De Novo Assembly of an Allotetraploid Artemisia argyi Genome
Abstract
1. Introduction
2. Materials and Methods
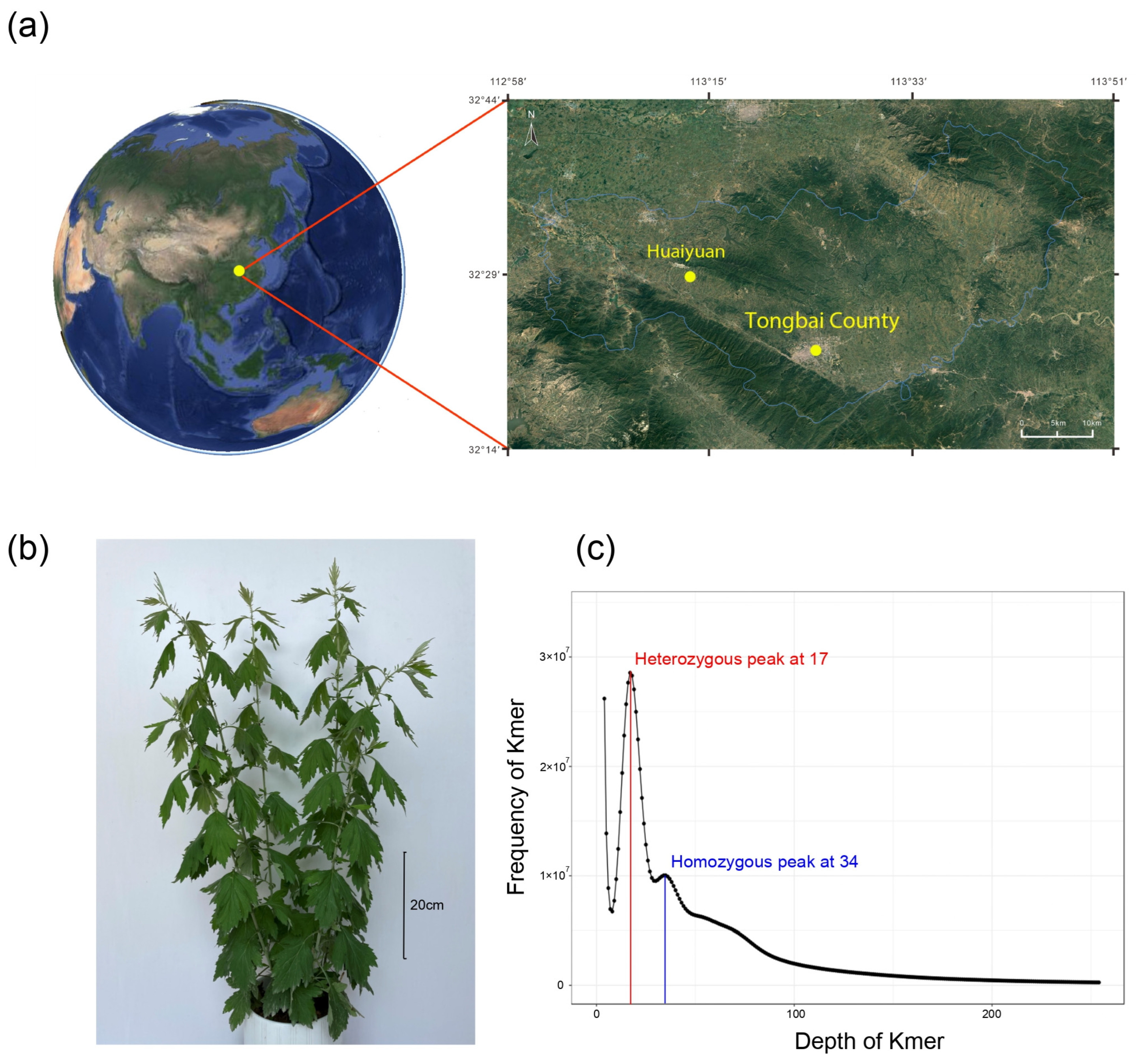
2.1. Plant Sample, Sequencing, and Genome Survey
2.2. Genome Assembly and Quality Evaluation
2.3. De Novo Genome Annotation
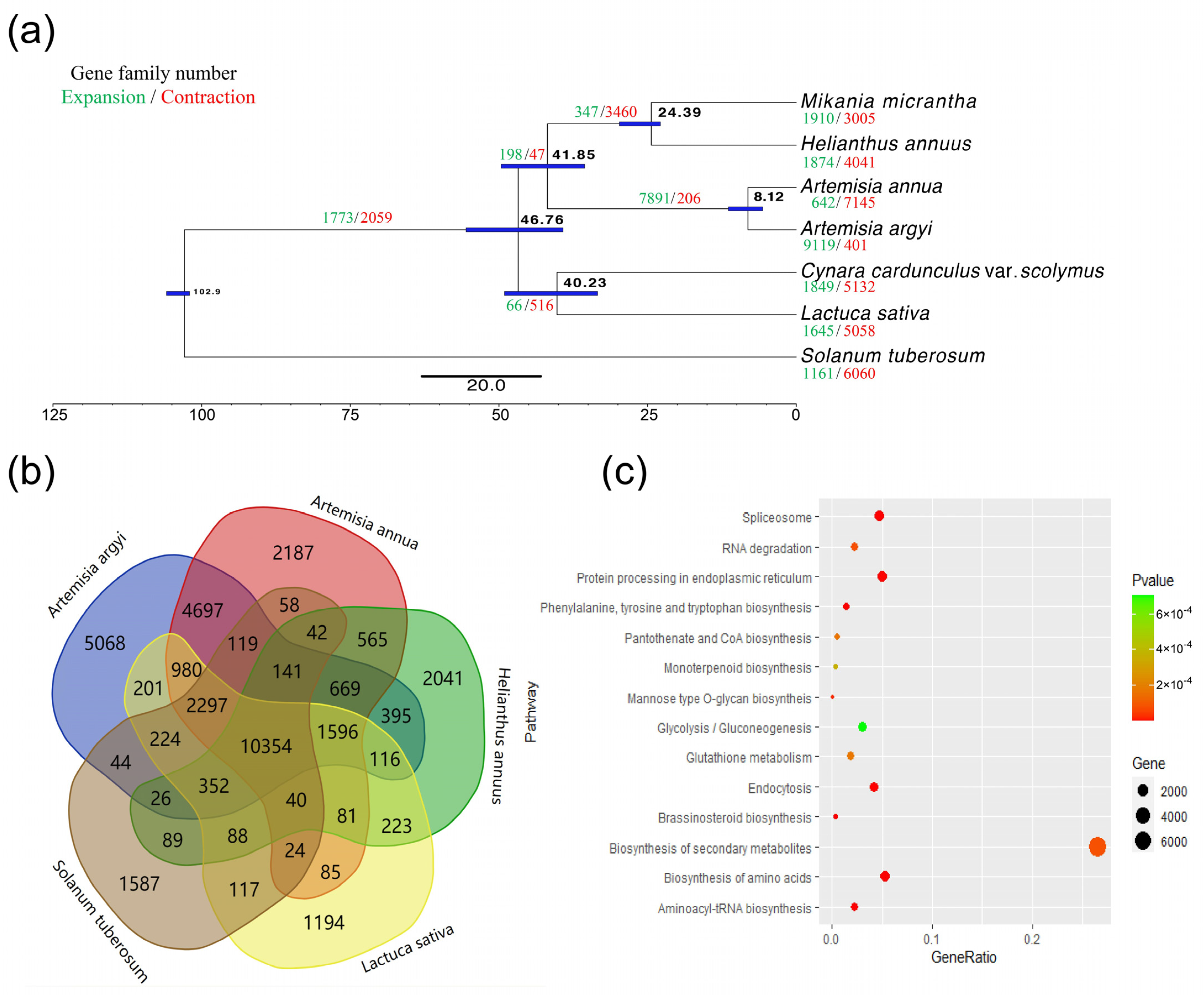
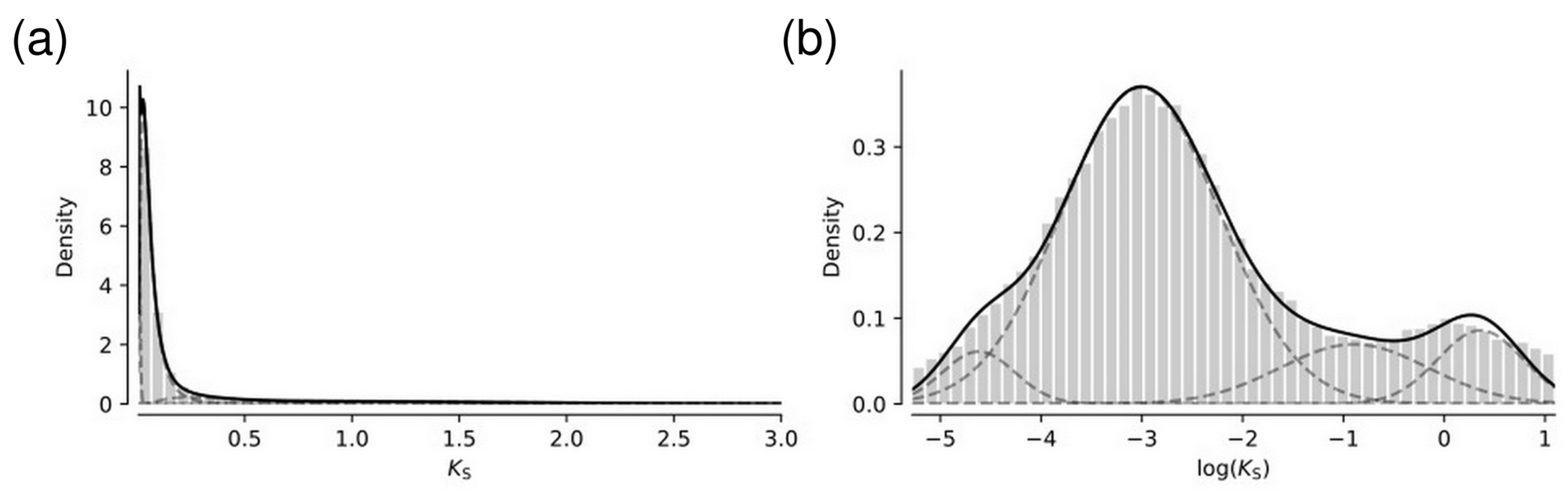
2.4. Evolutionary Analysis
3. Results
3.1. Genomic Characteristics of Wild A. argyi
3.2. De Novo Assembly and Annotation of the A. argyi Genome
3.3. Comparative Genome Analysis of Asteraceae Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shi, X.S.; Song, Y.P.; Meng, L.H.; Yang, S.Q.; Wang, D.J.; Zhou, X.W.; Ji, N.Y.; Wang, B.G.; Li, X.M. Isolation and characterization of antibacterial carotene sesquiterpenes from Artemisia argyi associated endophytic Trichoderma virens QA-8. Antibiotics 2021, 10, 213. [Google Scholar] [CrossRef]
- Liu, M.; Zhu, J.; Wu, S.; Wang, C.; Guo, X.; Wu, J.; Zhou, M. De novo assembly and analysis of the Artemisia argyi transcriptome and identification of genes involved in terpenoid biosynthesis. Sci. Rep. 2018, 8, 5824. [Google Scholar] [CrossRef] [PubMed]
- Madani, H.; Escrich, A.; Hosseini, B.; Sanchez-Munoz, R.; Khojasteh, A.; Palazon, J. Effect of polyploidy induction on natural metabolite production in medicinal plants. Biomolecules 2021, 11, 899. [Google Scholar] [CrossRef] [PubMed]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Mo, R.; Li, H.; Ni, Z.; Sun, Q.; Liu, Z. The transcriptional and splicing changes caused by hybridization can be globally recovered by genome doubling during allopolyploidization. Mol. Biol. Evol. 2021, 38, 2513–2519. [Google Scholar] [CrossRef] [PubMed]
- Alix, K.; Gerard, P.R.; Schwarzacher, T.; Heslop-Harrison, J.S.P. Polyploidy and interspecific hybridization: Partners for adaptation, speciation and evolution in plants. Ann. Bot. 2017, 120, 183–194. [Google Scholar] [CrossRef]
- Qi, X.Y.; Wang, H.B.; Song, A.P.; Jiang, J.F.; Chen, S.M.; Chen, F.D. Genomic and transcriptomic alterations following intergeneric hybridization and polyploidization in the Chrysanthemum nankingense × Tanacetum vulgare hybrid and allopolyploid (Asteraceae). Hortic. Res. 2018, 5, 5. [Google Scholar] [CrossRef]
