Identifying Critical Regulators in the Viral Stress Response of Wheat (Triticum aestivum L.) Using Large-Scale Transcriptomics Data
Abstract
1. Introduction
2. Materials and Methods
2.1. The RNA-Seq
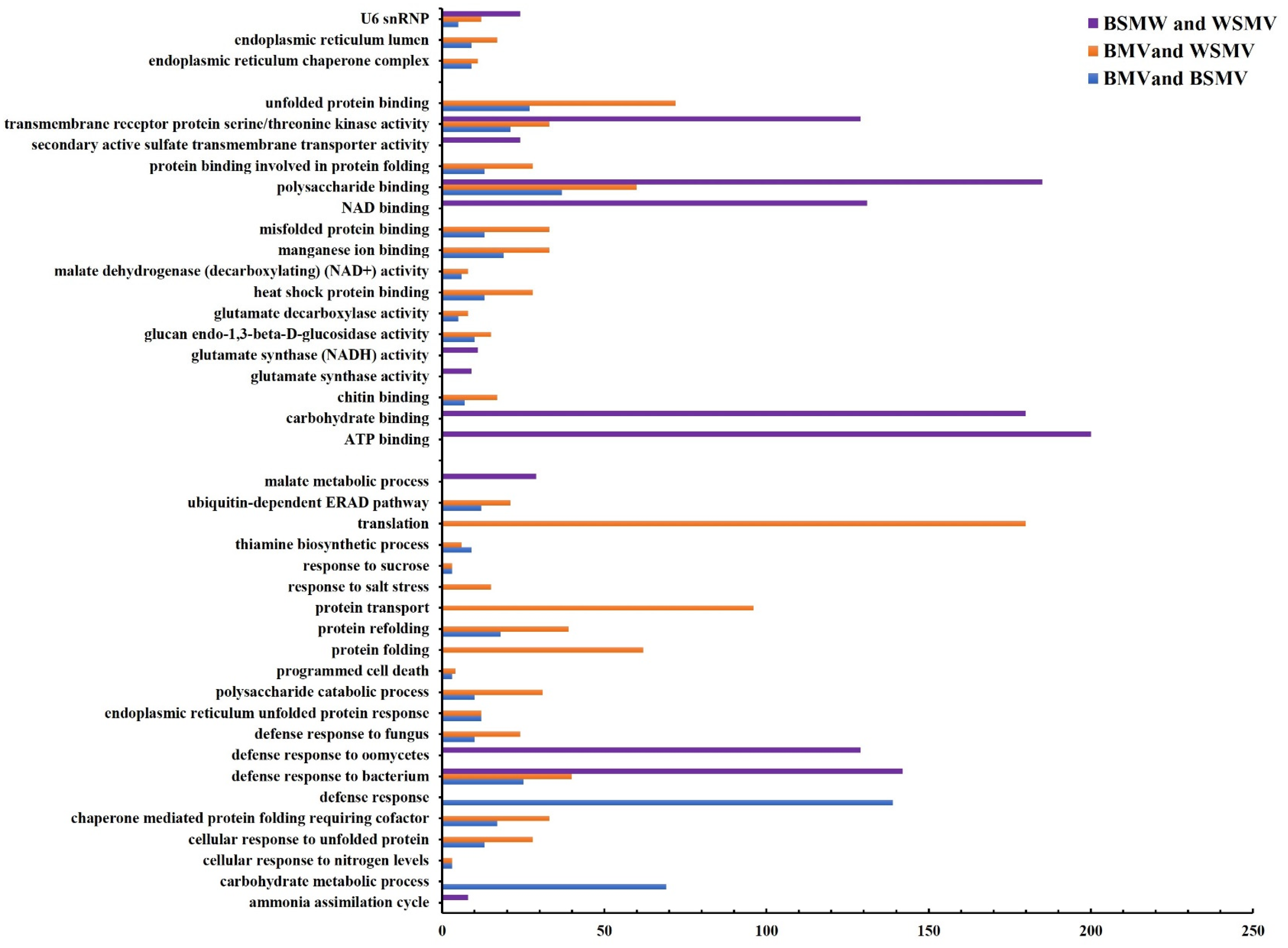
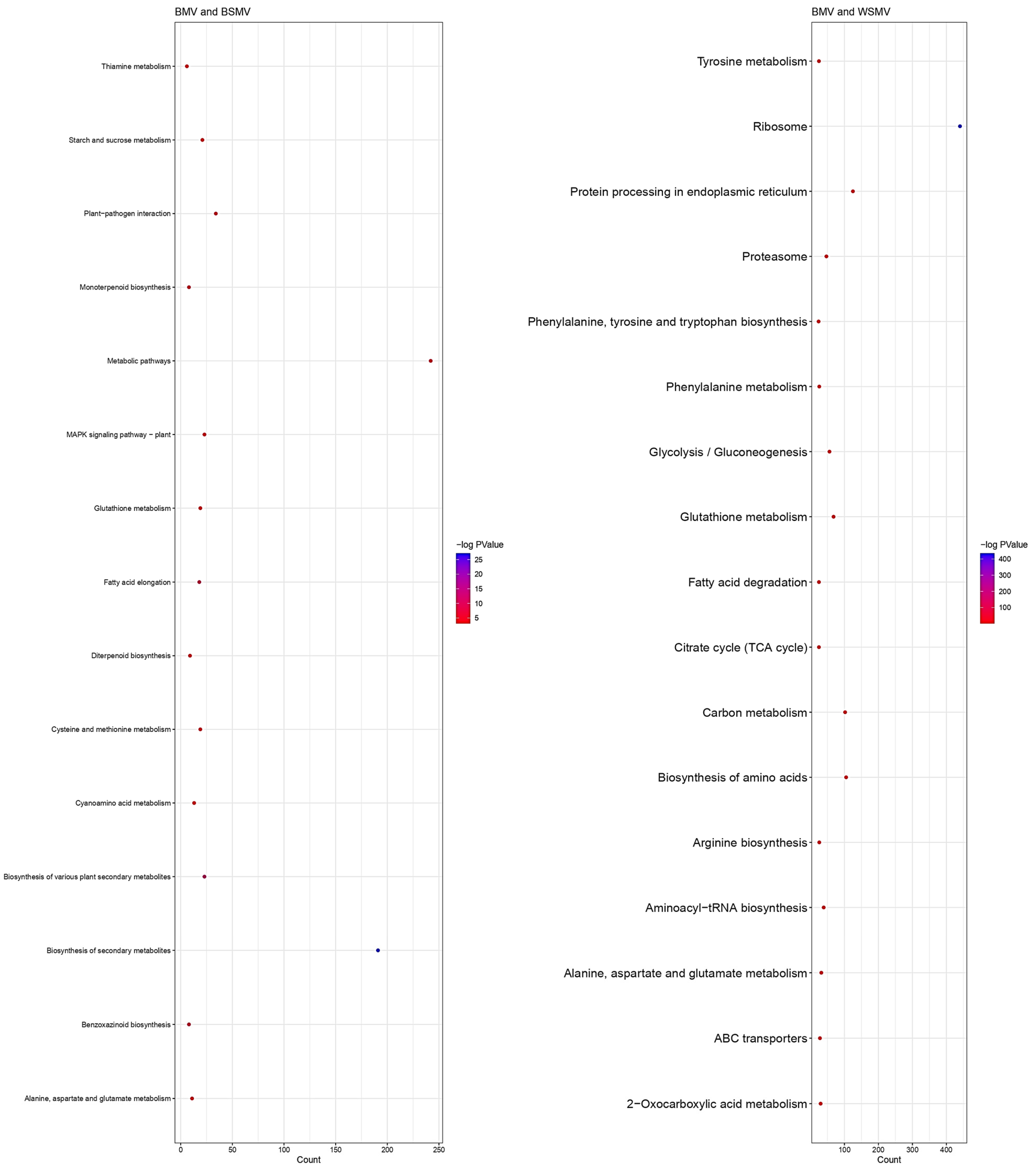
2.2. Functional Enrichment Analysis of the DEGs
2.3. The Analysis of Transcription Factors, Protein Kinases, and Promoters
2.4. Validation Analysis
2.5. Validation Analysis of Shared DEGs
2.6. Homology Analysis of Overlapping DEGs
2.7. Gene Network Analysis
3. Results
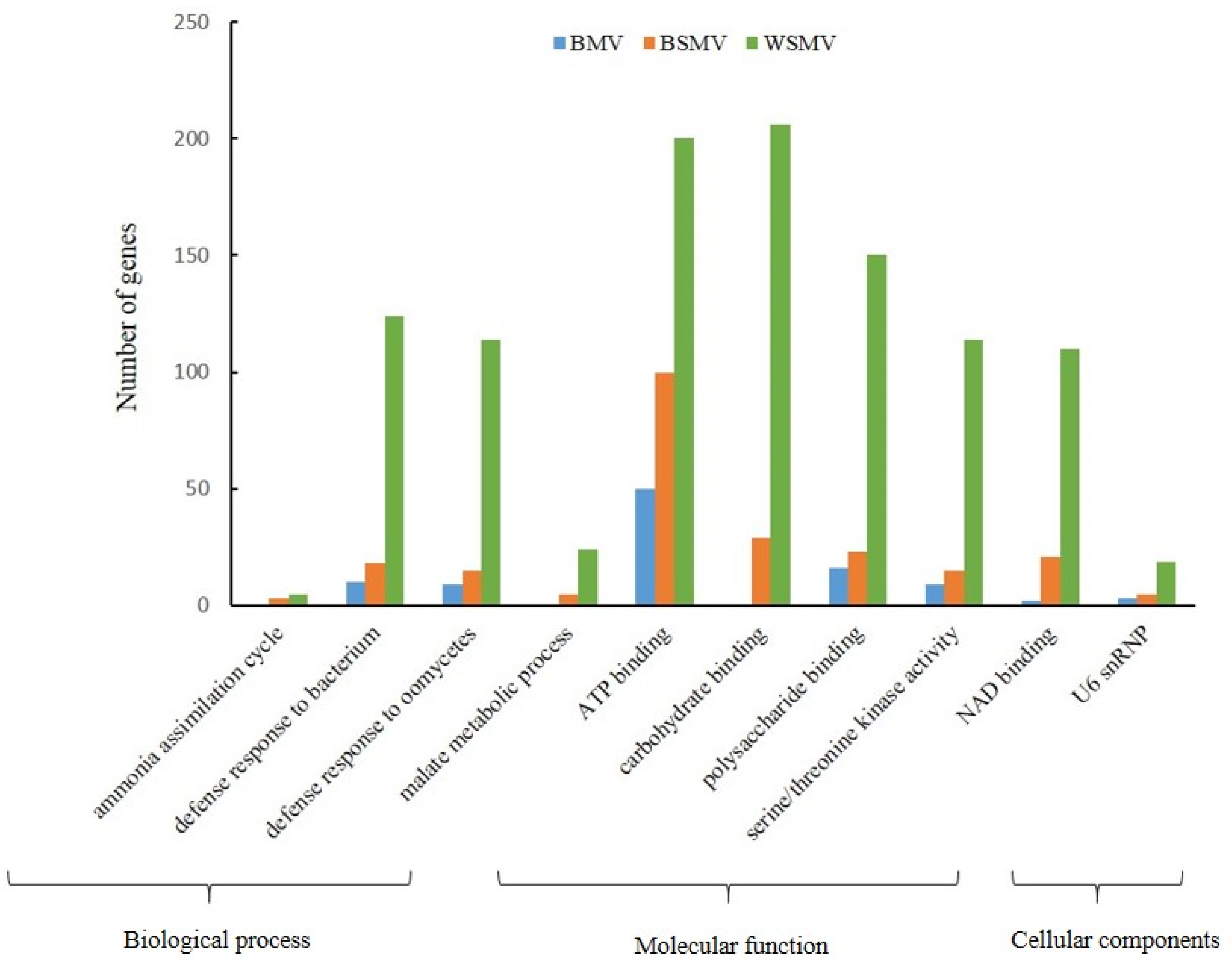
3.1. DEG Analysis
3.2. TF and PK Assays
3.3. Motif Analysis
3.4. Validation Analysis of Shared DEGs
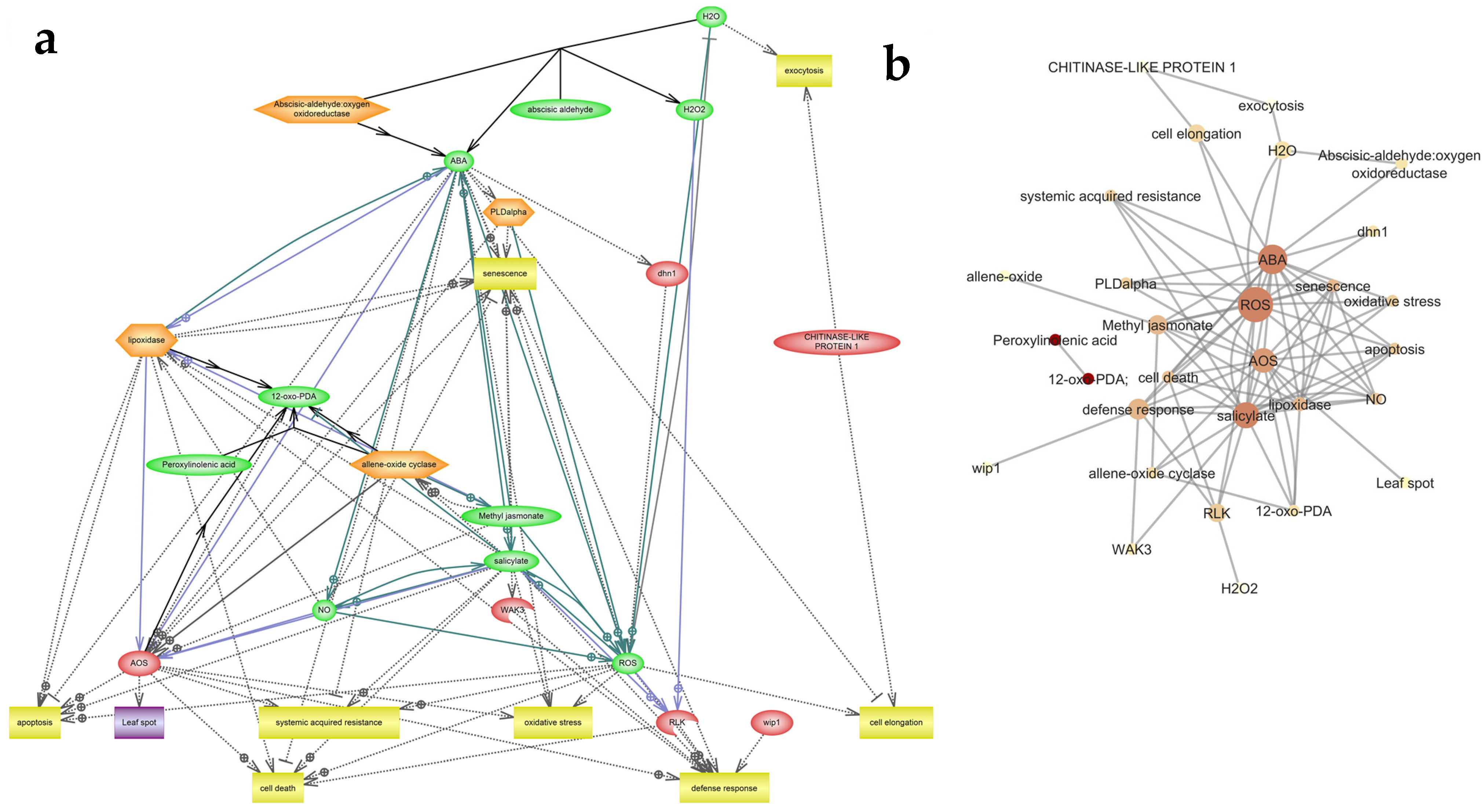
3.5. Homology and Gene Network Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Goodin, M.; Verchot, J. Introduction to Special Issue of Plant Virus Emergence. Viruses 2021, 13, 55. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.I.; Rao, G.P. Characterization of plant viruses. In Springer Protocols Handbooks; Springer: Berlin/Heidelberg, Germany, 2020; Volume 23. [Google Scholar]
- Sastry, K.S.; Mandal, B.; Hammond, J.; Scott, S.W.; Briddon, R.W. Encyclopedia of Plant Viruses and Viroids; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Inoue-Nagata, A.K.; Jordan, R.; Kreuze, J.; Li, F.; López-Moya, J.J.; Mäkinen, K.; Ohshima, K.; Wylie, S.J.; Consortium, I.R. ICTV virus taxonomy profile: Potyviridae 2022. J. Gen. Virol. 2022, 103, 001738. [Google Scholar] [CrossRef] [PubMed]
- Hadi, B.; Langham, M.; Osborne, L.; Tilmon, K. Wheat streak mosaic virus on wheat: Biology and management. J. Integr. Pest Manag. 2011, 2, J1–J5. [Google Scholar] [CrossRef]
- Singh, K.; Wegulo, S.N.; Skoracka, A.; Kundu, J.K. Wheat streak mosaic virus: A century old virus with rising importance worldwide. Mol. Plant Pathol. 2018, 19, 2193–2206. [Google Scholar] [CrossRef]
- Byamukama, E.; Wegulo, S.; Tatineni, S.; Hein, G.; Graybosch, R.; Baenziger, P.S.; French, R. Quantification of yield loss caused by Triticum mosaic virus and Wheat streak mosaic virus in winter wheat under field conditions. Plant Dis. 2014, 98, 127–133. [Google Scholar] [CrossRef]
- Adams, M.J.; Adkins, S.; Bragard, C.; Gilmer, D.; Li, D.; MacFarlane, S.A.; Wong, S.-M.; Melcher, U.; Ratti, C.; Ryu, K.H. ICTV virus taxonomy profile: Virgaviridae. J. Gen. Virol. 2017, 98, 1999–2000. [Google Scholar] [CrossRef]
- Murray, T.D.; Parry, D.W.; Cattlin, N.D. Diseases of Small Grain Cereal Crops: A Colour Handbook; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Kendall, A.; Williams, D.; Bian, W.; Stewart, P.L.; Stubbs, G. Barley stripe mosaic virus: Structure and relationship to the tobamoviruses. Virol. J. 2013, 443, 265–270. [Google Scholar] [CrossRef]
- Timian, R.G. Barley stripe mosaic virus in North Dakota. Farm. Res. 1971, 28, 5. [Google Scholar]
- Virus, B.S.M. Grains Industry Biosecurity Plan Threat Specific Contingency Plan. Plan Health 2009. [Google Scholar]
