Modeling Callus Induction and Regeneration in Hypocotyl Explant of Fodder Pea (Pisum sativum var. arvense L.) Using Machine Learning Algorithm Method
Abstract
:1. Introduction
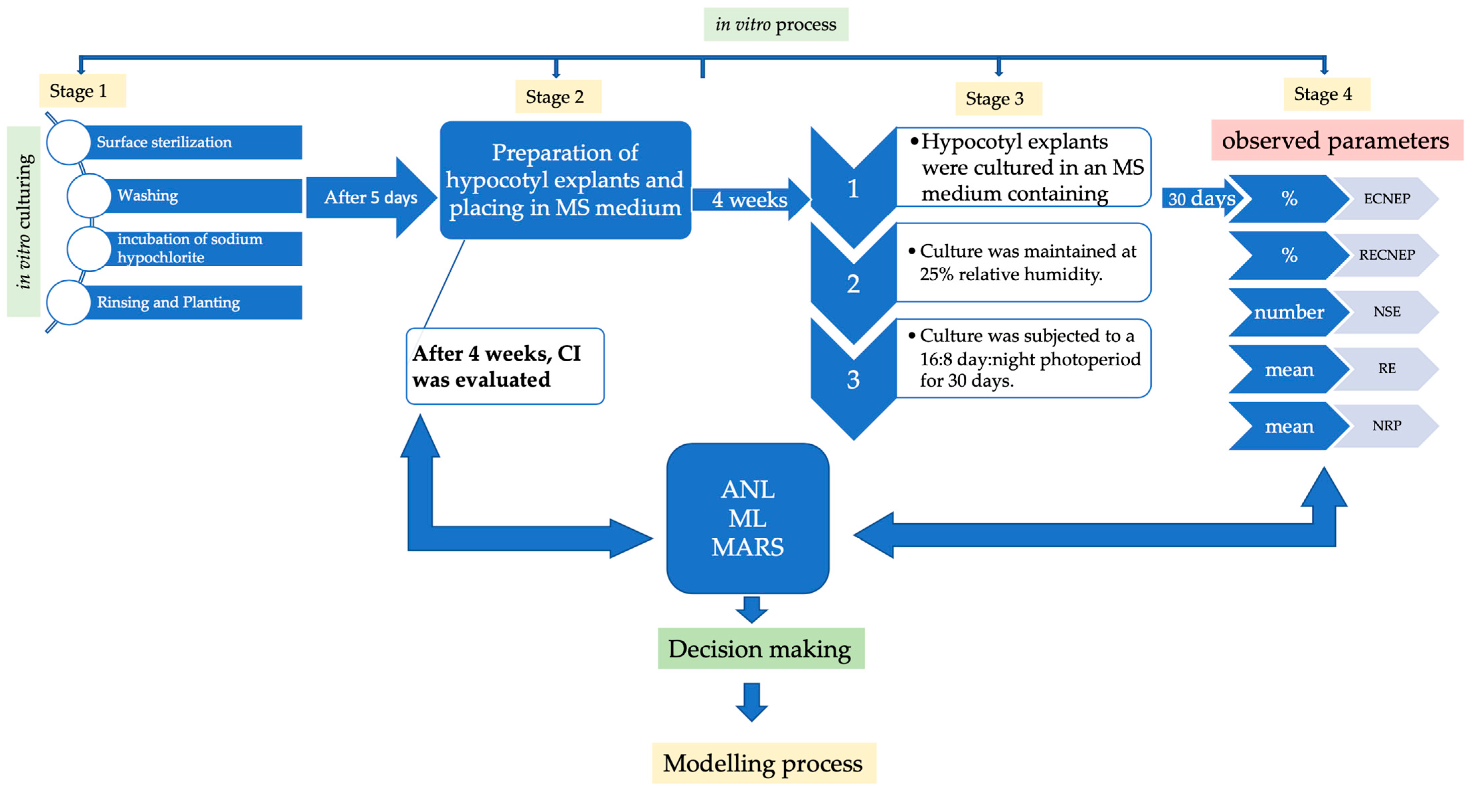
2. Materials and Methods
2.1. Plant Material, Callus Initiation, Formation of Embryogenic Calluses, and Plant Regeneration and Rooting Condition
2.2. Statistical Analysis
2.3. Modeling Using Machine Learning Algorithms
3. Results
3.1. In Vitro Parameters
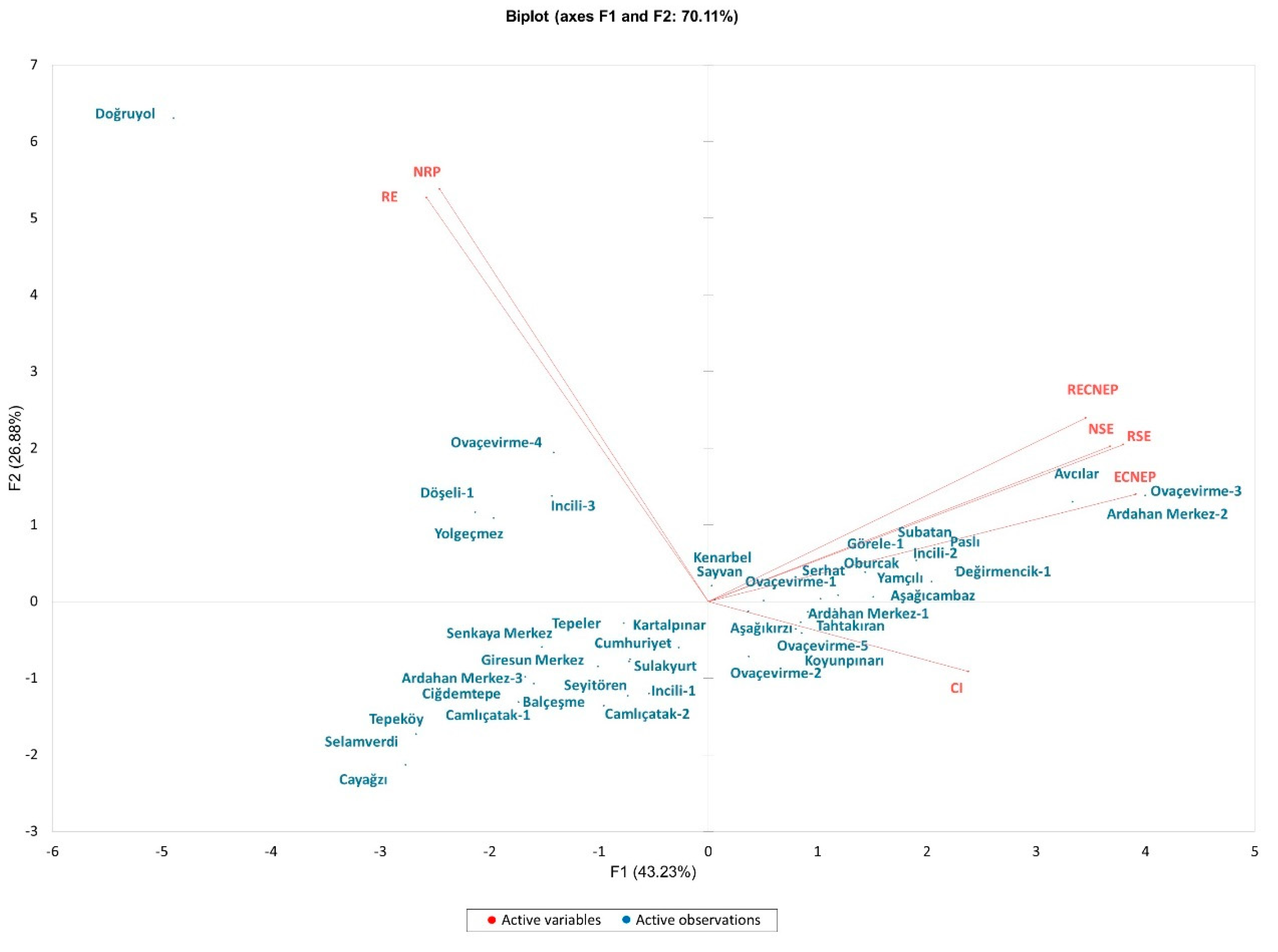
3.2. Principal Component Analysis
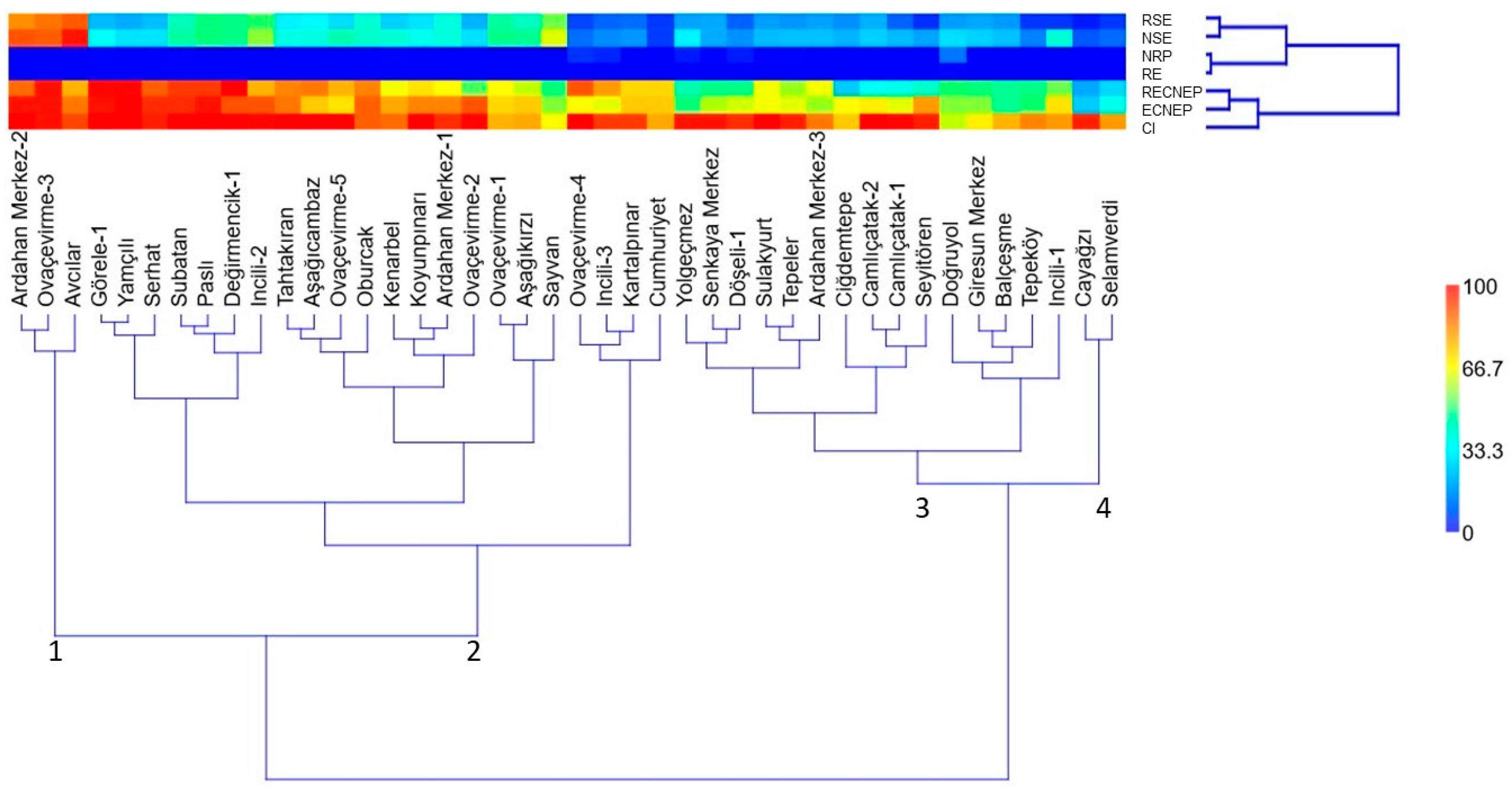
3.3. Cluster Analysis
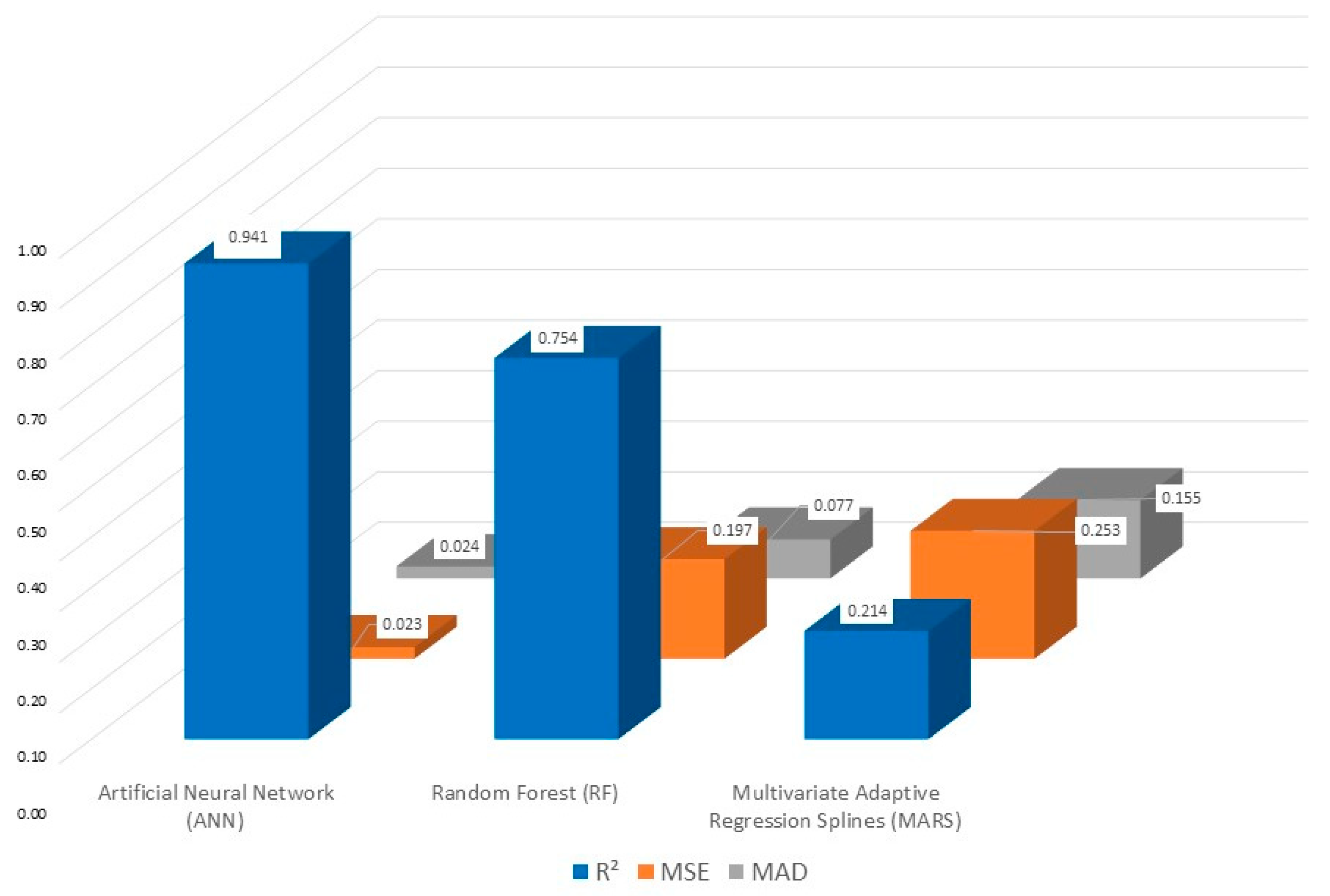
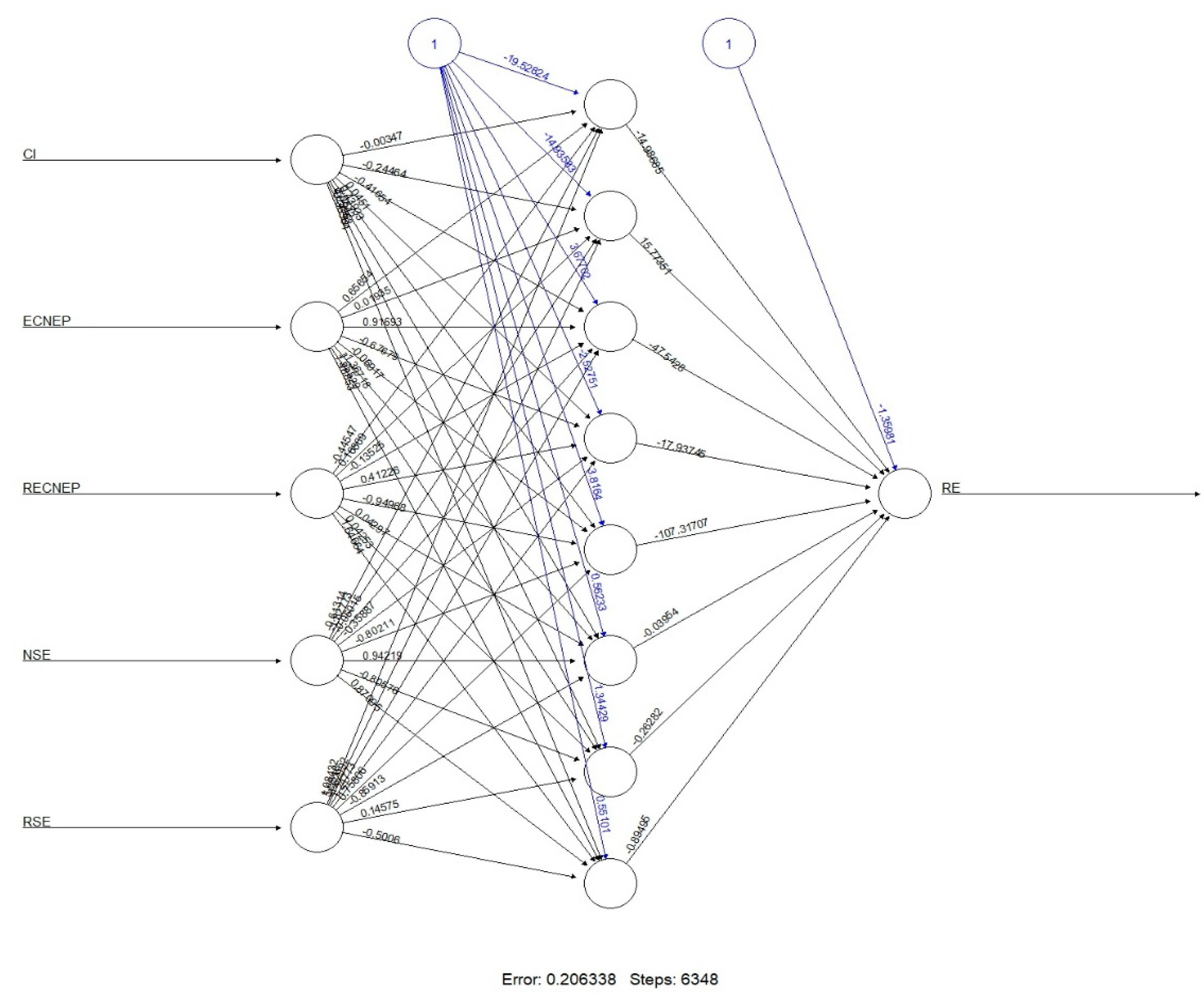
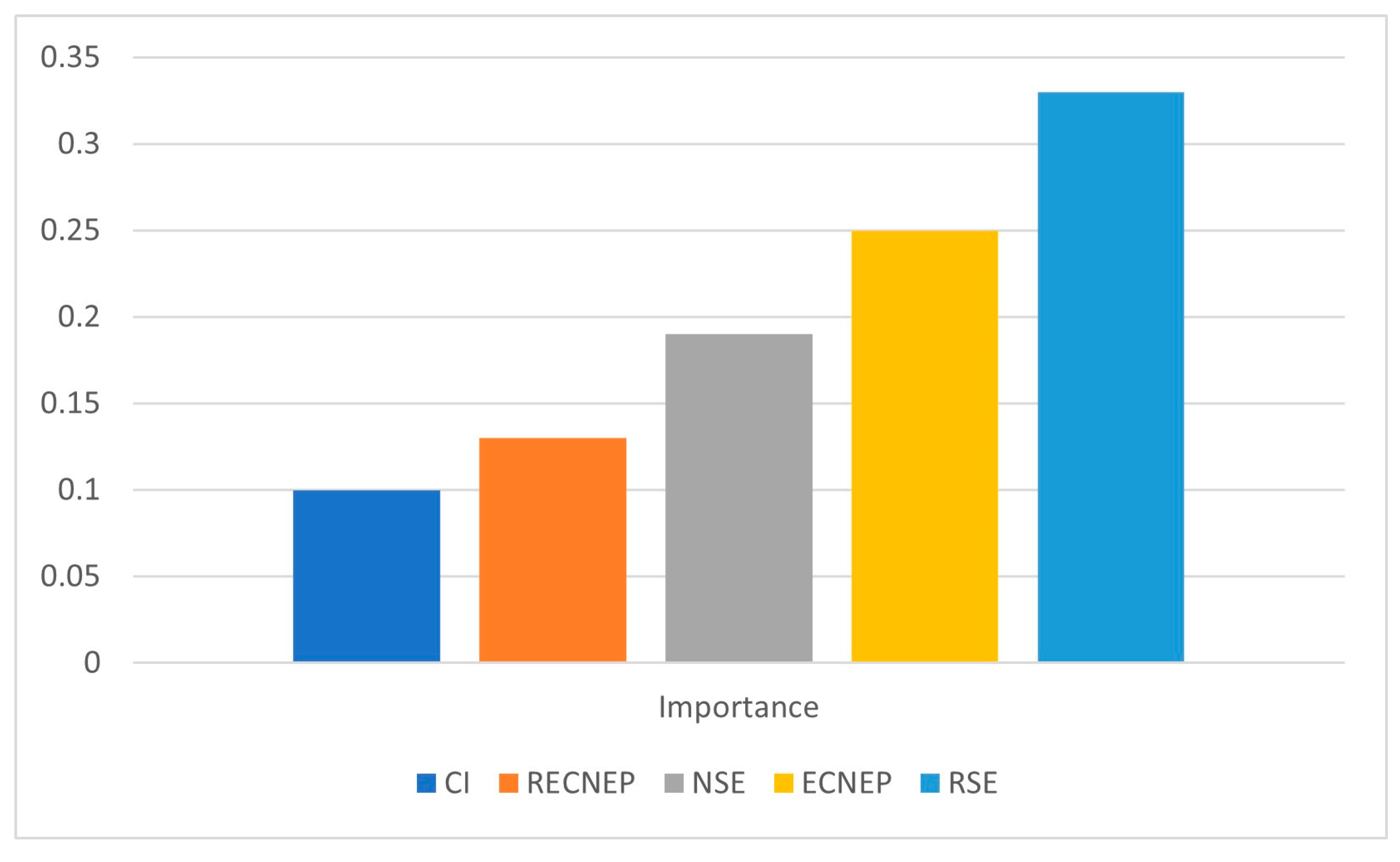
3.4. Machine Learning (ML) Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Vidyagina, E.O.; Kharchenko, N.N.; Shestibratov, K.A. Efficient cryopreservation of Populus tremula by in vitro-grown axillary buds and genetic stability of recovered plants. Plants 2021, 10, 77. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, K.; Huyop, F.Z. An easy and reliable method for establishment and maintenance of tissue cultures of Nicotiana tabacum cv TAPM 26. Int. J. Sci. Lett. 2020, 2, 62–71. [Google Scholar]
- Espinosa-Leal, C.A.; Puente-Garza, C.A.; García-Lara, S. In vitro plant tissue culture: Means for production of biologically active compounds. Planta 2018, 248, 1–18. [Google Scholar] [CrossRef]
- Altpeter, F.; Springer, N.M.; Bartley, L.E.; Blechl, A.E.; Brutnell, T.P.; Citovsky, V.; Conrad, L.J.; Gelvin, S.B.; Jackson, D.P.; Kausch, A.P. Advancing crop transformation in the era of genome editing. Plant Cell 2016, 28, 1510–1520. [Google Scholar] [CrossRef] [PubMed]
- Verdeil, J.-L.; Alemanno, L.; Niemenak, N.; Tranbarger, T.J. Pluripotent versus totipotent plant stem cells: Dependence versus autonomy? Trends Plant Sci. 2007, 12, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Efferth, T. Biotechnology applications of plant callus cultures. Engineering 2019, 5, 50–59. [Google Scholar] [CrossRef]
- Huang, H.; Wei, Y.; Zhai, Y.; Ouyang, K.; Chen, X.; Bai, L. High frequency regeneration of plants via callus-mediated organogenesis from cotyledon and hypocotyl cultures in a multipurpose tropical tree (Neolamarkia cadamba). Sci. Rep. 2020, 10, 4558. [Google Scholar] [CrossRef] [PubMed]
- Kaviani, B.; Hesar, A.A.; Kharabian-Masouleh, A. In vitro propagation of Matthiola incana (Brassicaceae)—An ornamental plant. Plant Omics 2011, 4, 435–440. [Google Scholar]
