Genetic Variation for Traits Related to Phosphorus Use Efficiency in Vigna Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growing Condition
2.2. Trait Measurement
2.3. Estimation of Phosphorus Concentration
2.4. Estimation of Phosphorus-Use Efficiency
2.5. Statistical Analysis
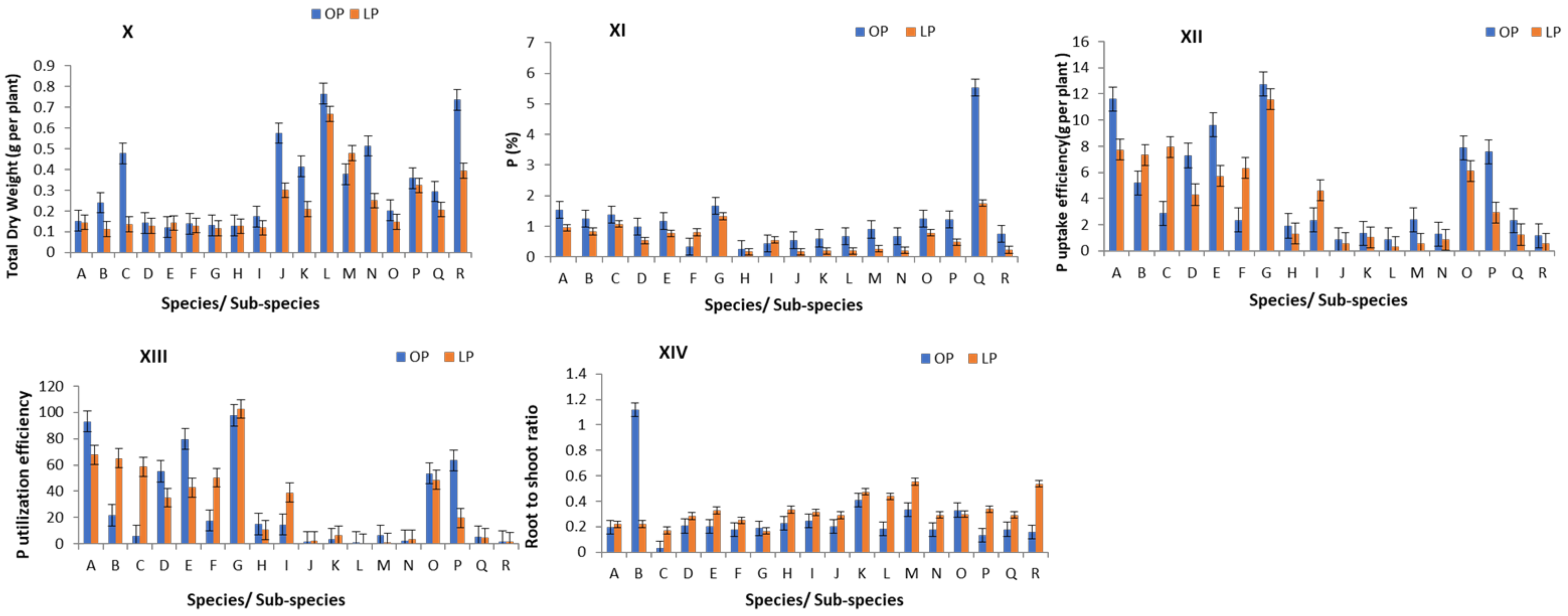
3. Results
3.1. Descriptive Statistics of Measured Traits
3.2. Variation of PUE Traits in Studied Vigna Species
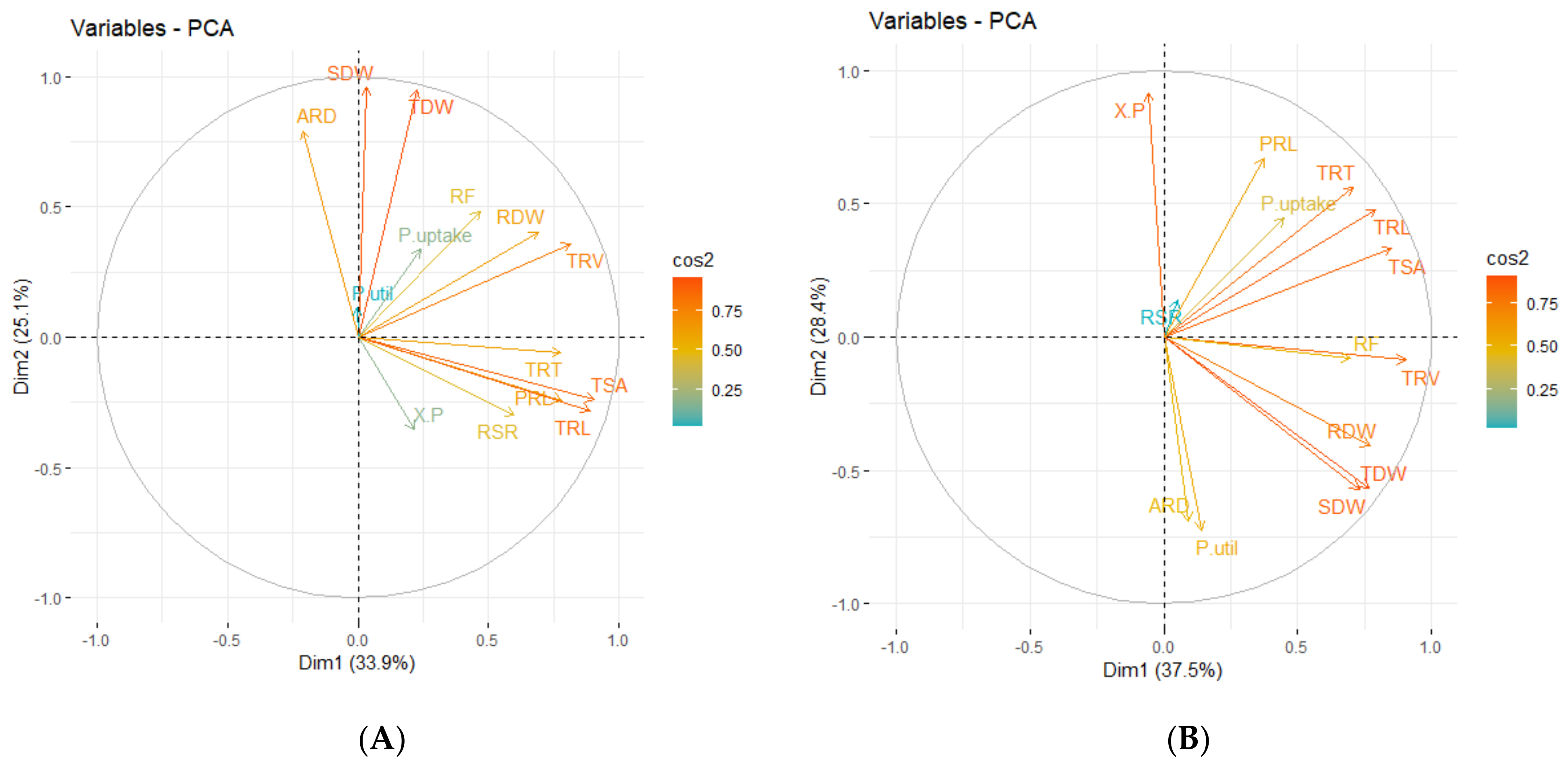
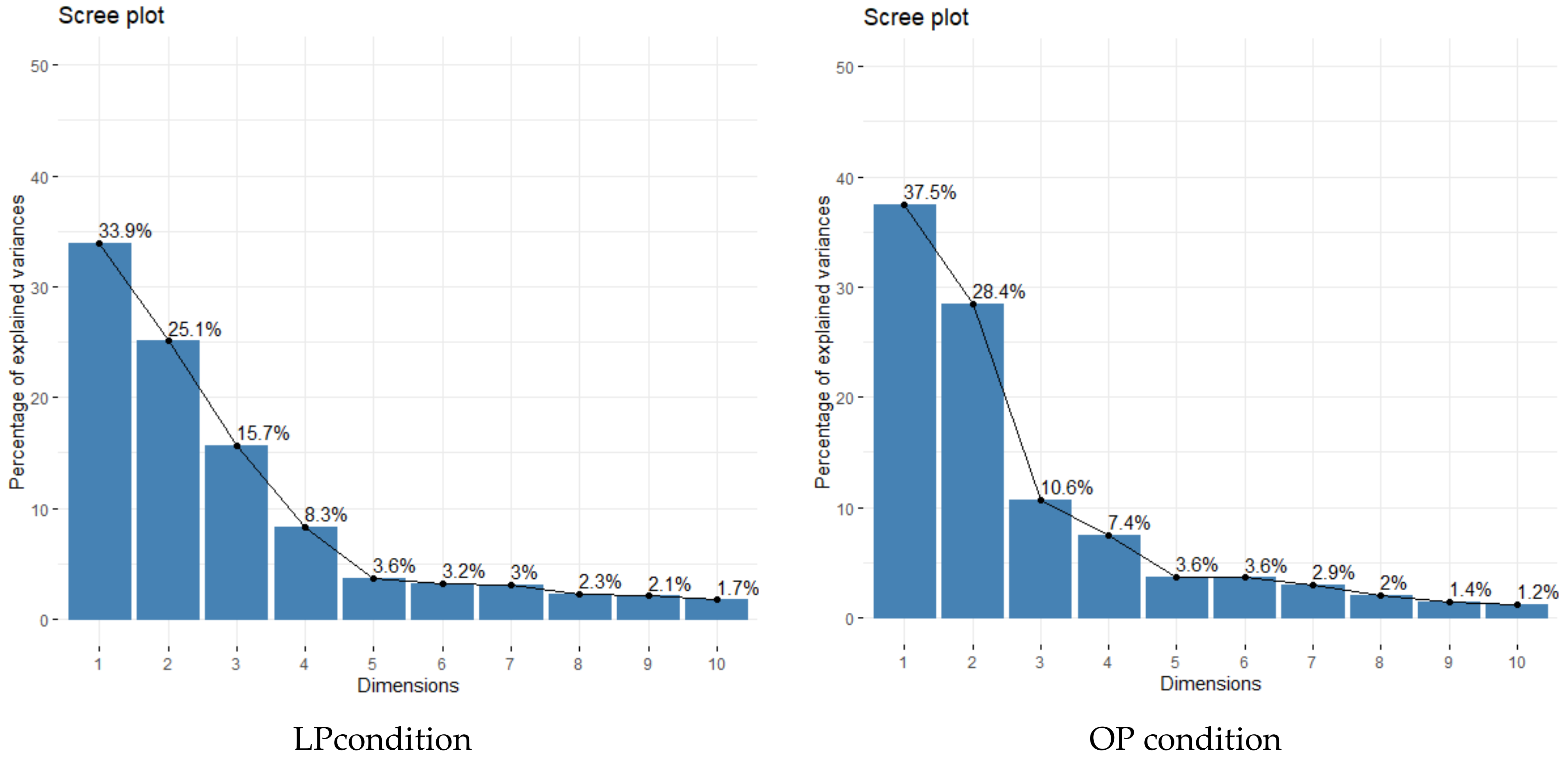
3.3. Correlation Coefficients between Measured Traits
3.4. Identification of Superior Genotypes for Important PUE Traits in Vigna Species
3.5. Categorization of Vigna Genotypes for PUE
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maxted, N. African Vigna: An Ecogeographic Study; IPGRI: Rome, Italy, 2004; Volume 11, ISBN 9290436379. [Google Scholar]
- Tomooka, N.; Kaga, A.; Isemura, T.; Vaughan, D. Vigna. In Wild Crop Relatives: Genomic and Breeding Resources; Springer: Berlin/Heidelberg, Germany, 2011; pp. 291–311. [Google Scholar]
- Nair, R.M.; Pandey, A.K.; War, A.R.; Hanumantharao, B.; Shwe, T.; Alam, A.; Pratap, A.; Malik, S.R.; Karimi, R.; Mbeyagala, E.K. Biotic and Abiotic Constraints in Mungbean Production—Progress in Genetic Improvement. Front. Plant Sci. 2019, 10, 1340. [Google Scholar] [CrossRef] [PubMed]
- Sandarani, M.; Kulathunga, K. A brief review: Lectins, protease inhibitors and saponins in cereals and legumes. Asian Food Sci. J. 2019, 10, 1–4. [Google Scholar] [CrossRef]
- Kaur, S.; Kumari, A.; Singh, P.; Kaur, L.; Sharma, N.; Garg, M. Biofortification in pulses. In Advances in Agri-Food Biotechnology; Tilak Raj, S., Rupesh, D., Humira, S., Eds.; Springer: New York, NY, USA, 2020; pp. 85–103. [Google Scholar]
- Dahiya, P.K.; Linnemann, A.R.; van Boekel, M.; Khetarpaul, N.; Grewal, R.B.; Nout, M.J.R. Mung Bean: Technological and Nutritional Potential. Crit. Rev. Food Sci. Nutr. 2015, 55, 670–688. [Google Scholar] [CrossRef] [PubMed]
- Warschefsky, E.J.; Rieseberg, L.H. Laying the Groundwork for Crop Wild Relative Conservation in the United States. Proc. Natl. Acad. Sci. USA 2021, 118, e2024375118. [Google Scholar] [CrossRef] [PubMed]
- Syers, J.K.; Johnston, A.E.; Curtin, D. Efficiency of Soil and Fertilizer Phosphorus Use. In FAO Fertilizer and Plant Nutrition Bulletin; FAO: Rome, Italy, 2008; Volume 18. [Google Scholar]
- Heuer, S.; Gaxiola, R.; Schilling, R.; Herrera-Estrella, L.; López-Arredondo, D.; Wissuwa, M.; Delhaize, E.; Rouached, H. Improving Phosphorus Use Efficiency: A Complex Trait with Emerging Opportunities. Plant J. 2017, 90, 868–885. [Google Scholar] [CrossRef]
- van Kauwenbergh, S.J.; Stewart, M.; Mikkelsen, R. World Reserves of Phosphate Rock a Dynamic and Unfolding Story. Better Crops Plant Food 2013, 97, 18–20. [Google Scholar]
- Ulrich, A.E.; Frossard, E. On the History of a Reoccurring Concept: Phosphorus Scarcity. Sci. Total Environ. 2014, 490, 694–707. [Google Scholar] [CrossRef]
- Niu, Y.F.; Chai, R.S.; Jin, G.L.; Wang, H.; Tang, C.X.; Zhang, Y.S. Responses of Root Architecture Development to Low Phosphorus Availability: A Review. Ann. Bot. 2013, 112, 391–408. [Google Scholar] [CrossRef]
- Batjes, N.H. A World Dataset of Derived Soil Properties by FAO–UNESCO Soil Unit for Global Modelling. Soil Use Manag. 1997, 13, 9–16. [Google Scholar] [CrossRef]
- Hammond, J.P.; Broadley, M.R.; White, P.J.; King, G.J.; Bowen, H.C.; Hayden, R.; Meacham, M.C.; Mead, A.; Overs, T.; Spracklen, W.P. Shoot Yield Drives Phosphorus Use Efficiency in Brassica Oleracea and Correlates with Root Architecture Traits. J. Exp. Bot. 2009, 60, 1953–1968. [Google Scholar] [CrossRef]
- Plaxton, W.C.; Tran, H.T. Metabolic Adaptations of Phosphate-Starved Plants. Plant Physiol. 2011, 156, 1006–1015. [Google Scholar] [CrossRef]
- Ramaekers, L.; Remans, R.; Rao, I.M.; Blair, M.W.; Vanderleyden, J. Strategies for Improving Phosphorus Acquisition Efficiency of Crop Plants. Field Crops Res. 2010, 117, 169–176. [Google Scholar] [CrossRef]
