Identification of Differential-Expressed Genes in Banana-Biostimulant Interaction Using Suppression Subtractive Hybridization
Abstract
1. Introduction
2. Results
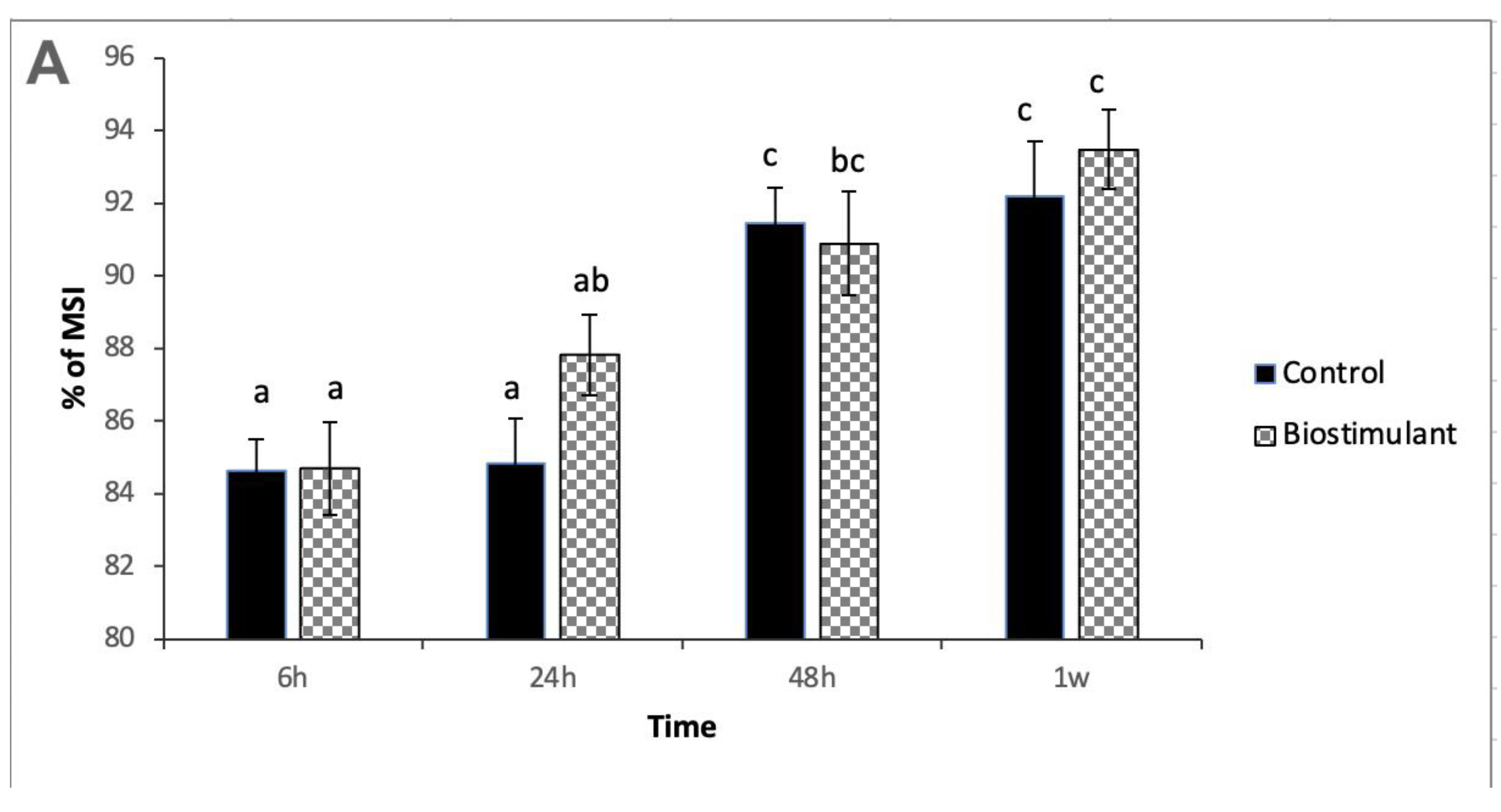
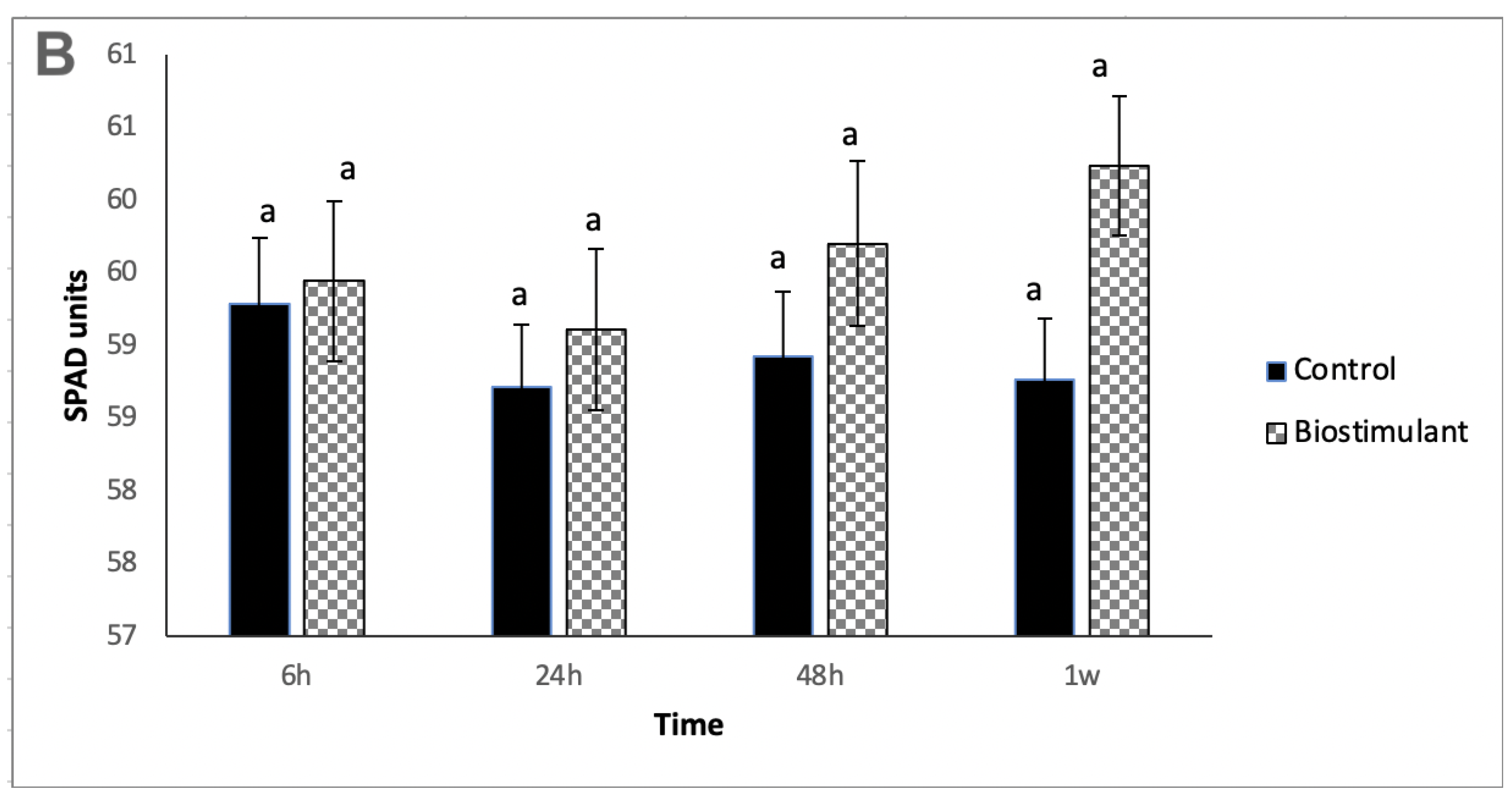
2.1. Physiological Analyses
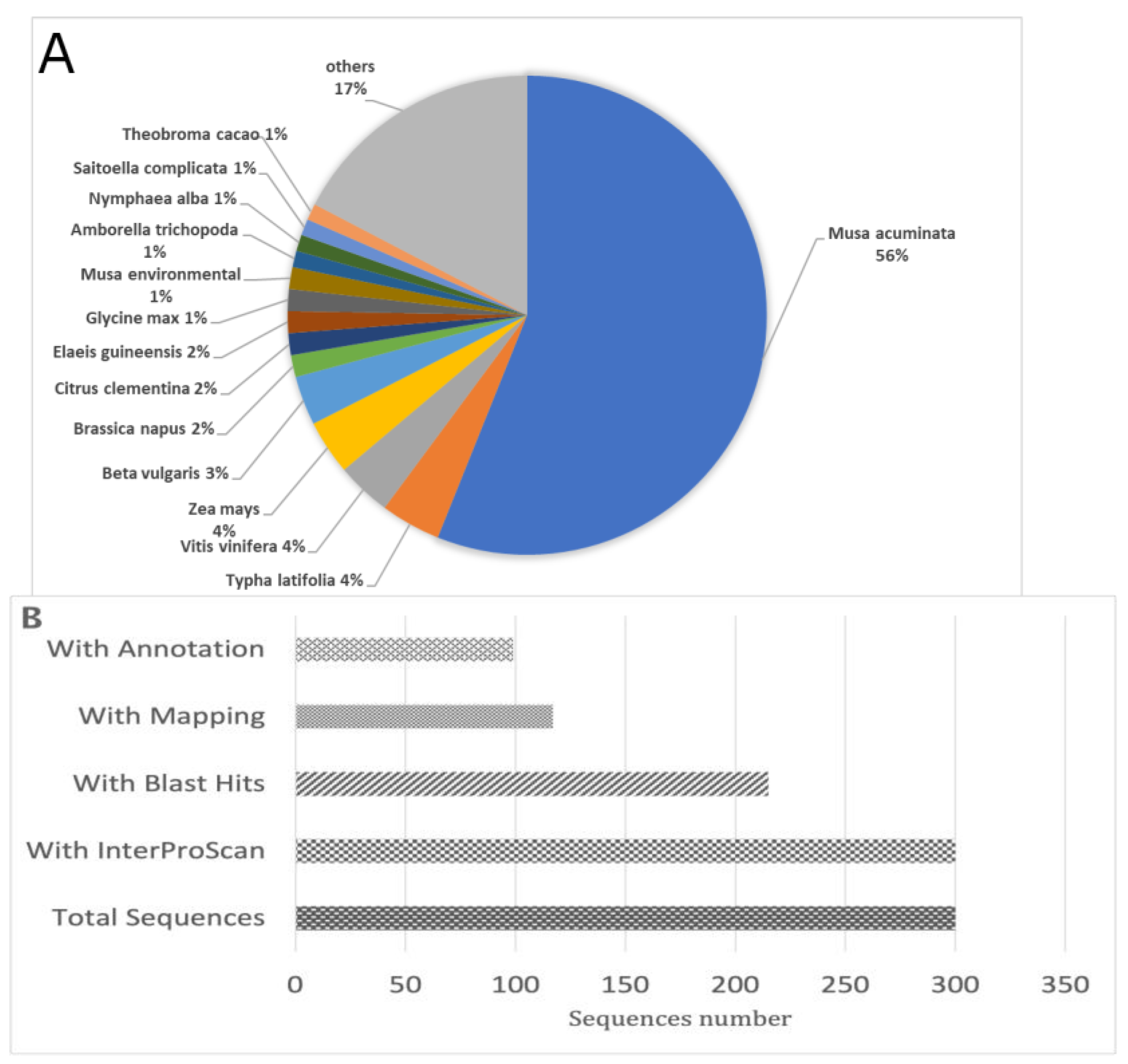
2.2. Genes Identified in the Subtractive Library