- Pramanik, D.; Shelake, R.M.; Park, J.; Kim, M.J.; Hwang, I.; Park, Y.; Kim, J.-Y. CRISPR/Cas9-mediated generation of pathogen-resistant tomato against tomato yellow leaf curl virus and powdery mildew. Int. J. Mol. Sci. 2021, 22, 1878. [Google Scholar] [CrossRef]
- Dalla Costa, L.; Malnoy, M.; Gribaudo, I. Breeding next generation tree fruits: Technical and legal challenges. Hortic. Res. 2017, 4, 17067. [Google Scholar] [CrossRef]
- Klap, C.; Yeshayahou, E.; Bolger, A.M.; Arazi, T.; Gupta, S.K.; Shabtai, S.; Usadel, B.; Salts, Y.; Barg, R. Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnol. J. 2017, 15, 634–647. [Google Scholar] [CrossRef] [PubMed]
- Palaz, E.; Ugur, R.; Yaman, M. Micropropagation Protocols of New Prunus Hybrids with Significant Rootstock Potential in Fruit Breeding and Cultivation. Erwerbs-Obstbau 2023, 65, 1359–1364. [Google Scholar] [CrossRef]
- Dinneny, J.R.; Benfey, P.N. Plant stem cell niches: Standing the test of time. Cell 2008, 132, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Trick, H.N. Recent advances in soybean transformation. Plant Tissue Cult. Biotechnol. 1997, 3, 9–26. [Google Scholar]
- Jain, S.M.; Gupta, P.K. Step-Wise Protocols for Somatic Embryogenesis of Important Woody Plants; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Niazian, M.; Niedbała, G. Machine learning for plant breeding and biotechnology. Agriculture 2020, 10, 436. [Google Scholar] [CrossRef]
- Farhadi, S.; Salehi, M.; Moieni, A.; Safaie, N.; Sabet, M.S. Modeling of paclitaxel biosynthesis elicitation in Corylus avellana cell culture using adaptive neuro-fuzzy inference system-genetic algorithm (ANFIS-GA) and multiple regression methods. PLoS ONE 2020, 15, e0237478. [Google Scholar] [CrossRef]
- Hesami, M.; Condori-Apfata, J.A.; Valderrama Valencia, M.; Mohammadi, M. Application of artificial neural network for modeling and studying in vitro genotype-independent shoot regeneration in wheat. Appl. Sci. 2020, 10, 5370. [Google Scholar] [CrossRef]
- Kirtis, A.; Aasim, M.; Katırcı, R. Application of artificial neural network and machine learning algorithms for modeling the in vitro regeneration of chickpea (Cicer arietinum L.). Plant Cell Tissue Organ Cult. PCTOC 2022, 150, 141–152. [Google Scholar] [CrossRef]
- Gökmen, F.; Uygur, V.; Sukuşu, E. Extreme Gradient Boosting Regression Model for Soil Available Boron. Eurasian Soil Sci. 2023, 56, 738–746. [Google Scholar] [CrossRef]