- Pant, B.; Erban, A.; Huhman, D.; Kopka, J.; Scheible, W. Identification of Primary and Secondary Metabolites with Phosphorus Status-dependent Abundance in A Rabidopsis, and of the Transcription Factor PHR 1 as a Major Regulator of Metabolic Changes during Phosphorus Limitation. Plant Cell Environ. 2015, 38, 172–187. [Google Scholar] [CrossRef]
- Pandey, R.; Meena, S.K.; Krishnapriya, V.; Ahmad, A.; Kishora, N. Root Carboxylate Exudation Capacity under Phosphorus Stress Does Not Improve Grain Yield in Green Gram. Plant Cell Rep. 2014, 33, 919–928. [Google Scholar] [CrossRef]
- Jakkeral, S.A.; Kajjidoni, S.T.; Koti, R. v Genotypic Variation for Root Traits to Phosphorus Deficiency in Blackgram (Vigna mungo L. Hepper). Karnataka J. Agric. Sci. 2009, 22, 946–950. [Google Scholar]
- Chen, Y.L.; Dunbabin, V.M.; Diggle, A.J.; Siddique, K.H.M.; Rengel, Z. Phosphorus Starvation Boosts Carboxylate Secretion in P-Deficient Genotypes of Lupinus Angustifolius with Contrasting Root Structure. Crop Pasture Sci. 2013, 64, 588–599. [Google Scholar] [CrossRef]
- Sarker, B.C.; Karmoker, J.L. Effects of Phosphorus Deficiency on the Root Growth of Lentil Seedlings Grown in Rhizobox. Bangladesh J. Bot. 2009, 38, 215–218. [Google Scholar] [CrossRef]
- He, J.; Jin, Y.; Du, Y.-L.; Wang, T.; Turner, N.C.; Yang, R.-P.; Siddique, K.H.M.; Li, F.-M. Genotypic Variation in Yield, Yield Components, Root Morphology and Architecture, in Soybean in Relation to Water and Phosphorus Supply. Front. Plant Sci. 2017, 8, 1499. [Google Scholar] [CrossRef]
- Borch, K.; Bouma, T.J.; Lynch, J.P.; Brown, K.M. Ethylene: A Regulator of Root Architectural Responses to Soil Phosphorus Availability. Plant Cell Environ. 1999, 22, 425–431. [Google Scholar] [CrossRef]
- Vejchasarn, P.; Lynch, J.P.; Brown, K.M. Genetic Variability in Phosphorus Responses of Rice Root Phenotypes. Rice 2016, 9, 29. [Google Scholar] [CrossRef]
- Shen, Q.; Wen, Z.; Dong, Y.; Li, H.; Miao, Y.; Shen, J. The Responses of Root Morphology and Phosphorus-Mobilizing Exudations in Wheat to Increasing Shoot Phosphorus Concentration. AoB Plants 2018, 10, ply054. [Google Scholar] [CrossRef] [PubMed]
- Fageria, N.K.; Moreira, A.; dos Santos, A.B. Phosphorus Uptake and Use Efficiency in Field Crops. J. Plant Nutr. 2013, 36, 2013–2022. [Google Scholar] [CrossRef]
- Thudi, M.; Chen, Y.; Pang, J.; Kalavikatte, D.; Bajaj, P.; Roorkiwal, M.; Chitikineni, A.; Ryan, M.H.; Lambers, H.; Siddique, K.H.M. Novel Genes and Genetic Loci Associated with Root Morphological Traits, Phosphorus-Acquisition Efficiency and Phosphorus-Use Efficiency in Chickpea. Front. Plant Sci. 2021, 12, 636973. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.R.P.; Das, S.; Dikshit, H.K.; Mishra, G.P.; Aski, M.; Meena, S.K.; Singh, A.; Pandey, R.; Singh, M.P.; Tripathi, K. Genome-Wide Association Analysis for Phosphorus Use Efficiency Traits in Mungbean (Vigna radiata L. Wilczek) Using Genotyping by Sequencing Approach. Front. Plant Sci. 2020, 11, 537766. [Google Scholar] [CrossRef] [PubMed]
- Kadirimangalam, S.R.; Jadhav, Y.; Nagamadhuri, K.V.; Putta, L.; Murugesan, T.; Variath, M.T.; Vemula, A.K.; Manohar, S.S.; Chaudhari, S.; Choudhary, S. Genetic Approaches for Assessment of Phosphorus Use Efficiency in Groundnut (Arachis hypogaea L.). Sci. Rep. 2022, 12, 21552. [Google Scholar] [CrossRef]
- Yan, X.; Liao, H.; Beebe, S.E.; Blair, M.W.; Lynch, J.P. QTL Mapping of Root Hair and Acid Exudation Traits and Their Relationship to Phosphorus Uptake in Common Bean. Plant Soil 2004, 265, 17–29. [Google Scholar] [CrossRef]
- Liang, Q.; Cheng, X.; Mei, M.; Yan, X.; Liao, H. QTL Analysis of Root Traits as Related to Phosphorus Efficiency in Soybean. Ann. Bot. 2010, 106, 223–234. [Google Scholar] [CrossRef]
- Su, J.-Y.; Zheng, Q.; Li, H.-W.; Li, B.; Jing, R.-L.; Tong, Y.-P.; Li, Z.-S. Detection of QTLs for Phosphorus Use Efficiency in Relation to Agronomic Performance of Wheat Grown under Phosphorus Sufficient and Limited Conditions. Plant Sci. 2009, 176, 824–836. [Google Scholar] [CrossRef]