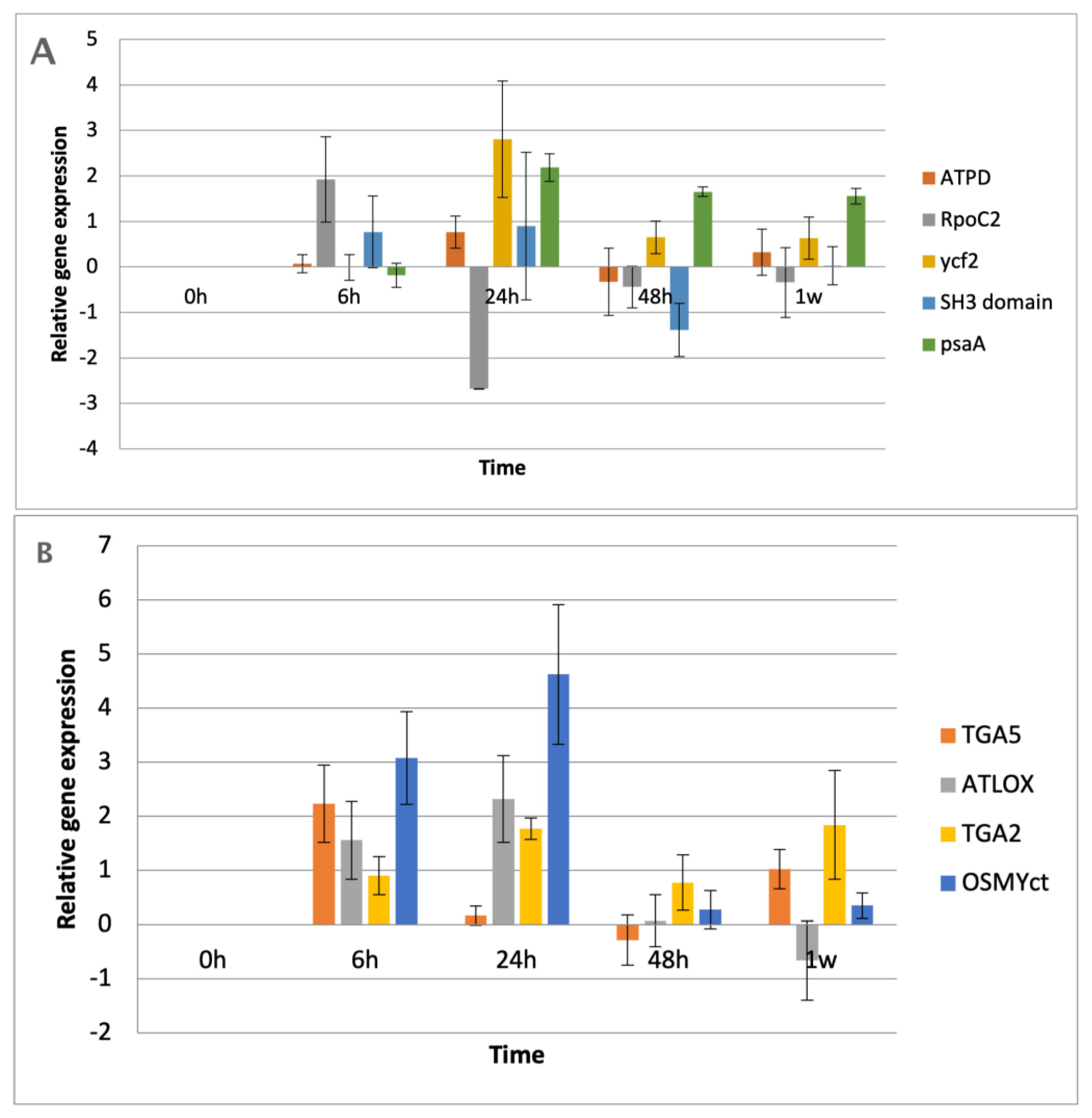
2.3. Upregulated Genes after Biostimulant Application
3. Discussion
4. Materials and Methods
4.1. Plant Conditions
4.2. Biostimulant Conditions
4.3. Physiological Parameters Analyzed
4.4. Total RNA Isolation and cDNA Synthesis
4.5. Library Generation and Analysis
4.6. Pathway Activation and Gene Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. FAOSTAT [Internet]. Rome, Economic and Social Development Department. 2022. Available online: https://www.fao.org/faostat/es/#data/FBS (accessed on 21 November 2014).
- INEC. Análisis del Sistema Agroalimentario del Banano en el Ecuador, 2–33. 2009. Available online: www.inec.gob.ec (accessed on 13 January 2023).
