identification of R2R3-MYB transcription factors in barley with their boron responsive expression analysis. Mol. Genet. Genom. MGG 2013, 288, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, A.; Takano, J.; Kobayashi, M.; von Wirén, N.; Fujiwara, T. Roles of BOR1, DUR3, and FPS1 in boron transport and tolerance in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2006, 262, 216–222. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef]
- Öz, M.T.; Yilmaz, R.; Eyidoğan, F.; Graaff, L.D.; Yücel, M.; Öktem, H.A. Microarray Analysis of Late Response to Boron Toxicity in Barley (Hordeum vulgare L.) Leaves. Turk. J. Agric. For. 2009, 33, 191–202. [Google Scholar] [CrossRef]
- Ochiai, K.; Shimizu, A.; Okumoto, Y.; Fujiwara, T.; Matoh, T. Suppression of a NAC-like transcription factor gene improves boron-toxicity tolerance in rice. Plant Physiol. 2011, 156, 1457–1463. [Google Scholar] [CrossRef]
- Liu, C.; Xu, X.; Kan, J.; Ming Cheng, Z.; Chang, Y.; Lin, J.; Li, H. Genome-wide analysis of the C3H zinc finger family reveals its functions in salt stress responses of Pyrus betulaefolia. PeerJ 2020, 8, e9328. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Xu, Y.; Han, G.; Zhou, L.; Ali, A.; Zhu, S.; Li, X. Molecular Evolution and Genetic Variation of G2-Like Transcription Factor Genes in Maize. PLoS ONE 2016, 11, e0161763. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wu, Q.; Teng, L.; Tang, F.; Pi, Z.; Shen, S. Transcriptional regulation of the paper mulberry under cold stress as revealed by a comprehensive analysis of transcription factors. BMC Plant Biol. 2015, 15, 108. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Guo, L.; Wang, R.; Zhang, Q.; Yao, H. Genome-Wide Identification and Characterization of G2-Like Transcription Factor Genes in Moso Bamboo (Phyllostachys edulis). Molecules 2022, 27, 5491. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-y.; Zhao, S.; Liu, J.-F.; Zhao, H.-y.; Sun, X.-y.; Wu, T.-r.; Pei, T.; Wang, Y.; Liu, Q.-F.; Yang, H.-H.; et al. Genome-wide identification of Tomato Golden 2-Like transcription factors and abiotic stress related members screening. BMC Plant Biol. 2022, 22, 82. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Kakkar, M.; Gahlaut, V.; Jaiswal, V.; Kumar, S. Multifarious roles of GRAS transcription factors in plants. Preprints 2021, 2021030066. [Google Scholar]
- Chen, Y.; Tai, S.; Wang, D.; Ding, A.; Sun, T.; Wang, W.; Sun, Y. Homology-based analysis of the GRAS gene family in tobacco. Genet. Mol. Res. 2015, 14, 15188–15200. [Google Scholar] [CrossRef]
- Öztürk, S.E.; Göktay, M.; Has, C.; Babaoğlu, M.; Allmer, J.; Doğanlar, S.; Frary, A. Transcriptomic analysis of boron hyperaccumulation mechanisms in Puccinellia distans. Chemosphere 2018, 199, 390–401. [Google Scholar] [CrossRef]
- Miwa, K.; Fujiwara, T. Boron transport in plants: Co-ordinated regulation of transporters. Ann. Bot. 2010, 105, 1103–1108. [Google Scholar] [CrossRef]
- Kayihan, C.; Aksoy, E.; Mutlu, S.N. Boron toxicity induces sulfate transporters at transcriptional level in Arabidopsis thaliana. Turk. J. Bot. 2023, 47, 1–22. [Google Scholar] [CrossRef]
- Padmanabhan, P.; Babaoglu, M.; Terry, N. A comparative transcriptomic analysis of the extremely boron tolerant plant Puccinellia distans with the moderately boron tolerant Gypsophila arrostil. Plant Cell Rep. 2012, 31, 1407–1413. [Google Scholar] [CrossRef] [PubMed]
- Pommerrenig, B.; Diehn, T.A.; Bienert, G.P. Metalloido-porins: Essentiality of Nodulin 26-like intrinsic proteins in metalloid transport. Plant Sci. 2015, 238, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Macho-Rivero, M.A.; Herrera-Rodríguez, M.B.; Brejcha, R.; Schäffner, A.R.; Tanaka, N.; Fujiwara, T.; González-Fontes, A.; Camacho-Cristóbal, J.J. Boron toxicity reduces water transport from root to shoot in Arabidopsis plants. Evidence for a reduced transpiration rate and expression of major PIP aquaporin genes. Plant Cell Physiol. 2018, 59, 841–849. [Google Scholar] [CrossRef]