- Lin, Y.; Ling, Y.; Humphries, C.J.; Gilbert, M.G. Artemisia. In Flora of China; Wu, Z., Raven, P.H., Eds.; Science Press: Beijing, China, 2011; pp. 20–21. [Google Scholar]
- Garcia, S.; Canela, M.A.; Garnatje, T.; Mcarthur, E.D.; Pellicer, J.; Sanderson, S.C.; Valles, J. Evolutionary and ecological implications of genome size in the North American endemic sagebrushes and allies (Artemisia, Asteraceae). Biol. J. Linn. Soc. 2008, 94, 631–649. [Google Scholar] [CrossRef]
- Pellicer, J.; Garcia, S.; Canela, M.A.; Garnatje, T.; Korobkov, A.A.; Twibell, J.D.; Valles, J. Genome size dynamics in Artemisia L. (Asteraceae): Following the track of polyploidy. Plant Biol. 2010, 12, 820–830. [Google Scholar] [CrossRef]
- Kang, S.H.; Kim, K.; Lee, J.H.; Ahn, B.O.; Won, S.Y.; Sohn, S.H.; Kim, J.S. The complete chloroplast genome sequence of medicinal plant, Artemisia argyi. Mitochondrial DNA Part B 2016, 1, 257–258. [Google Scholar] [CrossRef]
- Kim, G.B.; Lim, C.E.; Kim, J.S.; Kim, K.; Lee, J.H.; Yu, H.J.; Mun, J.H. Comparative chloroplast genome analysis of Artemisia (Asteraceae) in East Asia: Insights into evolutionary divergence and phylogenomic implications. BMC Genom. 2020, 21, 415. [Google Scholar] [CrossRef]
- Shen, Q.; Zhang, L.; Liao, Z.; Wang, S.; Yan, T.; Shi, P.; Liu, M.; Fu, X.; Pan, Q.; Wang, Y.; et al. The Genome of Artemisia annua provides insight into the evolution of Asteraceae family and artemisinin biosynthesis. Mol. Plant 2018, 11, 776–788. [Google Scholar] [CrossRef]
- Allen, G.C.; Flores-Vergara, M.A.; Krasynanski, S.; Kumar, S.; Thompson, W.F. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 2006, 1, 2320–2325. [Google Scholar] [CrossRef]
- Yang, X.; Liu, D.; Liu, F.; Wu, J.; Zou, J.; Xiao, X.; Zhao, F.; Zhu, B. HTQC: A fast quality control toolkit for Illumina sequencing data. BMC Bioinform. 2013, 14, 33. [Google Scholar] [CrossRef]
- Liu, B.; Shi, Y.; Yuan, J.; Hu, X.; Zhang, H.; Li, N.; Li, Z.; Chen, Y.; Mu, D.; Fan, W. Estimation of genomic characteristics by analyzing kmer frequency in de novo genome projects. arXiv 2013, arXiv:1308.2012. [Google Scholar]