- Bujarski, J.; Gallitelli, D.; García-Arenal, F.; Pallás, V.; Palukaitis, P.; Reddy, M.K.; Wang, A.; Consortium, I.R. ICTV virus taxonomy profile: Bromoviridae. J. Gen. Virol. 2019, 100, 1206–1207. [Google Scholar] [CrossRef]
- Bujarski, J.J. Bromoviruses (Bromoviridae). In Encyclopedia of Virology; Academic Press: Cambridge, MA, USA, 2021; Volume 260. [Google Scholar]
- Díaz-Cruz, G.; Smith, C.; Wiebe, K.; Charette, J.; Cassone, B. First report of brome mosaic virus infecting soybean, isolated in Manitoba, Canada. Plant Dis. 2018, 102, 460. [Google Scholar] [CrossRef]
- Hodge, B.; Salgado, J.; Paul, P.; Stewart, L. Characterization of an Ohio isolate of Brome mosaic virus and its impact on the development and yield of soft red winter wheat. Plant Dis. 2019, 103, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.B.; López-Moya, J.J. When viruses play team sports: Mixed infections in plants. J. Phytopathol. 2020, 110, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, T.; Zhu, Y.; Zhou, G. Co-infection of two reoviruses increases both viruses accumulation in rice by up-regulating of viroplasm components and movement proteins bilaterally and RNA silencing suppressor unilaterally. Virol. J. 2017, 14, 150. [Google Scholar] [CrossRef]
- Syller, J. Biological and molecular events associated with simultaneous transmission of plant viruses by invertebrate and fungal vectors. Mol. Plant Pathol. 2014, 15, 417–426. [Google Scholar] [CrossRef]
- Tatineni, S.; Graybosch, R.A.; Hein, G.L.; Wegulo, S.N.; French, R. Wheat cultivar-specific disease synergism and alteration of virus accumulation during co-infection with Wheat streak mosaic virus and Triticum mosaic virus. J. Phytopathol. 2010, 100, 230–238. [Google Scholar] [CrossRef]
- VALKONEN, J.P. Accumulation of potato virus Y is enhanced in Solatium brevidens also infected with tobacco mosaic virus or potato spindle tuber viroid. Ann. Appl. Biol. 1992, 121, 321–327. [Google Scholar] [CrossRef]
- Wintermantel, W.M.; Cortez, A.A.; Anchieta, A.G.; Gulati-Sakhuja, A.; Hladky, L.L. Co-infection by two criniviruses alters accumulation of each virus in a host-specific manner and influences efficiency of virus transmission. J. Phytopathol. 2008, 98, 1340–1345. [Google Scholar] [CrossRef]
- McLeish, M.J.; Zamfir, A.D.; Babalola, B.M.; Peláez, A.; Fraile, A.; García-Arenal, F. Metagenomics show high spatiotemporal virus diversity and ecological compartmentalisation: Virus infections of melon, Cucumis melo, crops, and adjacent wild communities. Virus Evol. 2022, 8, veac095. [Google Scholar] [CrossRef]
- Elena, S.F.; Carrera, J.; Rodrigo, G. A systems biology approach to the evolution of plant–virus interactions. Curr. Opin. Plant Biol. 2011, 14, 372–377. [Google Scholar] [CrossRef]
- Laliberté, J.-F.; Sanfaçon, H. Cellular remodeling during plant virus infection. Annu. Rev. Phytopathol. 2010, 48, 69–91. [Google Scholar] [CrossRef] [PubMed]
- Gong, Q.; Wang, Y.; Jin, Z.; Hong, Y.; Liu, Y. Transcriptional and post-transcriptional regulation of RNAi-related gene expression during plant-virus interactions. Stress Biol. 2022, 2, 33. [Google Scholar] [CrossRef] [PubMed]