- Aasim, M.; Katırcı, R.; Akgur, O.; Yildirim, B.; Mustafa, Z.; Nadeem, M.A.; Baloch, F.S.; Karakoy, T.; Yılmaz, G. Machine learning (ML) algorithms and artificial neural network for optimizing in vitro germination and growth indices of industrial hemp (Cannabis sativa L.). Ind. Crops Prod. 2022, 181, 114801. [Google Scholar] [CrossRef]
- Aasim, M.; Katirci, R.; Baloch, F.S.; Mustafa, Z.; Bakhsh, A.; Nadeem, M.A.; Ali, S.A.; Hatipoğlu, R.; Çiftçi, V.; Habyarimana, E. Innovation in the breeding of common bean through a combined approach of in vitro regeneration and machine learning algorithms. Front. Genet. 2022, 13, 897696. [Google Scholar] [CrossRef]
- Hesami, M.; Jones, A.M.P. Application of artificial intelligence models and optimization algorithms in plant cell and tissue culture. Appl. Microbiol. Biotechnol. 2020, 104, 9449–9485. [Google Scholar] [CrossRef] [PubMed]
- Salehi, M.; Farhadi, S.; Moieni, A.; Safaie, N.; Ahmadi, H. Mathematical modeling of growth and paclitaxel biosynthesis in Corylus avellana cell culture responding to fungal elicitors using multilayer perceptron-genetic algorithm. Front. Plant Sci. 2020, 11, 1148. [Google Scholar] [CrossRef] [PubMed]
- Niazian, M.; Shariatpanahi, M.E.; Abdipour, M.; Oroojloo, M. Modeling callus induction and regeneration in an anther culture of tomato (Lycopersicon esculentum L.) using image processing and artificial neural network method. Protoplasma 2019, 256, 1317–1332. [Google Scholar] [CrossRef] [PubMed]
- Genze, N.; Bharti, R.; Grieb, M.; Schultheiss, S.J.; Grimm, D.G. Accurate machine learning-based germination detection, prediction, and quality assessment of three grain crops. Plant Methods 2020, 16, 157. [Google Scholar] [CrossRef] [PubMed]
- Haliloglu, K.; Turkoglu, A.; Tan, M.; Poczai, P. SSR-based molecular identification and population structure analysis for forage pea (Pisum sativum var. arvense L.) landraces. Genes 2022, 13, 1086. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Bencheikh, M.; Gallais, A. Somatic embryogenesis in pea (Pisum sativum L. and Pisum arvense L.): Diallel analysis and genetic control. Euphytica 1996, 90, 257–264. [Google Scholar] [CrossRef]
- Lazzeri, P.A.; Hildebrand, D.F.; Collins, G.B. Soybean somatic embryogenesis: Effects of hormones and culture manipulations. Plant Cell Tissue Organ Cult. PCTOC 1987, 10, 197–208. [Google Scholar] [CrossRef]
- Demirel, F.; Gurcan, K.; Akar, T. Clustering analysis of morphological and phenological data in einkorn and emmer wheats collected from Kastamonu region. Int. J. Sci. Technol. Res. 2019, 5, 25–36. [Google Scholar]