- Ganugi, P.; Martinelli, E.; Lucini, L. Microbial biostimulants as a sustainable approach to improve the functional quality in plant-based foods: A review. Curr. Opin. Food Sci. 2021, 41, 217–223. [Google Scholar] [CrossRef]
- Skirycz, A.; Inzé, D. More from less: Plant growth under limited water. Curr. Opin. Biotechnol. 2010, 21, 197–203. [Google Scholar] [CrossRef]
- Cataldo, E.; Fucile, M.; Mattii, G.B. Biostimulants in Viticulture: A Sustainable Approach against Biotic and Abiotic Stresses. Plants 2022, 11, 162. [Google Scholar] [CrossRef]
- García, A.C.; Santos, L.A.; Izquierdo, F.G.; Sperandio, M.V.L.; Castro, R.N.; Berbara, R.L.L. Vermicompost humic acids as an ecological pathway to protect rice plant against oxidative stress. Ecol. Eng. 2012, 47, 203–208. [Google Scholar] [CrossRef]
- Van Oosten, M.J.; Pepe, O.; De Pascale, S.; Silletti, S.; Maggio, A. The role of biostimulants and bioeffectors as alleviators of abiotic stress in crop plants. Chem. Biol. Technol. Agric. 2017, 4, 5. [Google Scholar] [CrossRef]
- Desoky, E.S.M.; ElSayed, A.I.; Merwad, A.R.M.; Rady, M.M. Stimulating antioxidant defenses, antioxidant gene ex-pression, and salt tolerance in Pisum sativum seedling by pretreatment using licorice root extract (LRE) as an organic bi-ostimulant. Plant Physiol. Biochem. 2019, 142, 292–302. [Google Scholar] [CrossRef]
- Gutiérrez-Gamboa, G.; Romanazzi, G.; Garde-Cerdán, T.; Pérez-Álvarez, E.P. A review of the use of biostimulants in the vineyard for improved grape and wine quality: Effects on prevention of grapevine diseases. J. Sci. Food Agric. 2019, 99, 1001–1009. [Google Scholar] [CrossRef]
- Bulgari, R.; Cocetta, G.; Trivellini, A.; Vernieri, P.; Ferrante, A. Biostimulants and crop responses: A review. Biol. Agric. Hortic. 2015, 31, 1–17. [Google Scholar] [CrossRef]
- Vernieri, P.; Borghesi, E.; Ferrante, A.; Magnani, G. Application of biostimulants in floating system for improving rocket quality. J. Food Agric. Environ. 2005, 3, 86. [Google Scholar]
- Sharma, H.S.; Fleming, C.; Selby, C.; Rao, J.R.; Martin, T. Plant biostimulants: A review on the processing of macroalgae and use of extracts for crop management to reduce abiotic and biotic stresses. J. Appl. Phycol. 2014, 26, 465–490. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [PubMed]
- D’Addabbo, T.; Laquale, S.; Perniola, M.; Candido, V. Biostimulants for Plant Growth Promotion and Sustainable Management of Phytoparasitic Nematodes in Vegetable Crops. Agronomy 2019, 9, 616. [Google Scholar] [CrossRef]
- Drobek, M.; Frąc, M.; Cybulska, J. Plant biostimulants: Importance of the quality and yield of horticultural crops and the im-provement of plant tolerance to abiotic stress—A review. Agronomy 2019, 9, 335. [Google Scholar] [CrossRef]
- Bubici, G.; Kaushal, M.; Prigigallo, M.I.; Cabanás, C.G.-L.; Mercado-Blanco, J. Biological Control Agents Against Fusarium Wilt of Banana. Front. Microbiol. 2019, 10, 616. [Google Scholar] [CrossRef] [PubMed]
- Noar, R.D.; Thomas, E.; Daub, M.E. Genetic Characteristics and Metabolic Interactions between Pseudocercospora fijiensis and Banana: Progress toward Controlling Black Sigatoka. Plants 2022, 11, 948. [Google Scholar] [CrossRef]
- Bhardwaj, D.; Ansari, M.W.; Sahoo, R.K.; Tuteja, N. Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb. Cell Fact. 2014, 13, 66. [Google Scholar] [CrossRef]