- Yuan, Y.; Gao, M.; Zhang, M.; Zheng, H.; Zhou, X.; Guo, Y.; Zhao, Y.; Kong, F.; Li, S. QTL Mapping for Phosphorus Efficiency and Morphological Traits at Seedling and Maturity Stages in Wheat. Front. Plant Sci. 2017, 8, 614. [Google Scholar] [CrossRef]
- Luo, N.; Li, X.; Chen, A.Y.; Zhang, L.J.; Zhao, H.M.; Xiang, L.; Cai, Q.Y.; Mo, C.H.; Wong, M.H.; Li, H. Does Arbuscular Mycorrhizal Fungus Affect Cadmium Uptake and Chemical Forms in Rice at Different Growth Stages? Sci. Total Environ. 2017, 599, 1564–1572. [Google Scholar] [CrossRef]
- Mahender, A.; Anandan, A.; Pradhan, S.K.; Singh, O.N. Traits-Related QTLs and Genes and Their Potential Applications in Rice Improvement under Low Phosphorus Condition. Arch. Agron. Soil Sci. 2018, 64, 449–464. [Google Scholar] [CrossRef]
- Zhu, J.; Kaeppler, S.M.; Lynch, J.P. Mapping of QTLs for Lateral Root Branching and Length in Maize (Zea mays L.) under Differential Phosphorus Supply. Theoretical and Applied Genetics 2005, 111, 688–695. [Google Scholar] [CrossRef]
- Chen, J.; Xu, L.; Cai, Y.; Xu, J. QTL Mapping of Phosphorus Efficiency and Relative Biologic Characteristics in Maize (Zea mays L.) at Two Sites. Plant Soil 2008, 313, 251–266. [Google Scholar] [CrossRef]
- Sivasakthi, K.; Tharanya, M.; Kholová, J.; Wangari Muriuki, R.; Thirunalasundari, T.; Vadez, V. Chickpea Genotypes Contrasting for Vigor and Canopy Conductance Also Differ in Their Dependence on Different Water Transport Pathways. Front. Plant Sci. 2017, 8, 1663. [Google Scholar] [CrossRef]
- Reddy, V.R.P.; Aski, M.S.; Mishra, G.P.; Dikshit, H.K.; Singh, A.; Pandey, R.; Singh, M.P.; Ramtekey, V.; Rai, N.; Nair, R.M. Genetic Variation for Root Architectural Traits in Response to Phosphorus Deficiency in Mungbean at the Seedling Stage. PLoS ONE 2020, 15, e0221008. [Google Scholar] [CrossRef]
- Murphy, J.; Riley, J.P. A Modified Single Solution Method for the Determination of Phosphate in Natural Waters. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Irfan, M.; Abbas, M.; Shah, J.A.; Akram, M.A.; Depar, N.; Memon, M.Y. Categorization and Identification of Brassica Genotypes for Phosphorus Utilization Efficiency. Int. J. Agric. Biol. 2020, 23, 227–234. [Google Scholar]
- Neto, A.P.; Favarin, J.L.; Hammond, J.P.; Tezotto, T.; Couto, H.T.Z. Analysis of Phosphorus Use Efficiency Traits in Coffea Genotypes Reveals Coffea Arabica and Coffea Canephora Have Contrasting Phosphorus Uptake and Utilization Efficiencies. Front. Plant Sci. 2016, 7, 408. [Google Scholar] [CrossRef]
- Osborne, L.D.; Rengel, Z. Screening Cereals for Genotypic Variation in Efficiency of Phosphorus Uptake and Utilisation. Aust. J. Agric. Res. 2002, 53, 295–303. [Google Scholar] [CrossRef]
- Aziz, T.; Finnegan, P.M.; Lambers, H.; Jost, R. Organ-specific Phosphorus-allocation Patterns and Transcript Profiles Linked to Phosphorus Efficiency in Two Contrasting Wheat Genotypes. Plant Cell Environ. 2014, 37, 943–960. [Google Scholar] [CrossRef]
- Negarestani, M.; Tohidi-Nejad, E.; Khajoei-Nejad, G.; Nakhoda, B.; Mohammadi-Nejad, G. Comparison of Different Multivariate Statistical Methods for Screening the Drought Tolerant Genotypes of Pearl Millet (Pennisetum americanum L.) and Sorghum (Sorghum bicolor L.). Agronomy 2019, 9, 645. [Google Scholar] [CrossRef]
- Grzesiak, S.; Hordyńska, N.; Szczyrek, P.; Grzesiak, M.T.; Noga, A.; Szechyńska-Hebda, M. Variation among Wheat (Triticum easativum L.) Genotypes in Response to the Drought Stress: I–Selection Approaches. J. Plant Interact. 2019, 14, 30–44. [Google Scholar] [CrossRef]
- Gulles, A.A.; Bartolome, V.I.; Morantte, R.; Nora, L.A.; Relente, C.E.N.; Talay, D.T.; Caneda, A.A.; Ye, G. Randomization and Analysis of Data Using STAR [Statistical Tool for Agricultural Research]. Philipp. J. Crop Sci. 2014, 39, 137. [Google Scholar]