- Chen, S.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Xiao, C.-L.; Chen, Y.; Xie, S.-Q.; Chen, K.-N.; Wang, Y.; Han, Y.; Luo, F.; Xie, Z. MECAT: Fast mapping, error correction, and de novo assembly for single-molecule sequencing reads. Nat. Methods 2017, 14, 1072–1074. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.D.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Li, R.; Zhu, H.; Ruan, J.; Qian, W.; Fang, X.; Shi, Z.; Li, Y.; Li, S.; Shan, G.; Kristiansen, K.; et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010, 20, 265–272. [Google Scholar] [CrossRef]
- Dudchenko, O.; Batra, S.S.; Omer, A.D.; Nyquist, S.K.; Hoeger, M.; Durand, N.C.; Shamim, M.S.; Machol, I.; Lander, E.S.; Aiden, A.P.; et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 2017, 356, 92–95. [Google Scholar] [CrossRef]
- Durand, N.C.; Shamim, M.S.; Machol, I.; Rao, S.S.P.; Huntley, M.H.; Lander, E.S.; Aiden, E.L. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016, 3, 95–98. [Google Scholar] [CrossRef]
- Durand, N.C.; Robinson, J.T.; Shamim, M.S.; Machol, I.; Mesirov, J.P.; Lander, E.S.; Aiden, E.L. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 2016, 3, 99–101. [Google Scholar] [CrossRef]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness. Methods Mol. Biol. 2019, 1962, 227–245. [Google Scholar]
- Stanke, M.; Keller, O.; Gunduz, I.; Hayes, A.; Waack, S.; Morgenstern, B. AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006, 34, W435–W439. [Google Scholar] [CrossRef]
- Burge, C.; Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997, 268, 78–94. [Google Scholar] [CrossRef]
- Slater, G.S.; Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 2005, 6, 31. [Google Scholar]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Korf, I.; Robb, S.M.C.; Parra, G.; Ross, E.; Moore, B.; Holt, C.; Alvarado, A.S.; Yandell, M. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008, 18, 188–196. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef]