- González, R.; Butković, A.; Escaray, F.J.; Martínez-Latorre, J.; Melero, Í.; Pérez-Parets, E.; Gómez-Cadenas, A.; Carrasco, P.; Elena, S.F. Plant virus evolution under strong drought conditions results in a transition from parasitism to mutualism. Proc. Natl. Acad. Sci. USA 2021, 118, e2020990118. [Google Scholar] [CrossRef] [PubMed]
- Havelda, Z.; Várallyay, É.; Válóczi, A.; Burgyán, J. Plant virus infection-induced persistent host gene downregulation in systemically infected leaves. Plant J. 2008, 55, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.; Sharma, N.; Muthamilarasan, M.; Rana, S.; Prasad, M. Recent advances in small RNA mediated plant-virus interactions. Crit. Rev. Biotechnol. 2019, 39, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Tsalik, E.L.; Henao, R.; Montgomery, J.L.; Nawrocki, J.W.; Aydin, M.; Lydon, E.C.; Ko, E.R.; Petzold, E.; Nicholson, B.P.; Cairns, C.B. Discriminating bacterial and viral infection using a rapid host gene expression test. Crit. Care Med. 2021, 49, 1651–1663. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Maule, A.J. Inhibition of host gene expression associated with plant virus replication. Science 1995, 267, 229–231. [Google Scholar] [CrossRef] [PubMed]
- Whitham, S.A.; Yang, C.; Goodin, M.M. Global impact: Elucidating plant responses to viral infection. Mol. Plant Microbe Interact. 2006, 19, 1207–1215. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, Z.; Wan, Q.; Zhou, H.; Jiao, M.; Zheng, H.; Lu, Y.; Rao, S.; Wu, G.; Chen, J. Selection and validation of reference genes for RT-qPCR analysis of gene expression in Nicotiana benthamiana upon single infections by 11 positive-sense single-stranded RNA viruses from Four Genera. Plants 2023, 12, 857. [Google Scholar] [CrossRef]
- Zhao, S.; Li, Y. Current understanding of the interplays between host hormones and plant viral infections. PLoS Pathog. 2021, 17, e1009242. [Google Scholar] [CrossRef]
- Allie, F.; Pierce, E.J.; Okoniewski, M.J.; Rey, C. Transcriptional analysis of South African cassava mosaic virus-infected susceptible and tolerant landraces of cassava highlights differences in resistance, basal defense and cell wall associated genes during infection. BMC Genom. 2014, 15, 1006. [Google Scholar] [CrossRef]
- Bazzini, A.A.; Almasia, N.I.; Manacorda, C.A.; Mongelli, V.C.; Conti, G.; Maroniche, G.A.; Rodriguez, M.C.; Distéfano, A.J.; Hopp, H.E.; Del Vas, M. Virus infection elevates transcriptional activity of miR164a promoter in plants. BMC Plant Biol. 2009, 9, 152. [Google Scholar] [CrossRef] [PubMed]
- Marquez-Molins, J.; Juarez-Gonzalez, V.T.; Gomez, G.; Pallas, V.; Martinez, G. Occurrence of RNA post-transcriptional modifications in plant viruses and viroids and their correlation with structural and functional features. Virus Res. 2023, 323, 198958. [Google Scholar] [CrossRef] [PubMed]
- Javed, T.; Shabbir, R.; Ali, A.; Afzal, I.; Zaheer, U.; Gao, S.-J. Transcription factors in plant stress responses: Challenges and potential for sugarcane improvement. Plants 2020, 9, 491. [Google Scholar] [CrossRef] [PubMed]