- Du Jardin, P. Plant biostimulants: Definition, concept, main categories and regulation. Sci. Hortic. 2015, 196, 3–14. [Google Scholar] [CrossRef]
- Baltazar, M.; Correia, S.; Guinan, K.; Sujeeth, N.; Bragança, R.; Gonçalves, B. Recent Advances in the Molecular Effects of Biostimulants in Plants: An Overview. Biomolecules 2021, 11, 1096. [Google Scholar] [CrossRef]
- Jimenez, M.; Van der Veken, L.; Neirynck, H.; Rodríguez, H.; Ruiz, O.; Swennen, R. Organic banana production in Ecuador: Its implications on black Sigatoka development and plant–soil nutritional status. Renew. Agric. Food Syst. 2007, 22, 297–306. [Google Scholar] [CrossRef][Green Version]
- Rouphael, Y.; Colla, G. Editorial: Biostimulants in Agriculture. Front. Plant Sci. 2020, 11, 40. [Google Scholar] [CrossRef]
- Ali, O.; Ramsubhag, A.; Ramnarine, S.D.B.J.; Jayaraman, J. Transcriptomic changes induced by applications of a commercial extract of Ascophyllum nodosum on tomato plants. Sci. Rep. 2022, 12, 8042. [Google Scholar] [CrossRef] [PubMed]
- Nair, P.; Kandasamy, S.; Zhang, J.; Ji, X.; Kirby, C.; Benkel, B.; Hodges, M.D.; Critchley, A.T.; Hiltz, D.; Prithiviraj, B. Transcriptional and metabolomic analysis of Ascophyllum nodosum mediated freezing tolerance in Arabidopsis thaliana. BMC Genom. 2012, 13, 22–43. [Google Scholar] [CrossRef] [PubMed]
- Alharby, H.F.; Alzahrani, Y.M.; Rady, M.M. Seeds pretreatment with zeatins or maize grain-derived organic biostimulant improved hormonal contents, polyamine gene expression, and salinity and drought tolerance of wheat. Int. J. Agric. Biol. 2020, 24, 714–724. [Google Scholar]
- González-Morales, S.; Solís-Gaona, S.; Valdés-Caballero, M.V.; Juárez-Maldonado, A.; Loredo-Treviño, A.; Benavides-Mendoza, A. Transcriptomics of Biostimulation of Plants Under Abiotic Stress. Front. Genet. 2021, 12, 583888. [Google Scholar] [CrossRef] [PubMed]
- Della Lucia, M.C.; Baghdadi, A.; Mangione, F.; Borella, M.; Zegada-Lizarazu, W.; Ravi, S.; Deb, S.; Broccanello, C.; Concheri, G.; Monti, A.; et al. Transcriptional and Physiological Analyses to Assess the Effects of a Novel Biostimulant in Tomato. Front. Plant Sci. 2022, 12, 781993. [Google Scholar] [CrossRef]
- Islam, M.T.; Gan, H.M.; Ziemann, M.; Hussain, H.I.; Arioli, T.; Cahill, D. Phaeophyceaean (Brown Algal) Extracts Activate Plant Defense Systems in Arabidopsis thaliana Challenged with Phytophthora cinnamomi. Front. Plant Sci. 2020, 11, 852. [Google Scholar] [CrossRef]
- Wu, S.; Cao, Z.; Li, Z.; Cheung, K.; Wong, M. Effects of biofertilizer containing N-fixer, P and K solubilizers and AM fungi on maize growth: A greenhouse trial. Geoderma 2005, 125, 155–166. [Google Scholar] [CrossRef]
- Singh, J.S.; Pandey, V.C.; Singh, D. Efficient soil microorganisms: A new dimension for sustainable agriculture and environmental development. Agric. Ecosyst. Environ. 2011, 140, 339–353. [Google Scholar] [CrossRef]
- Sahoo, R.K.; Ansari, M.W.; Dangar, T.K.; Mohanty, S.; Tuteja, N. Phenotypic and molecular characterisation of efficient nitrogen-fixing Azotobacter strains from rice fields for crop improvement. Protoplasma 2013, 251, 511–523. [Google Scholar] [CrossRef]
- Ritika, B.; Utpal, D. Biofertilizer, a way towards organic agriculture: A review. Afr. J. Microbiol. Res. 2014, 8, 2332–2343. [Google Scholar] [CrossRef]