- Gago, J.; Martínez-Núñez, L.; Landín, M.; Gallego, P. Artificial neural networks as an alternative to the traditional statistical methodology in plant research. J. Plant Physiol. 2010, 167, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Demirel, F.; Eren, B.; Yilmaz, A.; Türkoğlu, A.; Haliloğlu, K.; Niedbała, G.; Bujak, H.; Jamshidi, B.; Pour-Aboughadareh, A.; Bocianowski, J. Prediction of Grain Yield in Wheat by CHAID and MARS Algorithms Analyses. Agronomy 2023, 13, 1438. [Google Scholar] [CrossRef]
- Eyduran, E.; Akin, M.; Eyduran, S. Application of Multivariate Adaptive Regression Splines through R Software; Nobel Academic Publishing: Ankara, Turkey, 2019. [Google Scholar]
- Eren, B.; Türkoğlu, A.; Haliloğlu, K.; Demirel, F.; Nowosad, K.; Özkan, G.; Niedbała, G.; Pour-Aboughadareh, A.; Bujak, H.; Bocianowski, J. Investigation of the Influence of Polyamines on Mature Embryo Culture and DNA Methylation of Wheat (Triticum aestivum L.) Using the Machine Learning Algorithm Method. Plants 2023, 12, 3261. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M.; Wing, J.; Weston, S.; Williams, A.; Keefer, C.; Engelhardt, A.; Cooper, T.; Mayer, Z.; Kenkel, B.; Benesty, M. Caret: Classification and Regression Training: R Package; CRAN: Vienna, Austria, 2019. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2010. [Google Scholar]
- Hussain, I.; Ali, S.; Liu, W.; Awais, M.; Li, J.; Liao, Y.; Zhu, M.; Fu, C.; Liu, D.; Wang, F. Identification of heterotic groups and patterns based on genotypic and phenotypic characteristics among rice accessions of diverse origins. Front. Genet. 2022, 13, 811124. [Google Scholar] [CrossRef] [PubMed]
- Carsono, N.; Yoshida, T. Variation in spikelet-related traits of rice plants regenerated from mature seed-derived callus culture. Plant Prod. Sci. 2007, 10, 86–90. [Google Scholar] [CrossRef]
- Haliloglu, K.; Bezirganoğlu, İ.; Aydin, M.; Tosun, M.; Şahin, E. Response of current winter wheat cultivars grown in Turkey to immature embryo culture. Atatürk Uni. J. Agri. Fac. 2013, 44, 21–25. [Google Scholar]
- Kuznetsova, O.; Ash, O.; Hartina, G.; Gostimskij, S. RAPD and ISSR analyses of regenerated pea (Pisum sativum L.) plants. Russ. J. Genet. 2005, 41, 60–65. [Google Scholar] [CrossRef]
- Varshney, A.; Altpeter, F. Stable transformation and tissue culture response in current European winter wheats (Triticum aestivum L.). Mol. Plant Breed. 2002, 8, 295–309. [Google Scholar] [CrossRef]