- Hallauer, A.R.; Carena, M.J.; de Miranda Filho, J.B. Quantitative Genetics in Maize Breeding; Springer Science & Business Media: New York, NY, USA, 2010; Volume 6, ISBN 1441907661. [Google Scholar]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Weih, M.; Hamnér, K.; Pourazari, F. Analyzing Plant Nutrient Uptake and Utilization Efficiencies: Comparison between Crops and Approaches. Plant Soil 2018, 430, 7–21. [Google Scholar] [CrossRef]
- Liu, Z.; Gao, K.; Shan, S.; Gu, R.; Wang, Z.; Craft, E.J.; Mi, G.; Yuan, L.; Chen, F. Comparative Analysis of Root Traits and the Associated QTLs for Maize Seedlings Grown in Paper Roll, Hydroponics and Vermiculite Culture System. Front. Plant Sci. 2017, 8, 436. [Google Scholar] [CrossRef]
- da Silva, D.A.; de Esteves, J.A.F.; Gonçalves, J.G.R.; Azevedo, C.V.G.; Ribeiro, T.; Chiorato, A.F.; Carbonell, S.A.M. Evaluation of Common Bean Genotypes for Phosphorus Use Efficiency in Eutrophic Oxisol. Bragantia 2016, 75, 152–163. [Google Scholar] [CrossRef]
- Wang, Q.; Yuan, Y.; Liao, Z.; Jiang, Y.; Wang, Q.; Zhang, L.; Gao, S.; Wu, F.; Li, M.; Xie, W. Genome-Wide Association Study of 13 Traits in Maize Seedlings under Low Phosphorus Stress. Plant Genome 2019, 12, 190039. [Google Scholar] [CrossRef]
- Akhtar, M.S.; Oki, Y.; Adachi, T.; Murata, Y.; Khan, M.H.R. Relative Phosphorus Utilization Efficiency, Growth Response, and Phosphorus Uptake Kinetics of Brassica Cultivars under a Phosphorus Stress Environment. Commun. Soil Sci. Plant Anal. 2007, 38, 1061–1085. [Google Scholar] [CrossRef]
- Wissuwa, M.; Kondo, K.; Fukuda, T.; Mori, A.; Rose, M.T.; Pariasca-Tanaka, J.; Kretzschmar, T.; Haefele, S.M.; Rose, T.J. Unmasking Novel Loci for Internal Phosphorus Utilization Efficiency in Rice Germplasm through Genome-Wide Association Analysis. PLoS ONE 2015, 10, e0124215. [Google Scholar] [CrossRef]
- Baligar, V.C. Nutrient Use Efficiency in Acid Soils: Nutrient Management and Plant Use Efficiency. In Plant-Soil Interactions at Low pH; Brazilian Soil Science Society: Viçosa, Brazil, 1997; pp. 75–95. [Google Scholar]
- Kim, H.-J.; Li, X. Effects of Phosphorus on Shoot and Root Growth, Partitioning, and Phosphorus Utilization Efficiency in Lantana. HortScience 2016, 51, 1001–1009. [Google Scholar] [CrossRef]
- Ramtekey, V.; Bansal, R.; Aski, M.S.; Kothari, D.; Singh, A.; Pandey, R.; Tripathi, K.; Mishra, G.P.; Kumar, S.; Dikshit, H.K. Genetic Variation for Traits Related to Phosphorus Use Efficiency in Lens Species at the Seedling Stage. Plants 2021, 10, 2711. [Google Scholar] [CrossRef]
- Faez, A.; Adam, P.; David, J. Root Architecture and Genetic Variations Associated with Phosphorus Uptake in Rice. Int. J. Appl. Agric. Sci. 2015, 1, 1–10. [Google Scholar] [CrossRef]
- Li, P.; Chen, F.; Cai, H.; Liu, J.; Pan, Q.; Liu, Z.; Gu, R.; Mi, G.; Zhang, F.; Yuan, L. A Genetic Relationship between Nitrogen Use Efficiency and Seedling Root Traits in Maize as Revealed by QTL Analysis. J. Exp. Bot. 2015, 66, 3175–3188. [Google Scholar] [CrossRef]
- Pang, J.; Bansal, R.; Zhao, H.; Bohuon, E.; Lambers, H.; Ryan, M.H.; Ranathunge, K.; Siddique, K.H.M. The Carboxylate-releasing Phosphorus-mobilizing Strategy Can Be Proxied by Foliar Manganese Concentration in a Large Set of Chickpea Germplasm under Low Phosphorus Supply. New Phytol. 2018, 219, 518–529. [Google Scholar] [CrossRef]
- Blum, A.; Brumbarova, T.; Bauer, P.; Ivanov, R. Hormone Influence on the Spatial Regulation of IRT1 Expression in Iron-Deficient Arabidopsis Thaliana Roots. Plant Signal Behav. 2014, 9, e28787. [Google Scholar] [CrossRef]
- Gill, M.A.; Sabir, M.; Ashraf, S.; Rahmatullah, A.T.; Aziz, T. Effect of P-Stress on Growth, Phosphorus Uptake and Utilization Efficiency of Different Cotton Cultivars. Pak. J. Agric. Sci. 2005, 42, 42–47. [Google Scholar]
- Thuynsma, R.; Kleinert, A.; Kossmann, J.; Valentine, A.J.; Hills, P.N. The Effects of Limiting Phosphate on Photosynthesis and Growth of Lotus Japonicus. S. Afr. J. Bot. 2016, 104, 244–248. [Google Scholar] [CrossRef]
- Singh, S.K.; Badgujar, G.B.; Reddy, V.R.; Fleisher, D.H.; Timlin, D.J. Effect of Phosphorus Nutrition on Growth and Physiology of Cotton under Ambient and Elevated Carbon Dioxide. J. Agron. Crop Sci. 2013, 199, 436–448. [Google Scholar] [CrossRef]