- Meraj, T.A.; Fu, J.; Raza, M.A.; Zhu, C.; Shen, Q.; Xu, D.; Wang, Q. Transcriptional factors regulate plant stress responses through mediating secondary metabolism. Genes 2020, 11, 346. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, H.; Cai, J.; Li, D.; Song, F. NAC transcription factors in plant immunity. Phytopathol. Res. 2019, 1, 3. [Google Scholar] [CrossRef]
- Tahmasebi, A.; Khahani, B.; Tavakol, E.; Afsharifar, A.; Shahid, M.S. Microarray analysis of Arabidopsis thaliana exposed to single and mixed infections with Cucumber mosaic virus and turnip viruses. Physiol. Mol. Biol. Plants 2021, 27, 11–27. [Google Scholar] [CrossRef]
- Tatineni, S.; Alexander, J.; Qu, F. Differential synergistic interactions among four different wheat-infecting viruses. Front. Microbiol. 2022, 12, 800318. [Google Scholar] [CrossRef]
- Zanardo, L.G.; de Souza, G.B.; Alves, M.S. Transcriptomics of plant–virus interactions: A review. Theor. Exp. Plant Physiol. 2019, 31, 103–125. [Google Scholar] [CrossRef]
- Xie, Y.; Nachappa, P.; Nalam, V.J.; Pearce, S. Genomic and molecular characterization of wheat streak mosaic virus resistance locus 2 (Wsm2) in common wheat (Triticum aestivum L.). Front. Plant Sci. 2022, 13, 928949. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2. 0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef]
- Shahriari, A.G.; Soltani, Z.; Tahmasebi, A.; Poczai, P. Integrative System Biology Analysis of Transcriptomic Responses to Drought Stress in Soybean (Glycine max L.). Genes 2022, 13, 1732. [Google Scholar] [CrossRef]
- Lorenzon, R.; Mariotti-Ferrandiz, E.; Aheng, C.; Ribet, C.; Toumi, F.; Pitoiset, F.; Chaara, W.; Derian, N.; Johanet, C.; Drakos, I. Clinical and multi-omics cross-phenotyping of patients with autoimmune and autoinflammatory diseases: The observational TRANSIMMUNOM protocol. BMJ Open 2018, 8, e021037. [Google Scholar] [CrossRef]
- Bolser, D.M.; Staines, D.M.; Perry, E.; Kersey, P.J. Ensembl plants: Integrating tools for visualizing, mining, and analyzing plant genomic data. In Plant Genomics Databases: Methods and Protocols; Spring: Berlin/Heidelberg, Germany, 2017; pp. 1–31. [Google Scholar]
- Nikitin, A.; Egorov, S.; Daraselia, N.; Mazo, I. Pathway studio—The analysis and navigation of molecular networks. Bioinformatics 2003, 19, 2155–2157. [Google Scholar] [CrossRef]
- Scardoni, G.; Petterlini, M.; Laudanna, C. Analyzing biological network parameters with CentiScaPe. Bioinformatics 2009, 25, 2857–2859. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Boccaletti, S.; Latora, V.; Moreno, Y.; Chavez, M.; Hwang, D. Complex Networks: Structure and Dynamics. Phys. Rep. 2006, 424, 175–308. [Google Scholar] [CrossRef]
- Girvan, M.; Newman, M.E. Community structure in social and biological networks. Proc. Natl. Acad. Sci. USA 2002, 99, 7821–7826. [Google Scholar] [CrossRef] [PubMed]
- Alcaide, C.; Rabadán, M.P.; Moreno-Perez, M.G.; Gómez, P. Implications of mixed viral infections on plant disease ecology and evolution. Adv. Virus Res. 2020, 106, 145–169. [Google Scholar] [PubMed]