- Nagar, D.S.; Jha, S.K.; Jani, J. Direct adventitious shoot bud formation on hypocotyls explants in Millettia pinnata (L.) Panigrahi —A biodiesel-producing medicinal tree species. Physiol. Mol. Biol. Plants 2015, 21, 287–292. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Z.; Ren, Y.; Feng, Y.; Guo, Q.; Dong, L.; Sun, Y.; Li, Y. Induction and early identification of tetraploid black locust by hypocotyl in vitro. Vitr. Cell. Dev. Biol. 2021, 57, 372–379. [Google Scholar] [CrossRef]
- Cardi, T.; Monti, L. Optimization of callus culture in pea (Pisum sativum L.). Ann. Della Fac. Di Sci. Agrar. Della Univ. Degli Studi Di Napoli Portici 1990, 24, 11–18. [Google Scholar]
- Bolouri, P.; Hosseinpour, A.; Jannati, G.; Haliloğlu, K. Genetic diversity of Pea (Pisum arvense L.) genotypes according to the tissue culture traits. Atatürk Uni. J. Agri. Fac. 2019, 50, 11–17. [Google Scholar]
- Chakraborty, N.; Chakraborty, P.; Sen, M.; Bandopadhyay, R. Choice of explant for plant genetic transformation. In Biolistic DNA Delivery in Plants: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2020; pp. 107–123. [Google Scholar]
- Koufan, M.; Belkoura, I.; Mazri, M.A. In vitro propagation of caper (Capparis spinosa L.): A review. Horticulturae 2022, 8, 737. [Google Scholar] [CrossRef]
- Nascimento, L.E.S.; Arriola, N.D.A.; da Silva, L.A.L.; Faqueti, L.G.; Sandjo, L.P.; de Araújo, C.E.S.; Biavatti, M.W.; Barcelos-Oliveira, J.L.; Amboni, R.D.M.C. Phytochemical profile of different anatomical parts of jambu (Acmella oleracea (L.) RK Jansen): A comparison between hydroponic and conventional cultivation using PCA and cluster analysis. Food Chem. 2020, 332, 127393. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Qu, H.; Liu, X.; Li, Q.; Liu, Y.; Wang, W.; Chen, D.; Xiao, L.; Gu, R. Comparison and selection of probiotic Lactobacillus from the human intestinal tract and traditional fermented food in vitro via PCA, unsupervised clustering algorithm, and heat-map analysis. Food Sci. Nutr. 2022, 10, 4247–4257. [Google Scholar] [CrossRef] [PubMed]
- Esan, V.I.; Oke, G.O.; Ogunbode, T.O.; Obisesan, I.A. AMMI and GGE biplot analyses of Bambara groundnut (Vigna ubterranean (L.) Verdc.) for agronomic performances under three environmental conditions. Front. Plant Sci. 2023, 13, 997429. [Google Scholar] [CrossRef]
- Fuller, D.Q.; Denham, T.; Allaby, R. Plant domestication and agricultural ecologies. Curr. Biol. 2023, 33, R636–R649. [Google Scholar] [CrossRef]