- Gill, H.S.; Singh, A.; Sethi, S.K.; Behl, R.K. Phosphorus Uptake and Use Efficiency in Different Varieties of Bread Wheat (Triticum aestivum L.). Arch. Agron. Soil Sci. 2004, 50, 563–572. [Google Scholar] [CrossRef]
- Kosar, H.S.; Gill, M.A.; Aziz, T.; Tahir, M.A. Relative Phosphorus Utilization Efficiency of Wheat Genotypes in Hydroponics. Pak. J. Agric. Sci. 2003, 40, 28–32. [Google Scholar]
- Gunes, A.; Inal, A.; Alpaslan, M.; Çakmak, İ. Genotypic Variation in Phosphorus Efficiency between Wheat Cultivars Grown under Greenhouse and Field Conditions. Soil Sci. Plant Nutr. 2006, 52, 470–478. [Google Scholar] [CrossRef]
- Bilal, H.M.; Aziz, T.; Maqsood, M.A.; Farooq, M.; Yan, G. Categorization of Wheat Genotypes for Phosphorus Efficiency. PLoS ONE 2018, 13, e0205471. [Google Scholar] [CrossRef] [PubMed]
- Aziz, T.; Rahmatullah; Maqsood, M.A.; Sabir, M.; Kanwal, S. Categorization of Brassica Cultivars for Phosphorus Acquisition from Phosphate Rock on Basis of Growth and Ionic Parameters. J. Plant Nutr. 2011, 34, 522–533. [Google Scholar] [CrossRef]
- Manske, G.G.B.; Ortiz-Monasterio, J.I.; van Ginkel, M.; Gonzalez, R.M.; Rajaram, S.; Molina, E.; Vlek, P.L.G. Traits Associated with Improved P-Uptake Efficiency in CIMMYT’s Semidwarf Spring Bread Wheat Grown on an Acid Andisol in Mexico. Plant Soil 2000, 221, 189–204. [Google Scholar] [CrossRef]
- Hammond, J.P.; White, P.J. Sucrose Transport in the Phloem: Integrating Root Responses to Phosphorus Starvation. J. Exp. Bot. 2008, 59, 93–109. [Google Scholar] [CrossRef]
- Fageria, N.K.; Baligar, V.C. Screening Crop Genotypes for Mineral Stresses; Workshop on Adaption of Plants to Soil Stresses; University of Nebrasca: Lincoln, NE, USA, 1993. [Google Scholar]
- Abbas, M.; Irfan, M.; Shah, J.A.; Memon, M.Y. AJAB. Asian J. Agric. Biol. 2018, 6, 35–45. [Google Scholar]
- Abbas, M.; Aslam, M.; Shah, J.A.; Depar, N.; Memon, M.Y. Relative Growth Response of Hydroponically Grown Wheat Genotypes to Deficient and Adequate Phosphorus Levels. Pak. J. Agric. Agric. Eng. Vet. Sci. 2016, 32, 169–181. [Google Scholar]
- Thiry, A.A.; Chavez Dulanto, P.N.; Reynolds, M.P.; Davies, W.J. How Can We Improve Crop Genotypes to Increase Stress Resilience and Productivity in a Future Climate? A New Crop Screening Method Based on Productivity and Resistance to Abiotic Stress. J. Exp. Bot. 2016, 67, 5593–5603. [Google Scholar] [CrossRef]
- Sareen, S.; Tyagi, B.S.; Tiwari, V.; Sharma, I. Response Estimation of Wheat Synthetic Lines to Terminal Heat Stress Using Stress Indices. J. Agric. Sci. 2012, 4, 97. [Google Scholar] [CrossRef]
- Khodarahmpour, Z.; Choukan, R.; Bihamta, M.R.; MAJIDI, H.E. Determination of the Best Heat Stress Tolerance Indices in Maize (Zea mays L.) Inbred Lines and Hybrids under Khuzestan Province Conditions. J. Agric. Sci. Technol. 2011, 13, 111–121. [Google Scholar]
- Mohammadi, M.; Karimizadeh, R.; Abdipour, M. Evaluation of Drought Tolerance in Bread Wheat Genotypes under Dryland and Supplemental Irrigation Conditions. Aust. J. Crop Sci. 2011, 5, 487–493. [Google Scholar]
- Khayatnezhad, M.; Zaeifizadeh, M.; Gholamin, R. Investigation and Selection Index for Drought Stress. Aust. J. Basic Appl. Sci. 2010, 4, 4815–4822. [Google Scholar]

| Traits | Optimum Phosphorus | Low Phosphorus | % Reduction of the Mean | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Max. | Min. | Mean | CV(%) | SE | Max. | Min. | Mean | CV(%) | SE | ||
| PRL | 55.70 | 10.1 | 30.47 | 9.74 | 0.25 | 60.20 | 10.10 | 32.88 | 9.02 | 0.31 | 7.91 |
| TRL | 1857.58 | 53.0644 | 707.84 | 11.28 | 12.54 | 2071.42 | 39.66 | 634.08 | 12.16 | 11.31 | −10.42 |
| TSA | 249.42 | 9.9568 | 73.59 | 12.00 | 1.44 | 234.52 | 6.91 | 66.85 | 12.59 | 1.27 | −9.16 |
| ARD | 0.69 | 0.2567 | 0.35 | 6.87 | 0.00 | 0.59 | 0.24 | 0.37 | 6.34 | 0.00 | 5.71 |
| TRV | 2.89 | 0.05 | 0.67 | 11.28 | 0.01 | 2.90 | 0.05 | 0.74 | 11.07 | 0.01 | 10.45 |
| TRT | 2495.00 | 65 | 765.06 | 11.53 | 12.35 | 2986.00 | 19.00 | 685.46 | 12.09 | 11.20 | −10.40 |