- Liu, Y.; Liu, Y.; Spetz, C.; Li, L.; Wang, X. Comparative transcriptome analysis in Triticum aestivum infecting wheat dwarf virus reveals the effects of viral infection on phytohormone and photosynthesis metabolism pathways. Phytopathol. Res. 2020, 2, 3. [Google Scholar] [CrossRef]
- Hull, R. Plant Virology; Academic Press: Cambridge, MA, USA, 2013. [Google Scholar]
- Alazem, M.; Lin, N.S. Roles of plant hormones in the regulation of host–virus interactions. Mol. Plant Pathol. 2015, 16, 529–540. [Google Scholar] [CrossRef]
- Lam, E.; Kato, N.; Lawton, M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature 2001, 411, 848–853. [Google Scholar] [CrossRef]
- Rojas, C.M.; Senthil-Kumar, M.; Tzin, V.; Mysore, K.S. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014, 5, 17. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Redox regulation in photosynthetic organisms: Signaling, acclimation, and practical implications. Antioxid. Redox Signal. 2009, 11, 861–905. [Google Scholar] [CrossRef]
- Batoko, H.; Dagdas, Y.; Baluska, F.; Sirko, A. Understanding and exploiting autophagy signaling in plants. Essays Biochem. 2017, 61, 675–685. [Google Scholar]
- Hafrén, A.; Macia, J.-L.; Love, A.J.; Milner, J.J.; Drucker, M.; Hofius, D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 2017, 114, E2026–E2035. [Google Scholar] [CrossRef]
- Dagdas, Y.F.; Belhaj, K.; Maqbool, A.; Chaparro-Garcia, A.; Pandey, P.; Petre, B.; Tabassum, N.; Cruz-Mireles, N.; Hughes, R.K.; Sklenar, J. An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. Elife 2016, 5, e10856. [Google Scholar] [CrossRef] [PubMed]
- Kabbage, M.; Williams, B.; Dickman, M.B. Cell death control: The interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 2013, 9, e1003287. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, W.; Zhao, Y.; Gong, X.; Guo, L.; Zhu, G.; Wang, X.; Gong, Z.; Schumaker, K.S.; Guo, Y. SAD2, an importin β-like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell. 2007, 19, 3805–3818. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012, 13, 89–102. [Google Scholar] [CrossRef]
- Verchot, J. The ER quality control and ER associated degradation machineries are vital for viral pathogenesis. Front. Plant Sci. 2014, 5, 66. [Google Scholar] [CrossRef]
- Vierstra, R.D. The ubiquitin–26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef]
- Citovsky, V.; Zaltsman, A.; Kozlovsky, S.V.; Gafni, Y.; Krichevsky, A. Proteasomal degradation in plant–pathogen interactions. In Seminars in Cell & Developmental Biology; Academic Press: Cambridge, MA, USA, 2009; pp. 1048–1054. [Google Scholar]
- DIELEN, A.S.; Badaoui, S.; Candresse, T.; GERMAN-RETANA, S. The ubiquitin/26S proteasome system in plant–pathogen interactions: A never-ending hide-and-seek game. Mol. Plant Pathol. 2010, 11, 293–308. [Google Scholar] [CrossRef]
- Trujillo, M.; Shirasu, K. Ubiquitination in plant immunity. Curr. Opin. Plant Biol. 2010, 13, 402–408. [Google Scholar] [CrossRef]