- Mohamed, A.; Aka Kaçar, Y.; Toklu, F.; Dönmez, D.; Erol, M.; Şimşek, Ö.; Biçen, B.; Karaköy, T.; Tlahig, S.; Loumerem, M. Evaluation of the Genetic Relationships of Some Endangered Tunisian Peas Adapted to Arid Regions and Turkish Accessions Revealed by Inter Simple Sequence Repeat (ISSR) Markers. Pol. J. Environ. Stud. 2023, 32, 2251–2260. [Google Scholar] [CrossRef]
- Silva, J.C.F.; Teixeira, R.M.; Silva, F.F.; Brommonschenkel, S.H.; Fontes, E.P. Machine learning approaches and their current application in plant molecular biology: A systematic review. Plant Sci. 2019, 284, 37–47. [Google Scholar] [CrossRef]
- Hesami, M.; Pepe, M.; Monthony, A.S.; Baiton, A.; Jones, A.M.P. Modeling and optimizing in vitro seed germination of industrial hemp (Cannabis sativa L.). Ind. Crops Prod. 2021, 170, 113753. [Google Scholar] [CrossRef]
- Zhang, Q.; Deng, D.; Dai, W.; Li, J.; Jin, X. Optimization of culture conditions for differentiation of melon based on artificial neural network and genetic algorithm. Sci. Rep. 2020, 10, 3524. [Google Scholar] [CrossRef]
- Wei, L.; Tian, W.; Silva, E.A.; Choudhary, R.; Meng, Q.; Yang, S. Comparative study on machine learning for urban building energy analysis. Procedia Eng. 2015, 121, 285–292. [Google Scholar] [CrossRef]
- Çeli, Ş.; Çaçan, E.; Yaryab, S. Prediction of stem weight in selected alfalfa varieties by artificial neural networks, multivariate adaptive regression splines, and multiple regression analysis. J. Anim. Plant Sci. 2023, 33, 1006–1020. [Google Scholar]

| Genotype | CI% * % | ECNEP % | RECNEP % | NSE (Number) | RSE (Number) | RE (Number) | NRP (Number) |
|---|---|---|---|---|---|---|---|
| Ardahan Merkez-1 | 93.75 a–d ** | 82.81 a–f | 67.19 e–m | 37.75 c–j | 37.50 b–i | 0.00 c | 0.00 d |
| Ardahan Merkez-2 | 100.00 a | 96.88 ab | 85.94 a–f | 89.25 a | 81.250 a | 0.00 c | 0.00 d |
| Ardahan Merkez-3 | 84.37 c–i | 57.81 h–k | 65.63 e–m | 25.25 g–n | 21.00 g.n | 0.00 c | 0.00 d |
| Aşağıcambaz | 100.00 a | 73.44 d–h | 84.38 a–g | 37.25 d–j | 32.75 c–j | 0.03 c | 0.25 d |
| Aşağıkırzı | 78.13 g–j | 73.44 d–h | 60.94 f–n | 47.50 b–e | 41.25 b–h | 0.00 c | 0.00 d |
| Avcılar | 90.63 a–g | 84.38 a–e | 76.56 a–i | 96.75 a | 88.25 a | 0.02 c | 0.25 d |
| Balçeşme | 76.56 h–j | 46.88 kl | 46.88 k–r | 25.25 g–n | 20.00 g–n | 0.00 c | 0.00 d |
| Camlıçatak-1 | 98.44 ab | 64.06 g–j | 35.94 n–r | 24.50 g–n | 12.75 j–n | 0.00 c | 0.00 d |
| Camlıçatak-2 | 100.00 a | 68.75 e–h | 34.38 o–r | 27.00 f–n | 23.00 g–n | 0.00 c | 0.00 d |
| Cayağzı | 95.31 a–d | 26.56 m | 21.88 r | 11.75 mn | 4.00 n | 0.00 c | 0.00 d |
| Ciğdemtepe | 73.44 ij | 67.19 e–i | 26.56 qr | 23.25 h–n | 18.75 I–n | 0.05 c | 0.25 d |