| RF | 9790.00 | 78 | 2474.44 | 9.52 | 39.69 | 8724.00 | 122.00 | 2005.40 | 10.99 | 36.53 | −18.96 |
| SDW | 1.10 | 0.074 | 0.25 | 8.41 | 0.01 | 0.55 | 0.05 | 0.17 | 8.47 | 0.00 | −32.00 |
| RDW | 0.33 | 0.005 | 0.048 | 10.01 | 0.00 | 0.23 | 0.01 | 0.048 | 11.91 | 0.00 | 0.00 |
| TDW | 1.36 | 0.098 | 0.30 | 7.69 | 0.01 | 0.69 | 0.08 | 0.21 | 7.59 | 0.00 | −30.00 |
| RSR | 1.18 | 0.024145 | 0.22 | 12.10 | 0.00 | 1.05 | 0.04 | 0.33 | 11.55 | 0.01 | 50.00 |
| % P | 12.96 | 0.339 | 5.74 | 4.83 | 0.08 | 4.95 | 0.03 | 1.60 | 8.49 | 0.03 | −72.13 |
| PUpE | 4.75 | 0.067689 | 1.45 | 9.94 | 0.02 | 1.74 | 0.00 | 0.31 | 11.65 | 0.01 | −78.62 |
| PUtiE | 2.95 | 0.07716 | 0.28 | 7.14 | 0.01 | 31.25 | 0.20 | 1.41 | 13.63 | 0.08 | 403.57 |
| Trait | Optimum Phosphorus | Low Phosphorus | Combined Analysis | Hcom | |||
|---|---|---|---|---|---|---|---|
| Genotypes | H | Genotypes | H | Genotypes | Genotypes × P levels | ||
| PRL | 173.32 ** | 0.86 | 259.32 ** | 0.90 | 326.23 ** | 106.41 ** | 0.67 |
| TRL | 450,566.87 ** | 0.96 | 360,584.61 ** | 0.95 | 740,774.95 ** | 70,375.99 ** | 0.90 |
| TSA | 5960.86 ** | 0.96 | 4611.21 ** | 0.96 | 9708.32 ** | 863.76 ** | 0.91 |
| ARD | 0.0076 ns | 0.80 | 0.0089 ** | 0.85 | 0.0137 ** | 0.0027 ** | 0.80 |
| TRV | 0.4106 ** | 0.96 | 0.356 ** | 0.94 | 0.6040 ** | 0.1626 ** | 0.73 |
| TRT | 434,595.05 ** | 0.95 | 347,816.83 ** | 0.94 | 613,095.14 ** | 169,316.74 ** | 0.72 |
| RF | 4,517,922.02 ** | 0.96 | 3,691,107.09 ** | 0.96 | 5,761,460.68 ** | 2,447,568.43 ** | 0.58 |
| SDW | 0.0966 ** | 0.99 | 0.0249 ** | 0.98 | 0.0861 ** | 0.0354 ** | 0.59 |
| RDW | 0.0040 ** | 1.00 | 0.0027 ** | 1.00 | 0.0051 ** | 0.0017 ** | 0.67 |
| TDW | 0.1301 ** | 0.99 | 0.0328 ** | 0.97 | 0.1190 ** | 0.0440 ** | 0.63 |
| RSR | 0.0466 ** | 0.96 | 0.0756 ** | 0.94 | 0.0761 ** | 0.0462 ** | 0.39 |
| % P | 17.11 ** | 0.99 | 2.2658 ** | 0.98 | 14.6616 ** | 4.7055 ** | 0.68 |
| PUpE | 1.3064 ** | 0.95 | 0.1116 ** | 0.97 | 0.9491 ** | 0.4688 ** | 0.51 |
| PUtiE | 0.3141 ** | 1.00 | 17.1371 ** | 0.99 | 10.8076 ** | 6.6428 ** | 0.39 |
| PRL | TRL | TSA | ARD | TRV | TRT | RF | SDW | RDW | TDW | RSR | P | PupE | PutiE | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PRL | 1.00 | 0.58 ** | 0.49 ** | −0.40 ** | 0.30 ** | 0.63 ** | 0.14 ** | −0.13 ** | 0.03 ns | −0.10 ** | 0.17 ** | 0.47 ** | 0.33 ** | −0.34 ** |
| TRL | 0.74 ** | 1.00 | 0.95 ** | −0.33 ** | 0.62 ** | 0.86 ** | 0.46 ** | 0.26 ** | 0.42 ** | 0.30 ** | 0.16 ** | 0.34 ** | 0.39 ** | −0.11 ** |
| TSA | 0.74 ** | 0.97 ** | 1.00 | −0.15 ** | 0.70 ** | 0.79 ** | 0.50 ** | 0.37 ** | 0.53 ** | 0.41 ** | 0.13 ** | 0.23 ** | 0.38 ** | −0.04 ns |
| ARD | −0.33 | −0.43 | −0.35 | 1.00 | 0.22 ** | −0.37 ** | 0.05 ns | 0.42 ** | 0.36 ** | 0.43 ** | −0.05 ns | −0.53 ** | −0.07 ns | 0.27 ** |
| TRV | 0.54 ** | 0.59 ** | 0.62 ** | 0.12 ** | 1.00 | 0.57 ** | 0.63 ** | 0.67 ** | 0.70 ** | 0.70 ** | 0.05 ns | −0.12 ** | 0.39 ** | 0.15 ** |
| TRT | 0.56 ** | 0.71 ** | 0.68 ** | −0.28 | 0.59 ** | 1.00 | 0.44 ** | 0.16 ** | 0.28 ** | 0.19 ** | 0.12 ** | 0.39 ** | 0.44 ** | −0.21 ** |
| RF | 0.28 ** | 0.31 ** | 0.29 ** | 0.15 ** | 0.53 ** | 0.37 ** | 1.00 | 0.52 ** | 0.41 ** | 0.52 ** | −0.17 | −0.13 | 0.26 ** | 0.14 ** |
| SDW | −0.17 | −0.20 | −0.18 | 0.67 ** | 0.34 ** | 0.02 ** | 0.45 ** | 1.00 | 0.75 ** | 0.99 ** | −0.25 | −0.52 | 0.22 ** | 0.40 ** |
| RDW | 0.33 ** | 0.41 ** | 0.50 ** | 0.21 ** | 0.65 ** | 0.38 ** | 0.34 ** | 0.32 ** | 1.00 | 0.82 ** | 0.34 ** | −0.40 | 0.13 ** | 0.39 ** |
| TDW | −0.05 | −0.05 | −0.01 | 0.64 ** | 0.48 ** | 0.12 ** | 0.49 ** | 0.96 ** | 0.57 ** | 1.00 | −0.15 | −0.52 | 0.21 ** | 0.41 ** |
| RSR | 0.42 ** | 0.47 ** | 0.53 ** | −0.24 ** | 0.36 ** | 0.30 ** | −0.03 ns | −0.39 | 0.65 ** | −0.15 | 1.00 | 0.08 | −0.11 | 0.06 ** |
| % P | 0.21 ** | 0.22 ** | 0.21 ** | −0.18 | 0.05 ** | 0.13 ** | −0.20 | −0.30 | −0.09 | −0.29 | 0.21 ** | 1.00 | 0.57 ** | −0.77 |
| PupE | 0.06 ** | 0.05 ** | 0.07 ** | 0.30 ** | 0.29 ** | 0.08 ** | 0.07 ** | 0.37 ** | 0.26 ** | 0.40 ** | 0.07 ** | 0.68 | 1.00 | −0.49 |