- Abdelrahman, H.; ElHady, M.; Alcivar-Warren, A.; Allen, S.; Al-Tobasei, R.; Bao, L.; Beck, B.; Blackburn, H.; Bosworth, B.; Buchanan, J. Aquaculture genomics, genetics and breeding in the United States: Current status, challenges, and priorities for future research. BMC Genom. 2017, 18, 191. [Google Scholar]
- Alcaide-Loridan, C.; Jupin, I. Ubiquitin and plant viruses, let’s play together! Plant Physiol. 2012, 160, 72–82. [Google Scholar] [CrossRef]
- Verchot, J. Plant virus infection and the ubiquitin proteasome machinery: Arms race along the endoplasmic reticulum. Viruses 2016, 8, 314. [Google Scholar] [CrossRef] [PubMed]
- Aono, A.H.; Pimenta, R.J.G.; da Silva Dambroz, C.M.; Costa, F.C.L.; Kuroshu, R.M.; de Souza, A.P.; Pereira, W.A. Genome-wide characterization of the common bean kinome: Catalog and insights into expression patterns and genetic organization. Gene 2023, 855, 147127. [Google Scholar] [CrossRef] [PubMed]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.B.d.; Aono, A.H.; Francisco, F.R.; da Silva, C.C.; Souza, L.M.; Souza, A.P.d. The rubber tree kinome: Genome-wide characterization and insights into coexpression patterns associated with abiotic stress responses. Front. Plant Sci. 2023, 14, 1068202. [Google Scholar] [CrossRef]
- Jackson, S.; Xiong, Y. CRL4s: The CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 2009, 34, 562–570. [Google Scholar] [CrossRef]
- Babu, M.; Griffiths, J.S.; Huang, T.-S.; Wang, A. Altered gene expression changes in Arabidopsis leaf tissues and protoplasts in response to Plum pox virus infection. BMC Genom. 2008, 9, 325. [Google Scholar] [CrossRef]
- Kaur, H.; Yadav, C.B.; Alatar, A.A.; Faisal, M.; Jyothsna, P.; Malathi, V.; Praveen, S. Gene expression changes in tomato during symptom development in response to leaf curl virus infection. J. Plant Biochem. Biotechnol. 2015, 24, 347–354. [Google Scholar] [CrossRef]
- Pesti, R.; Kontra, L.; Paul, K.; Vass, I.; Csorba, T.; Havelda, Z.; Várallyay, É. Differential gene expression and physiological changes during acute or persistent plant virus interactions may contribute to viral symptom differences. PLoS ONE 2019, 14, e0216618. [Google Scholar] [CrossRef]
- Whitham, S.A.; Quan, S.; Chang, H.S.; Cooper, B.; Estes, B.; Zhu, T.; Wang, X.; Hou, Y.M. Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 2003, 33, 271–283. [Google Scholar] [CrossRef]
- Manghwar, H.; Hussain, A.; Ali, Q.; Liu, F. Brassinosteroids (BRs) role in plant development and coping with different stresses. Int. J. Mol. Sci. 2022, 23, 1012. [Google Scholar] [CrossRef]
- Nawaz, F.; Naeem, M.; Zulfiqar, B.; Akram, A.; Ashraf, M.Y.; Raheel, M.; Shabbir, R.N.; Hussain, R.A.; Anwar, I.; Aurangzaib, M. Understanding brassinosteroid-regulated mechanisms to improve stress tolerance in plants: A critical review. Environ. Sci. Pollut. Res. 2017, 24, 15959–15975. [Google Scholar] [CrossRef]