- Rajasekaran, S.; Sundaramoorthy, P.; Sankar Ganesh, K. Effect of FYM, N, P fertilizers and biofertilizers on germi-nation and growth of paddy (Oryza sativa. L). Int. Lett. Nat. Sci. 2015, 35, 59–65. [Google Scholar]
- Schmitz-Linneweber, C.; Maier, R.M.; Alcaraz, J.-P.; Cottet, A.; Herrmann, R.G.; Mache, R. The plastid chromosome of spinach (Spinacia oleracea): Complete nucleotide sequence and gene organization. Plant Mol. Biol. 2001, 45, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Hobo, T.; Kowyama, Y.; Hattori, T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA 1999, 96, 15348–15353. Available online: http://www.ncbi.nlm.nih.gov/pubmed/10611387 (accessed on 13 January 2023). [CrossRef]
- Lancien, M.; Martin, M.; Hsieh, M.-H.; Leustek, T.; Goodman, H.; Coruzzi, G.M. Arabidopsis glt1-T mutant defines a role for NADH-GOGAT in the non-photorespiratory ammonium assimilatory pathway. Plant J. 2002, 29, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Cournac, L.; Redding, K.; Ravenel, J.; Rumeau, D.; Josse, E.-M.; Kuntz, M.; Peltier, G. Electron Flow between Photosystem II and Oxygen in Chloroplasts of Photosystem I-deficient Algae Is Mediated by a Quinol Oxidase Involved in Chlororespiration. J. Biol. Chem. 2000, 275, 17256–17262. [Google Scholar] [CrossRef]
- Bonfante, P.; Genre, A. Mechanisms underlying beneficial plant–fungus interactions in mycorrhizal symbiosis. Nat. Commun. 2010, 1, 48. [Google Scholar] [CrossRef]
- Plett, J.M.; Kemppainen, M.; Kale, S.D.; Kohler, A.; Legué, V.; Brun, A.; Tyler, B.M.; Pardo, A.G.; Martin, F. A Secreted Effector Protein of Laccaria bicolor Is Required for Symbiosis Development. Curr. Biol. 2011, 21, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, P.; López-Juez, E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 2013, 14, 787–802. [Google Scholar] [CrossRef] [PubMed]
- Liebers, M.; Grübler, B.; Chevalier, F.; Lerbs-Mache, S.; Merendino, L.; Blanvillain, R.; Pfannschmidt, T. Regulatory Shifts in Plastid Transcription Play a Key Role in Morphological Conversions of Plastids during Plant Development. Front. Plant Sci. 2017, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Hajdukiewicz, P.T.; Allison, L.A.; Maliga, P. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 1997, 16, 4041–4048. [Google Scholar] [CrossRef]
- Ishizaki, Y.; Tsunoyama, Y.; Hatano, K.; Ando, K.; Kato, K.; Shinmyo, A.; Kobori, M.; Takeba, G.; Nakahira, Y.; Shiina, T. A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J. 2005, 42, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Loschelder, H.; Schweer, J.; Link, B.; Link, G. Dual Temporal Role of Plastid Sigma Factor 6 in Arabidopsis Development. Plant Physiol. 2006, 142, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Ghulam, M.M.; Zghidi-Abouzid, O.; Lambert, E.; Lerbs-Mache, S.; Merendino, L. Transcriptional organization of the large and the small ATP synthase operons, atpI/H/F/A and atpB/E, in Arabidopsis thaliana chloroplasts. Plant Mol. Biol. 2012, 79, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Woodson, J.D.; Perez-Ruiz, J.M.; Schmitz, R.J.; Ecker, J.R.; Chory, J. Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J. 2013, 73, 1–13. [Google Scholar] [CrossRef]
- Allison, L.A.; Simon, L.D.; Maliga, P. Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J. 1996, 15, 2802–2809. [Google Scholar] [CrossRef] [PubMed]