| PutiE | −0.05 ns | 0.01 ns | 0.03 ns | −0.04 ns | 0.03 ns | 0.003 ns | 0.11 ns | 0.06 ns | 0.15 ** | 0.10 ** | 0.02 ns | −0.60 ns | −0.49 ns | 1.00 |
| Genotypes | TDW (g) | Genotypes | SDW (g) | Genotypes | RDW (g) | Genotypes | PupE (g plant−1) | Genotypes | PutiE (g DW/mg P) |
|---|---|---|---|---|---|---|---|---|---|
| IC 343440 | 0.669 | V1003959BG | 0.516 | V1002190BG | 0.220 | IC 251950 | 18.21 | IC 251950 | 167.0 |
| V1002190BG | 0.652 | V1001162AG | 0.505 | IC 350347 | 0.210 | IC 585931 | 15.07 | IC 585931 | 140.7 |
| V1001162AG | 0.598 | V1001066BG | 0.497 | IC 343440 | 0.207 | IC 259502 | 13.05 | V1002532AG | 137.7 |
| V1003959BG | 0.544 | V1000470AG | 0.476 | IC 343436 | 0.170 | V1002532AG | 12.17 | IC 259502 | 114.6 |
| V1002872BG | 0.537 | V1001698BG | 0.470 | IC 146239 | 0.167 | IC 371653 | 12.15 | V1001400AG | 97.4 |
| V1001066BG | 0.535 | V1002872BG | 0.468 | KPS 1546 | 0.160 | IC 331615 | 11.30 | IC 371653 | 97.0 |
| V1002672AG | 0.534 | V1000380AG | 0.465 | SM 18-99 | 0.137 | V1001400AG | 11.06 | IC 331615 | 88.1 |
| V1001698BG | 0.524 | V1002672AG | 0.465 | IC 273244 | 0.120 | V1000532BG | 9.45 | V1000532BG | 75.0 |
| V1000470AG | 0.522 | IC 343440 | 0.462 | V1002195AG | 0.119 | V10002647AG | 9.07 | IC 583666 | 66.5 |
| IC 273244 | 0.502 | V1002190BG | 0.432 | IPM 02-3 | 0.113 | V1004734AG | 9.01 | IC 583665 | 65.2 |
| S.No. | Genotype | Species | Low Phosphorus | Optimum Phosphorus | Total Score /Out of 30 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RDW | SDW | RSR | TPU | PUtE | Total Score /Out of 15 | RDW | SDW | RSR | TPU | PUtE | Total Score /Out of 15 | ||||
| 1 | IC 251950 | V.radiata var. bourneae | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 2 | IC 277000 | V.radiata var. bourneae | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 3 | IC 25950 | V.radiata var. bourneae | M | M | M | E | E | 12 | M | M | E | E | E | 13 | 25 |
| 4 | IC 305153 | V.radiata var. bourneae | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 5 | IC 202643 | Vigna radiata var. sublobata | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 6 | IC 259502 | Vigna radiata var. sublobata | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 7 | IC 371653 | Vigna radiata var. sublobata | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 8 | IC 248206 | Vigna mungo var. silvestris | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 9 | IC 585931 | Vigna mungo var. silvestris | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 10 | IC 251419 | Vigna radiata var. setulosa | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 11 | IC 583670 | Vigna dalzelliana | M | M | M | E | E | 12 | M | M | E | E | E | 12 | 24 |
| 12 | IC 583662 | Vigna dalzelliana | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 13 | IC 583689 | Vigna dalzelliana | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 14 | IC 331450 | Vigna hainiana | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 15 | IC 331615 | Vigna hainiana | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 16 | IC 331452 | Vigna hainiana | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 17 | IC 583665 | Vigna trinervia var. bourneae | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 18 | IC 583664 | Vigna trinervia var. bourneae | M | M | M | E | E | 12 | M | M | E | E | E | 13 | 25 |
| 19 | V1001339AG | Vigna radiata | M | E | M | E | M | 12 | M | E | M | E | M | 12 | 24 |
| 20 | V10002647AG | Vigna radiata | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 21 | V1000532BG | Vigna radiata | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 22 | V1001400AG | Vigna radiata | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 23 | V1004734AG | Vigna radiata | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 24 | V1001806BG | Vigna radiata | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| 25 | V1002532AG | Vigna radiata | M | M | M | E | E | 12 | M | M | M | E | E | 12 | 24 |