- Zhang, D.-W.; Deng, X.-G.; Fu, F.-Q.; Lin, H.-H. Induction of plant virus defense response by brassinosteroids and brassinosteroid signaling in Arabidopsis thaliana. Planta 2015, 241, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Farmer, E.E.; Ryan, C.A. Octadecanoid precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell 1992, 4, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Mandadi, K.K.; Pyle, J.D.; Scholthof, K.-B.G. Comparative analysis of antiviral responses in Brachypodium distachyon and Setaria viridis reveals conserved and unique outcomes among C3 and C4 plant defenses. Mol. Plant Microbe Interact. 2014, 27, 1277–1290. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 2001, 13, 2191–2209. [Google Scholar] [CrossRef]
- Stenzel, I.; Hause, B.; Miersch, O.; Kurz, T.; Maucher, H.; Weichert, H.; Ziegler, J.; Feussner, I.; Wasternack, C. Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol. Biol. 2003, 51, 895–911. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.; Ponce, M. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Turner, J.G.; Ellis, C.; Devoto, A. The jasmonate signal pathway. Plant Cell 2002, 14, S153–S164. [Google Scholar] [CrossRef]
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.-M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350. [Google Scholar] [CrossRef] [PubMed]
- VanEtten, H.D.; Mansfield, J.W.; Bailey, J.A.; Farmer, E.E. Two classes of plant antibiotics: Phytoalexins versus “phytoanticipins”. Plant Cell 1994, 6, 1191. [Google Scholar] [CrossRef] [PubMed]
- Bera, S.; Blundell, R.; Liang, D.; Crowder, D.; Casteel, C. The oxylipin signaling pathway is required for increased aphid attraction and retention on virus-infected plants. J. Chem. Ecol. 2020, 46, 771–781. [Google Scholar] [CrossRef]
- Broekaert, W.F.; Delauré, S.L.; De Bolle, M.F.; Cammue, B.P. The role of ethylene in host-pathogen interactions. Annu. Rev. Phytopathol. 2006, 44, 393–416. [Google Scholar] [CrossRef]
- Lorenzo, O.; Chico, J.M.; Saénchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Laudert, D.; Weiler, E.W. Allene oxide synthase: A major control point in Arabidopsis thaliana octadecanoid signalling. Plant J. 1998, 15, 675–684. [Google Scholar] [CrossRef]
- Wasternack, C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef]
- Laudert, D.; Schaller, F.; Weiler, E.W. Transgenic Nicotiana tabacum and Arabidopsis thaliana plants overexpressing allene oxide synthase. Planta 2000, 211, 163–165. [Google Scholar] [CrossRef] [PubMed]
- Park, C.J.; Kim, K.J.; Shin, R.; Park, J.M.; Shin, Y.C.; Paek, K.H. Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. Plant J. 2004, 37, 186–198. [Google Scholar] [CrossRef] [PubMed]
- van Loon, L.C.; Rep, M.; Pieterse, C.M. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef]
- Campos, M.L.; Yoshida, Y.; Major, I.T.; de Oliveira Ferreira, D.; Weraduwage, S.M.; Froehlich, J.E.; Johnson, B.F.; Kramer, D.M.; Jander, G.; Sharkey, T.D. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 2016, 7, 12570. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, N.; Pichersky, E.; Gershenzon, J. Biochemistry of plant volatiles. Plant Physiol. 2004, 135, 1893–1902. [Google Scholar] [CrossRef] [PubMed]