- Martinez-Cuenca, M.-R.; Martinez-Alcantara, B.; Quinones, A.; Ruiz, M.; Iglesias, D.J.; Primo-Millo, E.; Forner-Giner, M.A. Physiological and molecular responses to excess boron in Citrus macrophylla W. PLoS ONE 2015, 10, e0134372. [Google Scholar] [CrossRef]
- Kumar, S.; Pandey, A.K. Chemistry and biological activities of flavonoids: An overview. Sci. World J. 2013, 2013, 162750. [Google Scholar] [CrossRef]
- Martinoia, E.; Maeshima, M.; Neuhaus, H.E. Vacuolar transporters and their essential role in plant metabolism. J. Exp. Bot. 2007, 58, 83–102. [Google Scholar] [CrossRef]
- Roytrakul, S.; Verpoorte, R. Role of vacuolar transporter proteins in plant secondary metabolism: Catharanthus roseus cell culture. Phytochem. Rev. 2007, 6, 383–396. [Google Scholar] [CrossRef]
- Yıldırım, K.; Uylaş, S. Genome-wide transcriptome profiling of black poplar (Populus nigra L.) under boron toxicity revealed candidate genes responsible in boron uptake, transport and detoxification. Plant Physiol. Biochem. 2016, 109, 146–155. [Google Scholar] [CrossRef]
- Fu, Y.; Zhang, H.; Mandal, S.N.; Wang, C.; Chen, C.; Ji, W. Quantitative proteomics reveals the central changes of wheat in response to powdery mildew. J. Proteom. 2016, 130, 108–119. [Google Scholar] [CrossRef]
- Fan, Y.; Chen, J.; Wang, Z.; Tan, T.; Li, S.; Li, J.; Wang, B.; Zhang, J.; Cheng, Y.; Wu, X. Soybean (Glycine max L. Merr.) seedlings response to shading: Leaf structure, photosynthesis and proteomic analysis. BMC Plant Biol. 2019, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Ruban, A.; Walters, R. Regulation of light harvesting in green plants. Annu. Rev. Plant. Biol. 1996, 47, 655–684. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zheng, Q.; Shen, W.; Cram, D.; Fowler, D.B.; Wei, Y.; Zou, J. Understanding the biochemical basis of temperature-induced lipid pathway adjustments in plants. Plant Cell 2015, 27, 86–103. [Google Scholar] [CrossRef] [PubMed]
- Arefian, M.; Vessal, S.; Malekzadeh-Shafaroudi, S.; Siddique, K.H.; Bagheri, A. Comparative proteomics and gene expression analyses revealed responsive proteins and mechanisms for salt tolerance in chickpea genotypes. BMC Plant Biol. 2019, 19, 300. [Google Scholar] [CrossRef]
- Lopes, K.L.; Rodrigues, R.A.O.; Silva, M.C.; Braga, W.G.S.; Silva-Filho, M.C. The Zinc-Finger Thylakoid-Membrane Protein FIP Is Involved With Abiotic Stress Response in Arabidopsis thaliana. Front. Plant Sci. 2018, 9, 504. [Google Scholar] [CrossRef]
- Seo, K.-I.; Lee, J.-H.; Nezames, C.D.; Zhong, S.; Song, E.; Byun, M.-O.; Deng, X.W. ABD1 Is an Arabidopsis DCAF Substrate Receptor for CUL4-DDB1–Based E3 Ligases That Acts as a Negative Regulator of Abscisic Acid Signaling. Plant Cell 2014, 26, 695–711. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, Y.; Wang, C.; Liu, M.; Li, H.; Fu, Y.; Wang, Y.; Nie, Y.; Liu, X.; Ji, W. Large-scale transcriptome comparison reveals distinct gene activations in wheat responding to stripe rust and powdery mildew. BMC Genom. 2014, 15, 898. [Google Scholar] [CrossRef]
| Gene Code | Selected Gene | Annotated Gene Information | log2 Fold Change RNA seq | Primer Type | Sequence (5’->3’) |
|---|---|---|---|---|---|
| TdG1 | TraesCS3B02G001500 | PREDICTED: Triticum dicoccoides swi5-dependent recombination DNA repair protein 1 homolog (LOC119274246), mRNA | 10.68 | Forward primer | CCACTGTAAACGGCGCTAGA |
| Reverse primer | GGATCGGTTGGGGTTGCTTT | ||||
| TdG2 | TraesCS3B02G252900 | PREDICTED: Triticum dicoccoides ATP-dependent zinc metalloprotease FTSH 9, chloroplastic/mitochondrial (LOC119276292), mRNA | 14.69 | Forward primer | GCTGAGAAGTGCATCACGCT |