| Cumhuriyet | 78.13 g–j | 73.44 d–h | 71.88 b–k | 8.25 n | 7.75 k–n | 0.00 c | 0.00 d |
| Değirmencik-1 | 100.00 a | 89.06 a–d | 100.00 a | 48.25 b–e | 48.25 b–e | 0.00 c | 0.00 d |
| Doğruyol | 60.94 k | 56.25 h–k | 51.56 I–q | 29.00 e–m | 20.25 g–n | 1.61 a | 15.00 a |
| Döşeli-1 | 93.75 a–d | 62.50 g–k | 48.44 k–q | 19.25 I–n | 12.75 j–n | 0.65 b | 4.25 b–d |
| Giresun Merkez | 68.75 jk | 56.25 h–k | 50.00 I–q | 27.25 f–n | 21.50 g–n | 0.05 c | 0.50 d |
| Görele-1 | 100.00 a | 96.88 ab | 98.44 ab | 34.25 d–l | 24.25 f–n | 0.06 c | 1.00 cd |
| Incili-1 | 79.69 e–j | 70.31 e–h | 54.69 h–o | 39.25 c–i | 7.25 l–n | 0.00 c | 0.00 d |
| Incili-2 | 98.44 ab | 89.06 a–d | 76.56 a–j | 56.75 bc | 52.00 bc | 0.00 c | 0.00 d |
| Incili-3 | 92.19 a–e | 64.06 g–j | 79.69 a–h | 18.25 j–n | 13.25 j–n | 0.43 bc | 5.25 bc |
| Kartalpınar | 92.19 a–f | 73.44 d–h | 71.88 c–k | 19.75 I–n | 14.25 j–n | 0.04 c | 0.50 d |
| Kenarbel | 85.94 b–i | 78.13 c–g | 65.63 e–m | 39.25 c–i | 27.25 e–m | 0.13 c | 1.25 cd |
| Koyunpınarı | 100.0 a | 82.81 a–f | 70.31 d–l | 35.75 d–k | 29.75 d–k | 0.00 c | 0.00 d |
| Oburcak | 87.50 a–h | 87.50 a–d | 82.81 a–g | 40.00 c–h | 39.00 b–i | 0.00 c | 0.00 d |
| Ovaçevirme-1 | 76.56 h–j | 71.88 d–h | 67.19 e–m | 48.25 b–e | 45.25 b–f | 0.00 c | 0.00 d |
| Ovaçevirme-2 | 98.44 ab | 89.06 a–d | 53.13 h–p | 29.25 e–m | 23.25 g–n | 0.00 c | 0.00 d |
| Ovaçevirme-3 | 96.88 a–c | 96.88 ab | 96.88 abcd | 88.00 a | 85.00 a | 0.00 c | 0.00 d |
| Ovaçevirme-4 | 100.00 a | 67.19 e–i | 89.06 a–e | 14.50 l–n | 9.00 k–n | 0.51 bc | 7.00 b |
| Ovaçevirme-5 | 100.00 a | 70.31 e–h | 76.56 a–j | 43.75 c–g | 29.00 d–l | 0.00 c | 0.00 d |
| Paslı | 100.00 a | 96.88 ab | 92.19 a–e | 49.75 b–d | 49.25 b–d | 0.00 c | 0.00 d |
| Sayvan | 67.19 jk | 56.25 h–k | 50.00 j–q | 63.50 b | 54.75 b | 0.00 c | 0.00 d |
| Selamverdi | 79.69 f–j | 35.94 lm | 28.13 p–r | 14.25 l–n | 7.50 l–n | 0.00 c | 0.00 d |
| Senkaya Merkez | 96.88 a–c | 59.38 h–k | 48.44 k–q | 22.50 h–n | 22.00 g–n | 0.15 c | 1.00 cd |
| Serhat | 95.31 a–d | 93.75 a–c | 87.50 a–f | 30.00 e–m | 24.75 f–n | 0.04 c | 0.50 d |
| Seyitören | 95.31 a–d | 79.69 b–g | 40.63 m–r | 21.25 h–n | 6.50 mn | 0.00 c | 0.00 d |
| Subatan | 100.00 a | 93.75 a–c | 92.19 a–e | 45.50 b–f | 42.00 b–g | 0.05 c | 0.75 cd |
| Sulakyurt | 100.00 a | 65.63 f–j | 65.63 e–m | 24.50 g–n | 20.50 g–n | 0.05 c | 0.75 cd |
| Tahtakıran | 100.00 a | 84.38 a–e | 82.81 a–g | 36.75 d–k | 31.75 c–j | 0.02 c | 0.25 d |
| Tepeköy | 82.81 d–i | 51.6 I–k | 48.44 k–q | 17.00 k–n | 7.25 l–n | 0.00 c | 0.00 d |