- Legen, J.; Kemp, S.; Krause, K.; Profanter, B.; Herrmann, R.G.; Maier, R.M. Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J. 2002, 31, 171–188. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Yakhin, O.I.; Lubyanov, A.A.; Yakhin, I.A.; Brown, P.H. Biostimulants in Plant Science: A Global Perspective. Front. Plant Sci. 2017, 7, 2049. [Google Scholar] [CrossRef] [PubMed]
- Tejada, M.; Benítez, C.; Parrado, J. Application of biostimulants in benzo(a)pyrene polluted soils: Short-time effects on soil biochemical properties. Appl. Soil Ecol. 2011, 50, 21–26. [Google Scholar] [CrossRef]
- Cheng, L.; Wang, Y.; He, Q.; Li, H.; Zhang, X.; Zhang, F. Comparative proteomics illustrates the complexity of drought resistance mechanisms in two wheat (Triticum aestivum L.) cultivars under dehydration and rehydration. BMC Plant Biol. 2016, 16, 188. [Google Scholar] [CrossRef]
- Feussner, I.; Wasternack, C. The lipoxygenase pathway. Annu. Rev. Plant Biol. 2002, 53, 275–297. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Han, S.-W.; Hwang, I.S.; Kim, D.S.; Hwang, B.K.; Lee, S.C. The Pepper Lipoxygenase CaLOX1 Plays a Role in Osmotic, Drought and High Salinity Stress Response. Plant Cell Physiol. 2015, 56, 930–942. [Google Scholar] [CrossRef]
- Marla, S.S.; Singh, V.K. LOX genes in blast fungus (Magnaporthe grisea) resistance in rice. Funct. Integr. Genom. 2012, 12, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. MYC2: The Master in Action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef] [PubMed]
- Woldemariam, M.G.; Dinh, S.T.; Oh, Y.; Gaquerel, E.; Baldwin, I.T.; Galis, I. NaMYC2 transcription factor regulates a subset of plant defense responses in Nicotiana attenuata. BMC Plant Biol. 2013, 13, 73. [Google Scholar] [CrossRef]
- Pozo, M.J.; Van Der Ent, S.; Van Loon, L.C.; Pieterse, C.M.J. Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol. 2008, 180, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Sanchez Timm, L.E. Stress Tolerance Enhancement of Rice by Genetic Manipulation of a bHLH-Myc2 Transcription Factor; Louisiana State University: Baton Rouge, LA, USA, 2015. [Google Scholar]
- Sánchez Timm, E.; Hidalgo Pardo, L.; Pacheco Coello, R.; Chávez Navarrete, T.; Navarrete Villegas, O.; Santos Ordóñez, E. Identification of Differentially-Expressed Genes in Response to Mycosphaerella fijiensis in the Resistant Musa Accession “Calcutta-4” Using Suppression Subtractive Hybridization. PLoS ONE 2016, 11, e0160083. [Google Scholar] [CrossRef]
- Zhou, J.-M.; Trifa, Y.; Silva, H.; Pontier, D.; Lam, E.; Shah, J.; Klessig, D.F. NPR1 Differentially Interacts with Members of the TGA/OBF Family of Transcription Factors That Bind an Element of the PR-1 Gene Required for Induction by Salicylic Acid. Mol. Plant-Microbe Interact. 2000, 13, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Karasov, T.L.; Chae, E.; Herman, J.J.; Bergelson, J. Mechanisms to Mitigate the Trade-Off between Growth and Defense. Plant Cell 2017, 29, 666–680. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.H.; Rosa, C.; Scully, E.D.; Peiffer, M.; Tooker, J.F.; Hoover, K.; Luthe, D.S.; Felton, G.W. Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc. Natl. Acad. Sci. USA 2013, 110, 15728–15733. [Google Scholar] [CrossRef]