- Soudy, M.; Anwar, A.M.; Ahmed, E.A.; Osama, A.; Ezzeldin, S.; Mahgoub, S.; Magdeldin, S. UniprotR: Retrieving and visualizing protein sequence and functional information from Universal Protein Resource (UniProt knowledgebase). J. Proteom. 2020, 213, 103613. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.H.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.Z.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Bateman, A.; Finn, R.D. Predicting active site residue annotations in the Pfam database. BMC Bioinform. 2007, 8, 298. [Google Scholar] [CrossRef]
- Mitchell, A.; Chang, H.Y.; Daugherty, L.; Fraser, M.; Hunter, S.; Lopez, R.; McAnulla, C.; McMenamin, C.; Nuka, G.; Pesseat, S.; et al. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2015, 43, D213–D221. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- De Bie, T.; Cristianini, N.; Demuth, J.P.; Hahn, M.W. CAFE: A computational tool for the study of gene family evolution. Bioinformatics 2006, 22, 1269–1271. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Yang, Z.; Rannala, B. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol. Biol. Evol. 2006, 23, 212–226. [Google Scholar] [CrossRef]
- Zwaenepoel, A.; Van de Peer, Y. Inference of ancient whole-genome duplications and the evolution of gene duplication and loss rates. Mol. Biol. Evol. 2019, 36, 1384–1404. [Google Scholar] [CrossRef] [PubMed]
- Proost, S.; Fostier, J.; De Witte, D.; Dhoedt, B.; Demeester, P.; VandePeer, Y.; Vandepoele, K. i-ADHoRe 3.0—Fast and sensitive detection of genomic homology in extremely large data sets. Nucleic Acids Res. 2012, 40, e11. [Google Scholar] [CrossRef] [PubMed]
- Yun, D.; Yoon, S.Y.; Park, S.J.; Park, Y.J. The anticancer effect of natural plant alkaloid isoquinolines. Int. J. Mol. Sci. 2021, 22, 1653. [Google Scholar] [CrossRef]
- Scaglione, D.; Reyes-Chin-Wo, S.; Acquadro, A.; Froenicke, L.; Portis, E.; Beitel, C.; Tirone, M.; Mauro, R.; Lo Monaco, A.; Mauromicale, G.; et al. The genome sequence of the outbreeding globe artichoke constructed de novo incorporating a phase-aware low-pass sequencing strategy of F1 progeny. Sci. Rep. 2016, 6, 19427. [Google Scholar] [CrossRef]