| S.No. | Genotype | Species | SSI | MPI | GMPI | HMI | STI | TI | SI | STC |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IC 277060 | V. minima | 5.612886 | 0.307 | 0.254743 | 0.211381 | 0.771628 | 0.34 | 0.283624 | 7.78 |
| 2 | IC 697141 | Vigna membranaceae | 4.956035 | 0.49 | 0.41637 | 0.353804 | 2.061402 | 0.52 | 0.309577 | 9.10 |
| 3 | IC 343440 | Vigna unguiculata | 0.322366 | 0.717167 | 0.715525 | 0.713887 | 6.087702 | 0.10 | 0.873313 | 9.53 |
| 4 | KPS 1546 | Vigna radiata | 5.026549 | 0.802667 | 0.680292 | 0.576575 | 5.502946 | 0.85 | 0.306565 | 13.75 |
| 5 | Pusa 0831 | Vigna radiata | 4.53159 | 0.686667 | 0.592734 | 0.51165 | 4.177566 | 0.69 | 0.329032 | 11.52 |
| 6 | Pusa 0871 | Vigna radiata | 4.213564 | 0.5 | 0.436794 | 0.381578 | 2.268596 | 0.49 | 0.345291 | 8.63 |
| 7 | Pusa 1371 | Vigna radiata | 7.852941 | 0.627167 | 0.482636 | 0.371413 | 2.769771 | 0.80 | 0.220564 | 13.13 |
| 8 | Pusa 1431 | Vigna radiata | 11.6595 | 0.748833 | 0.516533 | 0.356296 | 3.172493 | 1.08 | 0.160083 | 17.70 |
| 9 | Pusa 1641 | Vigna radiata | 5.574879 | 0.5185 | 0.430825 | 0.357975 | 2.207015 | 0.58 | 0.285006 | 9.95 |
| 10 | M 1319 | Vigna radiata | 2.684268 | 0.54 | 0.500255 | 0.463436 | 2.97569 | 0.41 | 0.452915 | 8.02 |
| 11 | M 209 | Vigna radiata | 5.303704 | 0.658 | 0.552087 | 0.463222 | 3.624257 | 0.72 | 0.295276 | 11.61 |
| 12 | V 6173 | Vigna radiata | 7.321839 | 0.511833 | 0.400663 | 0.313639 | 1.908812 | 0.64 | 0.232838 | 11.33 |
| 13 | EESM 18-163 | Vigna radiata | 14.08796 | 0.667167 | 0.433467 | 0.281629 | 2.234166 | 1.01 | 0.136248 | 18.85 |
| 14 | SM 18-93 | Vigna radiata | 6.697108 | 0.61 | 0.487499 | 0.389599 | 2.825869 | 0.73 | 0.249147 | 11.99 |
| 15 | SM 18-94 | Vigna radiata | 4.960876 | 0.500833 | 0.4255 | 0.361498 | 2.152794 | 0.53 | 0.309368 | 9.24 |
| 16 | SM 18-96 | Vigna radiata | 8.546296 | 0.467667 | 0.352212 | 0.26526 | 1.475069 | 0.62 | 0.206363 | 11.93 |
| 17 | SM 18-98 | Vigna radiata | 3.983539 | 0.512 | 0.451198 | 0.397617 | 2.42069 | 0.48 | 0.35809 | 8.61 |
| 18 | SM 18-99 | Vigna radiata | 5.00463 | 0.680333 | 0.577073 | 0.489486 | 3.95973 | 0.72 | 0.307495 | 11.74 |
| 19 | SM 18-101 | Vigna radiata | 2.535354 | 0.575833 | 0.536501 | 0.499855 | 3.422513 | 0.42 | 0.467091 | 8.46 |
| 20 | SM 18-102 | Vigna radiata | 6.229798 | 0.7045 | 0.572068 | 0.464531 | 3.891346 | 0.82 | 0.262922 | 12.95 |
| 21 | SM 18-106 | Vigna radiata | 4.666667 | 0.567167 | 0.487122 | 0.418374 | 2.821496 | 0.58 | 0.322581 | 9.86 |
| 22 | SM 18-9 | Vigna radiata | 4.746032 | 0.579 | 0.495823 | 0.424594 | 2.923187 | 0.60 | 0.318907 | 10.09 |
| 23 | SM 18-21 | Vigna radiata | 6.951311 | 0.760667 | 0.602758 | 0.477631 | 4.320069 | 0.93 | 0.242243 | 14.28 |
| 24 | IPM 02-3 | Vigna radiata | 2.840484 | 0.551833 | 0.508157 | 0.467937 | 3.070432 | 0.43 | 0.43894 | 8.31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kothari, D.; Pargaien, N.; Tewari, L.M.; Dikshit, H.K.; Mishra, G.P.; Aski, M.S.; Bansal, R.; Gupta, S.; Kumar, S.; Nair, R.M. Genetic Variation for Traits Related to Phosphorus Use Efficiency in Vigna Species. Agronomy 2023, 13, 305. https://doi.org/10.3390/agronomy13020305
Kothari D, Pargaien N, Tewari LM, Dikshit HK, Mishra GP, Aski MS, Bansal R, Gupta S, Kumar S, Nair RM. Genetic Variation for Traits Related to Phosphorus Use Efficiency in Vigna Species. Agronomy. 2023; 13(2):305. https://doi.org/10.3390/agronomy13020305
Chicago/Turabian StyleKothari, Deepali, Nirmala Pargaien, Lalit Mohan Tewari, Harsh Kumar Dikshit, Gyan Prakash Mishra, Muraleedhar S. Aski, Ruchi Bansal, Sanjeev Gupta, Shiv Kumar, and Ramakrishnan Madhavan Nair. 2023. "Genetic Variation for Traits Related to Phosphorus Use Efficiency in Vigna Species" Agronomy 13, no. 2: 305. https://doi.org/10.3390/agronomy13020305
APA StyleKothari, D., Pargaien, N., Tewari, L. M., Dikshit, H. K., Mishra, G. P., Aski, M. S., Bansal, R., Gupta, S., Kumar, S., & Nair, R. M. (2023). Genetic Variation for Traits Related to Phosphorus Use Efficiency in Vigna Species. Agronomy, 13(2), 305. https://doi.org/10.3390/agronomy13020305