- Chávez-Navarrete, T.P.; Sánchez-Timm, E.; Santos-Ordóñez, E. Dataset of suppression subtractive hybridization libraries of banana-biostimulant-Pseudocercospora fijiensis molecular interaction. Data Brief 2019, 27, 104557. [Google Scholar] [CrossRef] [PubMed]
- Zambrano-Mendoza, J.L.; Sangoquiza-Caiza, C.A.; Campaña-Cruz, D.F.; Yánez-Guzmán, C.F. Use of Biofertilizers in Agricultural Production. In Technology in Agriculture; Ahmad, F., Sultan, M., Eds.; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Caradonia, F.; Battaglia, V.; Righi, L.; Pascali, G.; La Torre, A. Plant Biostimulant Regulatory Framework: Prospects in Europe and Current Situation at International Level. J. Plant Growth Regul. 2019, 38, 438–448. [Google Scholar] [CrossRef]
- Basu, A.; Prasad, P.; Das, S.N.; Kalam, S.; Sayyed, R.Z.; Reddy, M.S.; El Enshasy, H. Plant Growth Promoting Rhizobacteria (PGPR) as Green Bioinoculants: Recent Developments, Constraints, and Prospects. Sustainability 2021, 13, 1140. [Google Scholar] [CrossRef]
- Sairam, R.; Shukla, D.; Saxena, D. Stress induced injury and antioxidant enzymes in relation to drought tolerance in wheat genotypes. Biol. Plant. 1997, 39, 357–364. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R. [Online]; RStudio, Inc.: Boston, MA, USA, 2016; Available online: http://www.rstudio.com (accessed on 13 January 2023). [CrossRef]
- Sambrook, J.; Russel, D.W. Molecular Cloning, 3-Volume Set: A Laboratory Manual; Cold Spring Harbour Laboratory Press: New York, NY, USA, 2000. [Google Scholar]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Droc, G.; Larivière, D.; Guignon, V.; Yahiaoui, N.; This, D.; Garsmeur, O.; Dereeper, A.; Hamelin, C.; Argout, X.; Dufayard, J.-F.; et al. The Banana Genome Hub. Database 2013, 2013, bat035. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
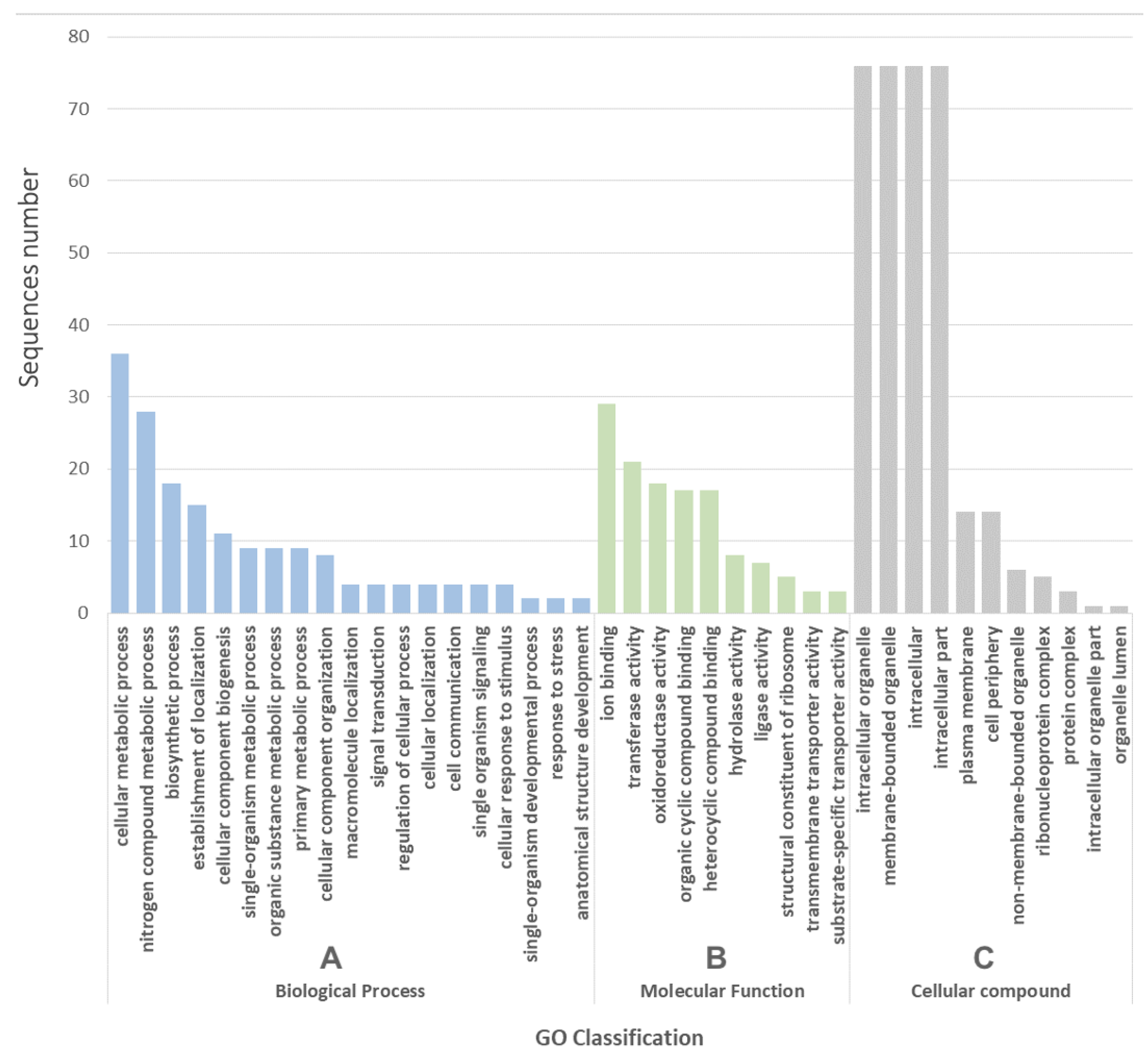
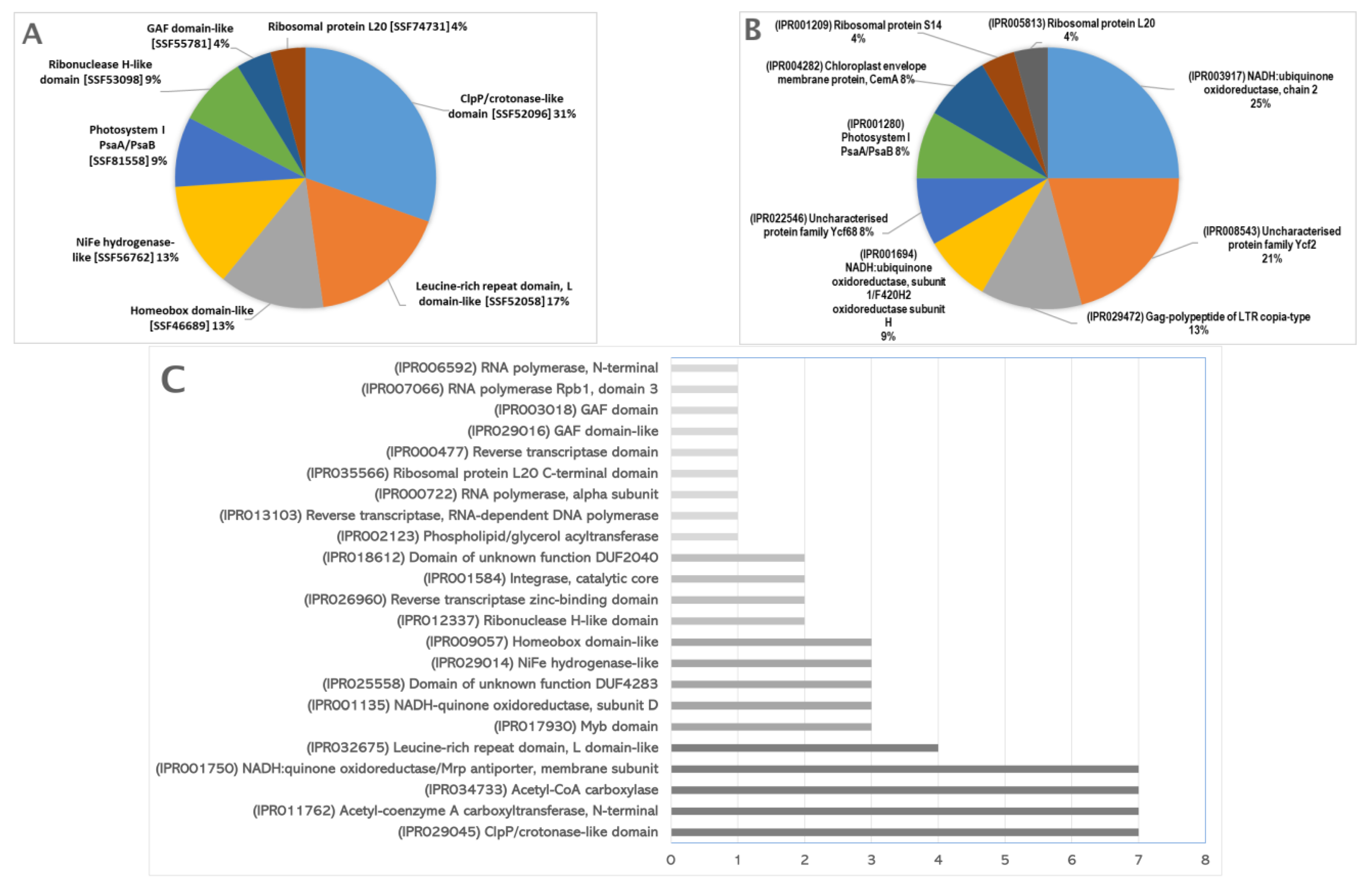
| Pathways | #Sequences | #Enzymes |
|---|---|---|
| Purine metabolism | 21 | 3 |
| Oxidative phosphorylation | 18 | 2 |
| Pyrimidine metabolism | 16 | 1 |
| Thiamine metabolism | 5 | 1 |
| Phenylpropanoid biosynthesis | 2 | 1 |
| Glutathione metabolism | 2 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chavez-Navarrete, T.; Sanchez-Timm, L.; Pacheco-Coello, R.; Baisakh, N.; Santos-Ordóñez, E. Identification of Differential-Expressed Genes in Banana-Biostimulant Interaction Using Suppression Subtractive Hybridization. Agronomy 2023, 13, 415. https://doi.org/10.3390/agronomy13020415
Chavez-Navarrete T, Sanchez-Timm L, Pacheco-Coello R, Baisakh N, Santos-Ordóñez E. Identification of Differential-Expressed Genes in Banana-Biostimulant Interaction Using Suppression Subtractive Hybridization. Agronomy. 2023; 13(2):415. https://doi.org/10.3390/agronomy13020415
Chicago/Turabian StyleChavez-Navarrete, Tatiana, Luis Sanchez-Timm, Ricardo Pacheco-Coello, Niranjan Baisakh, and Efrén Santos-Ordóñez. 2023. "Identification of Differential-Expressed Genes in Banana-Biostimulant Interaction Using Suppression Subtractive Hybridization" Agronomy 13, no. 2: 415. https://doi.org/10.3390/agronomy13020415
APA StyleChavez-Navarrete, T., Sanchez-Timm, L., Pacheco-Coello, R., Baisakh, N., & Santos-Ordóñez, E. (2023). Identification of Differential-Expressed Genes in Banana-Biostimulant Interaction Using Suppression Subtractive Hybridization. Agronomy, 13(2), 415. https://doi.org/10.3390/agronomy13020415