- Staton, S.E.; Bakken, B.H.; Blackman, B.K.; Chapman, M.A.; Kane, N.C.; Tang, S.; Ungerer, M.C.; Knapp, S.J.; Rieseberg, L.H.; Burke, J.M. The sunflower (Helianthus annuus L.) genome reflects a recent history of biased accumulation of transposable elements. Plant J. 2012, 72, 142–153. [Google Scholar] [CrossRef]
- Reyes-Chin-Wo, S.; Wang, Z.; Yang, X.; Kozik, A.; Arikit, S.; Song, C.; Xia, L.; Froenicke, L.; Lavelle, D.O.; Truco, M.-J.; et al. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 2017, 8, 14953. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Yan, J.; Li, W.; Yin, L.; Li, P.; Yu, H.; Xing, L.; Cai, M.; Wang, H.; Zhao, M.; et al. Mikania micrantha genome provides insights into the molecular mechanism of rapid growth. Nat. Commun. 2020, 11, 340. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Luo, D.; Zhao, T.; Du, H.; Liu, Z.; Xu, Z.; Guo, L.; Chen, C.; Peng, S.; Li, J.X.; et al. Genome sequencing reveals chromosome fusion and extensive expansion of genes related to secondary metabolism in Artemisia argyi. Plant Biotechnol. J. 2022, 20, 1902–1915. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Guo, M.; Dong, S.; Wu, X.; Zhang, G.; He, L.; Jiao, Y.; Chen, S.; Li, L.; Luo, H. A chromosome-scale genome assemblyof Artemisia argyi reveals unbiased subgenome evolution and key contributions of gene duplication to volatile terpenoid diversity. Plant Commun. 2023, 2, 100516. [Google Scholar] [CrossRef]
- Kyriakidou, M.; Tai, H.H.; Anglin, N.L.; Ellis, D.; Stromvik, M.V. Current strategies of polyploid plant genome sequence assembly. Front. Plant Sci. 2018, 9, 1660. [Google Scholar] [CrossRef]
- Xu, C.G.; Tang, T.X.; Chen, R.; Liang, C.H.; Liu, X.Y.; Wu, C.L.; Yang, Y.S.; Yang, D.P.; Wu, H. A comparative study of bioactive secondary metabolite production in diploid and tetraploid Echinacea purpurea (L.) Moench. Plant Cell Tissue Organ Cult. 2014, 116, 323–332. [Google Scholar] [CrossRef]
- Cheng, D.; Vrieling, K.; Klinkhamer, P.G. The effect of hybridization on secondary metabolites and herbivore resistance: Implications for the evolution of chemical diversity in plants. Phytochem. Rev. 2011, 10, 107–117. [Google Scholar] [CrossRef]