| Reverse primer | GTTTCCTTTAAACAATGGCGAGGCA | ||||
| TdG3 | TraesCS3B02G191500 | PREDICTED: Triticum dicoccoides DDB1- and CUL4-associated factor 13-like (LOC119275595), mRNA | 8.69 | Forward primer | CTTGCAAACCTTGGACAGCG |
| Reverse primer | ACAGCGATTGATTGACGGAGG | ||||
| TdG4 | TraesCS3B02G388100 | PREDICTED: Triticum dicoccoides UDP-glucuronic acid decarboxylase 1-like (LOC119277519), mRNA | 13.04 | Forward primer | GCCGCGTGGTTAGCAATTTT |
| Reverse primer | GCCATCAATCCAGCAACCAG |
| Accessions | PI94655 | Bolal 2973 |
|---|---|---|
| Traits/Species | Triticum turgidum subsp. dicoccum | Triticum aestivum subsp. aestivum |
| Root Length | −2 | −50 |
| Shoot Length | 4 | −38 |
| Root Fresh Weight | 3 | −108 |
| Shoot Fresh Weight | −7 | −30 |
| Root Dry Weight | 10 | −53 |
| Shoot Dry Weight | 16 | 0 |
| Parameters/Sample | Td_Control | Td_TB_ Treatment | Parameters/Sample | Td_Control | Td_TB_ Treatment |
|---|---|---|---|---|---|
| Total Clean Reads (M) | 76.92 | 75.27 | Uniquely Gene Mapping Ratio (%) | 51.28 | 48.54 |
| Total Clean Bases (Gb) | 7.69 | 7.53 | Total Gene Number | 43,852 | 44,921 |
| Clean Reads Q20 (%) | 97.43 | 97.42 | Known Gene Number | 42,227 | 43,309 |
| Clean Reads Q30 (%) | 90.39 | 90.30 | Novel Gene Number | 1625 | 1612 |
| Total Genome Mapping Ratio (%) | 14.69 | 16.68 | Total Transcript Number | 49,471 | 51,449 |
| Uniquely Genome Mapping Ratio (%) | 4.84 | 5.50 | Known Transcript Number | 43,458 | 45,256 |
| Total Gene Mapping Ratio (%) | 87.60 | 85.93 | Novel Transcript Number | 6013 | 6193 |
| Pathway | Pathway ID | No. of Genes Annotated in this Pathway | Level 1 | Level 2 |
|---|---|---|---|---|
| Metabolic pathways | ko01100 | 172 | Metabolism | Global and overview maps |
| Biosynthesis of secondary metabolites | ko01110 | 62 | Metabolism | Global and overview maps |
| Oxidative phosphorylation | ko00190 | 32 | Metabolism | Energy metabolism |
| Photosynthesis—antenna proteins | ko00196 | 21 | Metabolism | Energy metabolism |
| Ribosome | ko03010 | 19 | Genetic Information Processing | Translation |
| RNA transport | ko03013 | 19 | Genetic Information Processing | Translation |
| mRNA surveillance pathway | ko03015 | 18 | Genetic Information Processing | Translation |
| RNA polymerase | ko03020 | 17 | Genetic Information Processing | Transcription |
| Starch and sucrose metabolism | ko00500 | 17 | Metabolism | Carbohydrate metabolism |
| Phenylpropanoid biosynthesis | ko00940 | 16 | Metabolism | Biosynthesis of other secondary metabolites |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, M.K.; Pandey, A.; Hamurcu, M.; Rajpal, V.R.; Vyhnanek, T.; Topal, A.; Raina, S.N.; Gezgin, S. Insight into the Boron Toxicity Stress-Responsive Genes in Boron-Tolerant Triticum dicoccum Shoots Using RNA Sequencing. Agronomy 2023, 13, 631. https://doi.org/10.3390/agronomy13030631
Khan MK, Pandey A, Hamurcu M, Rajpal VR, Vyhnanek T, Topal A, Raina SN, Gezgin S. Insight into the Boron Toxicity Stress-Responsive Genes in Boron-Tolerant Triticum dicoccum Shoots Using RNA Sequencing. Agronomy. 2023; 13(3):631. https://doi.org/10.3390/agronomy13030631
Chicago/Turabian StyleKhan, Mohd. Kamran, Anamika Pandey, Mehmet Hamurcu, Vijay Rani Rajpal, Tomas Vyhnanek, Ali Topal, Soom Nath Raina, and Sait Gezgin. 2023. "Insight into the Boron Toxicity Stress-Responsive Genes in Boron-Tolerant Triticum dicoccum Shoots Using RNA Sequencing" Agronomy 13, no. 3: 631. https://doi.org/10.3390/agronomy13030631
APA StyleKhan, M. K., Pandey, A., Hamurcu, M., Rajpal, V. R., Vyhnanek, T., Topal, A., Raina, S. N., & Gezgin, S. (2023). Insight into the Boron Toxicity Stress-Responsive Genes in Boron-Tolerant Triticum dicoccum Shoots Using RNA Sequencing. Agronomy, 13(3), 631. https://doi.org/10.3390/agronomy13030631