| Tepeler | 95.31 a–d | 64.06 g–j | 57.81 g–o | 25.25 g–n | 19.50 h–n | 0.17 c | 1.25 cd |
| Yamçılı | 100.00 a | 100.00 a | 98.44 a–c | 29.75 e–m | 22.25 g–n | 0.02 c | 0.25 d |
| Yolgeçmez | 96.88 a–c | 50.00 j–l | 43.75 l–r | 32.00 d–l | 21.00 g–n | 0.62 b | 4.00 b–d |
| Means | 90.67 | 72.58 | 65.40 | 35.63 | 29.01 | 0.11 | 1.05 |
| F value | 7.82 *** | 11.83 *** | 7.62 *** | 11.95 *** | 10.46 *** | 4.03 *** | 3.64 *** |
| In Vitro Traits | F1 | F2 | F3 | F4 | F5 |
|---|---|---|---|---|---|
| CI | 0.256 | 0.020 | 0.484 | 0.240 | 0.000 |
| ECNEP | 0.691 | 0.047 | 0.086 | 0.063 | 0.113 |
| RECNEP | 0.538 | 0.138 | 0.141 | 0.099 | 0.084 |
| RE | 0.299 | 0.668 | 0.008 | 0.012 | 0.003 |
| NSE | 0.609 | 0.099 | 0.242 | 0.034 | 0.000 |
| RSE | 0.650 | 0.101 | 0.203 | 0.028 | 0.002 |
| NRP | 0.272 | 0.697 | 0.020 | 0.002 | 0.000 |
| Eigen value | 3.03 | 1.88 | 1.20 | 0.50 | 0.31 |
| Percent of Variance | 43.23 | 26.88 | 17.88 | 7.18 | 4.43 |
| Cumulative Percentage | 43.23 | 70.11 | 87.30 | 94.47 | 98.90 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Türkoğlu, A.; Bolouri, P.; Haliloğlu, K.; Eren, B.; Demirel, F.; Işık, M.İ.; Piekutowska, M.; Wojciechowski, T.; Niedbała, G. Modeling Callus Induction and Regeneration in Hypocotyl Explant of Fodder Pea (Pisum sativum var. arvense L.) Using Machine Learning Algorithm Method. Agronomy 2023, 13, 2835. https://doi.org/10.3390/agronomy13112835
Türkoğlu A, Bolouri P, Haliloğlu K, Eren B, Demirel F, Işık Mİ, Piekutowska M, Wojciechowski T, Niedbała G. Modeling Callus Induction and Regeneration in Hypocotyl Explant of Fodder Pea (Pisum sativum var. arvense L.) Using Machine Learning Algorithm Method. Agronomy. 2023; 13(11):2835. https://doi.org/10.3390/agronomy13112835
Chicago/Turabian StyleTürkoğlu, Aras, Parisa Bolouri, Kamil Haliloğlu, Barış Eren, Fatih Demirel, Muhammet İslam Işık, Magdalena Piekutowska, Tomasz Wojciechowski, and Gniewko Niedbała. 2023. "Modeling Callus Induction and Regeneration in Hypocotyl Explant of Fodder Pea (Pisum sativum var. arvense L.) Using Machine Learning Algorithm Method" Agronomy 13, no. 11: 2835. https://doi.org/10.3390/agronomy13112835
APA StyleTürkoğlu, A., Bolouri, P., Haliloğlu, K., Eren, B., Demirel, F., Işık, M. İ., Piekutowska, M., Wojciechowski, T., & Niedbała, G. (2023). Modeling Callus Induction and Regeneration in Hypocotyl Explant of Fodder Pea (Pisum sativum var. arvense L.) Using Machine Learning Algorithm Method. Agronomy, 13(11), 2835. https://doi.org/10.3390/agronomy13112835