- Wang, P.; Yu, J.; Jin, S.; Chen, S.; Yue, C.; Wang, W.; Gao, S.; Cao, H.; Zheng, Y.; Gu, M.; et al. Genetic basis of high aroma and stress tolerance in the oolong tea cultivar genome. Hortic. Res. 2021, 8, 107. [Google Scholar] [CrossRef]
- Ivanescu, B.; Burlec, A.F.; Crivoi, F.; Rosu, C.; Corciova, A. Secondary metabolites from Artemisia genus as biopesticides and innovative nano-based application strategies. Molecules 2021, 26, 3061. [Google Scholar] [CrossRef]
| Species | Genes | Families | Clustered Genes | Unclustered Genes | Specific Genes | Specific Families |
|---|---|---|---|---|---|---|
| A. argyi | 139,245 | 27,279 | 123,512 | 15,733 | 18,576 | 4858 |
| A. annua | 66,918 | 23,935 | 61,118 | 5800 | 7547 | 2030 |
| C. cardunculus | 38,406 | 16,749 | 37,567 | 839 | 1695 | 333 |
| H. annuus | 44,144 | 16,818 | 40,078 | 4066 | 5909 | 1656 |
| L. sativa | 45,243 | 17,972 | 43,605 | 1638 | 5612 | 1000 |
| M. micrantha | 46,351 | 17,554 | 43,088 | 3263 | 8059 | 1522 |
| S. tuberosum | 37,967 | 15,602 | 35,723 | 2244 | 7563 | 1541 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mei, Q.; Li, H.; Liu, Y.; Wu, F.; Liu, C.; Wang, K.; Liu, H.; Peng, C.; Wang, Z.; Wang, L.; et al. De Novo Assembly of an Allotetraploid Artemisia argyi Genome. Agronomy 2023, 13, 436. https://doi.org/10.3390/agronomy13020436
Mei Q, Li H, Liu Y, Wu F, Liu C, Wang K, Liu H, Peng C, Wang Z, Wang L, et al. De Novo Assembly of an Allotetraploid Artemisia argyi Genome. Agronomy. 2023; 13(2):436. https://doi.org/10.3390/agronomy13020436
Chicago/Turabian StyleMei, Qiming, Hanxiang Li, Yanbin Liu, Feng Wu, Chuang Liu, Keya Wang, Hongjun Liu, Cheng Peng, Zhengfeng Wang, Long Wang, and et al. 2023. "De Novo Assembly of an Allotetraploid Artemisia argyi Genome" Agronomy 13, no. 2: 436. https://doi.org/10.3390/agronomy13020436
APA StyleMei, Q., Li, H., Liu, Y., Wu, F., Liu, C., Wang, K., Liu, H., Peng, C., Wang, Z., Wang, L., Liu, Z., Yan, J., & Zhang, W. (2023). De Novo Assembly of an Allotetraploid Artemisia argyi Genome. Agronomy, 13(2), 436. https://doi.org/10.3390/agronomy13020436