- Huot, O.B.; Levy, J.G.; Tamborindeguy, C. Global gene regulation in tomato plant (Solanum lycopersicum) responding to vector (Bactericera cockerelli) feeding and pathogen (‘Candidatus Liberibacter solanacearum’) infection. Plant Mol. Biol. 2018, 97, 57–72. [Google Scholar] [CrossRef]
- Wang, X.-W.; Li, P.; Liu, S.-S. Whitefly interactions with plants. Curr. Opin. Insect Sci. 2017, 19, 70–75. [Google Scholar] [CrossRef]
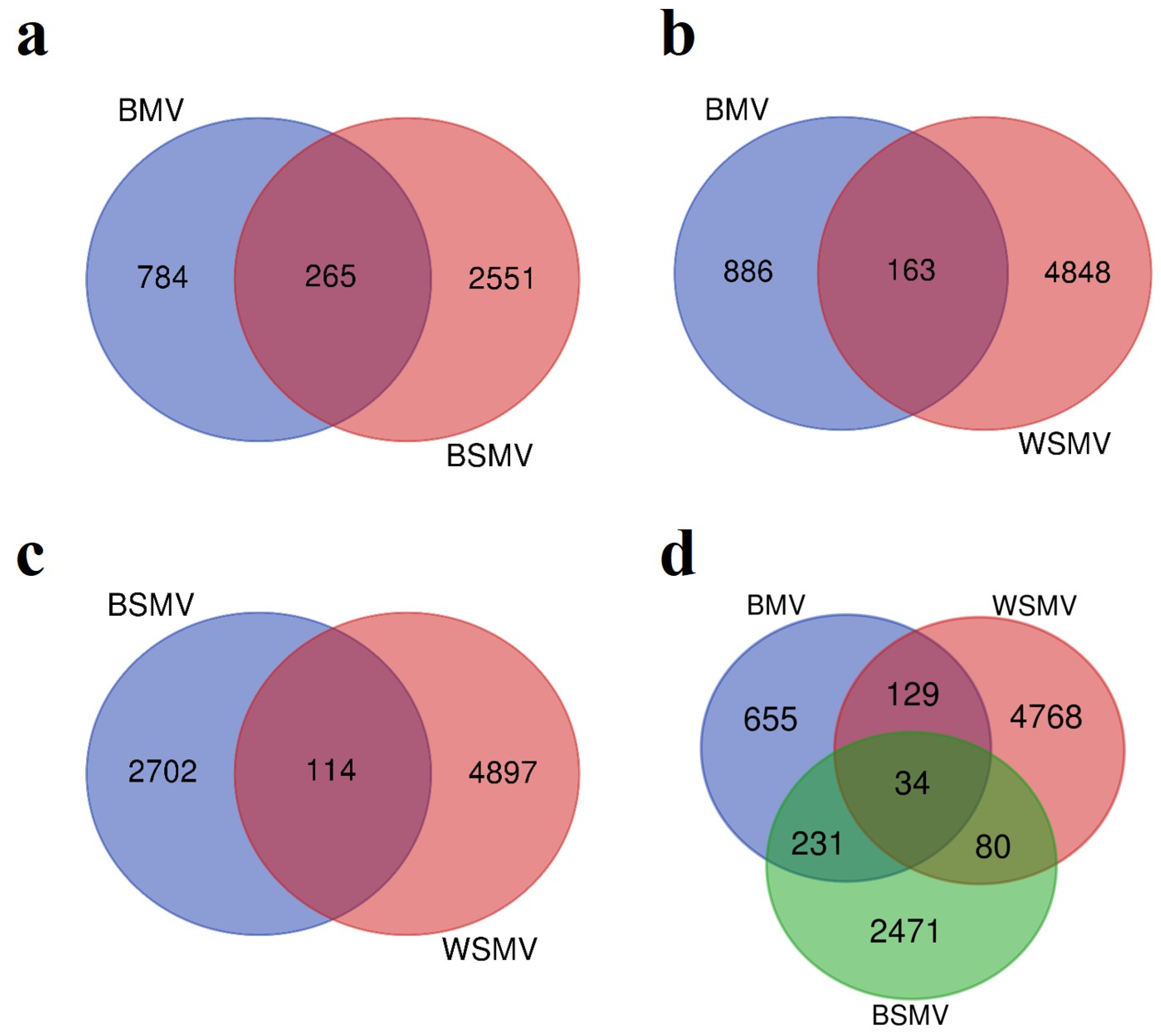
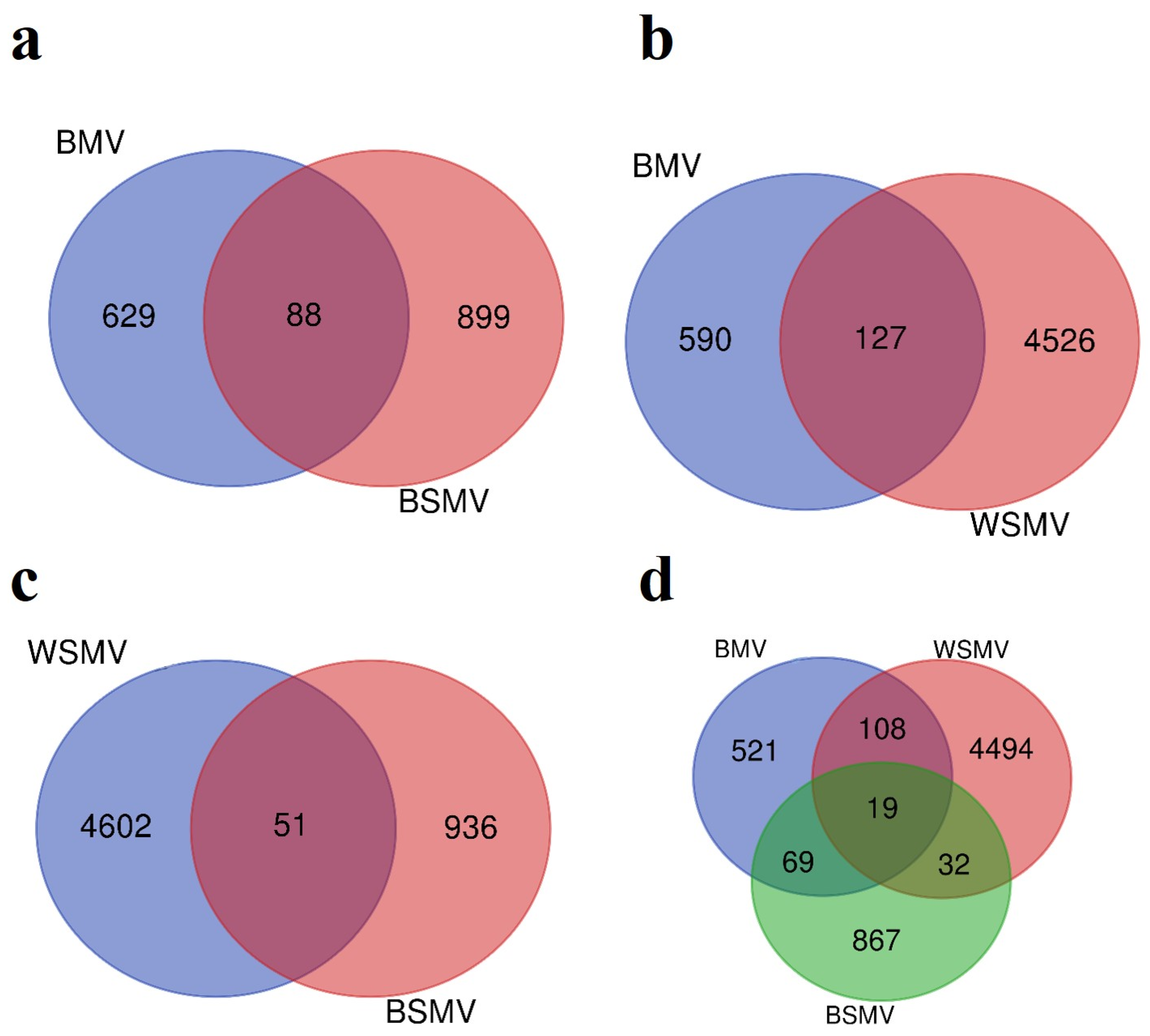
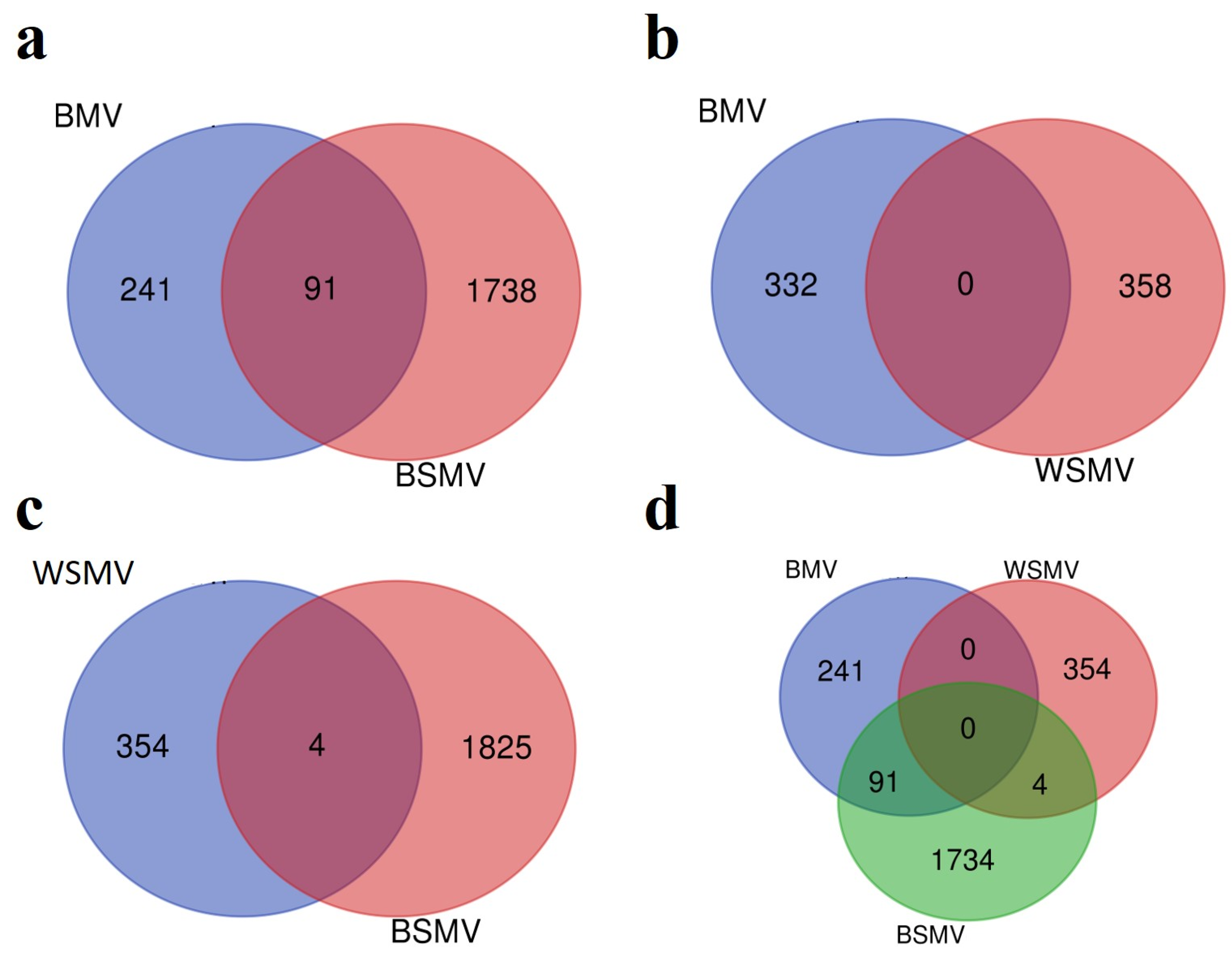
| Accession Number | Type | Platform | Sample Number | Tissue | Released | ||
|---|---|---|---|---|---|---|---|
| Control | Treatment | Total | |||||
| SRP349660 | WSMV infection | Illumina HiSeq 2000 | 8 | 8 | 16 | Leaf | 30 April 2022 |
| ERP128185 | BSMV infection | Illumina NovaSeq 6000 | 6 | 6 | 12 | Leaf & root | 6 June 2022 |
| ERP128185 | BMV infection | Illumina NovaSeq 6000 | 4 | 4 | 8 | Leaf & root | 6 June 2022 |
| TF Families | Genes Number | |||||
|---|---|---|---|---|---|---|
| WSMV | BMV | BSMV | BMV&WSMV | BMV&BSMV | BSMV&WSMV | |
| AP2/ERF-AP2 | ND | 2 | ND | 2 | 2 | ND |
| AP2/ERF-ERF | 23 | 3 | 10 | 30 | 12 | 27 |
| AP2/ERF-RAV | 1 | ND | ND | 2 | ND | 2 |
| B3 | 2 | 1 | 14 | 3 | 15 | 2 |
| bHLH | 6 | 7 | 30 | 13 | 34 | 7 |
| bZIP | 6 | ND | 5 | 9 | 5 | 9 |
| C2C2-CO-like | ND | 2 | ND | 2 | 2 | ND |
| C2C2-Dof | 4 | 5 | 1 | 9 | 6 | 4 |
| C2C2-GATA | 1 | ND | ND | 1 | ND | 1 |
| C2C2-LSD | 1 | ND | ND | 1 | ND | 1 |
| C2C2-YABBY | ND | ND | 3 | ND | 3 | ND |
| C2H2 | 28 | 6 | 8 | 36 | 14 | 30 |
| C3H | 5 | 1 | 4 | 9 | 4 | 8 |
| CPP | 5 | 1 | ND | 6 | 1 | 5 |
| CSD | 3 | ND | ND | 3 | ND | 3 |
| E2F-DP | 2 | ND | ND | 2 | ND | 2 |
| EIL | ND | ND | 1 | ND | 1 | ND |
| GARP-G2-like | ND | 2 | 2 | 4 | 2 | 2 |
| GeBP | ND | ND | 1 | ND | 1 | ND |
| GRAS | 8 | 3 | 2 | 12 | 5 | 9 |
| GRF | ND | ND | 3 | ND | 3 | ND |
| HB-BELL | 1 | ND | ND | 2 | ND | 2 |
| HB-HD-ZIP | 5 | ND | 7 | 5 | 7 | 5 |
| HB-other | 1 | 1 | 1 | 2 | 2 | 1 |
| HB-PHD | ND | ND | 1 | ND | 1 | ND |
| HB-WOX | ND | ND | 1 | ND | 1 | ND |
| HSF | 5 | ND | 1 | 6 | 1 | ND |
| LIM | 1 | ND | ND | 1 | ND | 1 |
| LOB | 3 | ND | 1 | 3 | 1 | 3 |
| MADS-MIKC | 2 | 1 | 8 | 3 | 9 | 2 |
| MADS-M-type | ND | ND | 1 | ND | 1 | ND |
| MYB | 5 | 3 | 6 | 11 | 8 | 8 |
| MYB-related | 4 | 3 | 7 | 7 | 10 | 4 |
| NAC | 38 | 13 | 10 | 61 | 22 | 57 |
| NF-X1 | 1 | ND | ND | 3 | ND | 3 |
| NF-YA | ND | 3 | 2 | 3 | 5 | ND |
| NF-YB | ND | ND | 1 | ND | 1 | ND |
| OFP | ND | 3 | ND | 3 | 3 | ND |
| RWP-RK | ND | ND | 2 | ND | 2 | ND |
| Tify | 4 | ND | ND | 4 | ND | 4 |
| Trihelix | 5 | 3 | ND | 7 | 3 | 5 |
| WRKY | 14 | 12 | 6 | 24 | 17 | 16 |
| PK Families | Genes Number | |||||
|---|---|---|---|---|---|---|
| WSMV | BMV | BSMV | BMV&WSMV | BMV&BSMV | BSMV&WSMV | |
| AGC-Pl | ND | ND | 2 | ND | 2 | ND |
| CAMK_AMPK | 1 | ND | ND | 1 | ND | 1 |
| CAMK_CAMKL-CHK1 | 4 | 1 | 2 | 5 | 3 | 4 |
| CAMK_CDPK | 9 | ND | 3 | 13 | 3 | 13 |
| CK1_CK1 | ND | ND | 2 | ND | 2 | ND |
| CMGC_CDK-CRK7-CDK9 | 3 | 3 | ND | 6 | 3 | 3 |
| CMGC_CDKL-Os | 1 | 1 | 2 | 2 | 3 | 1 |
| CMGC_GSK | 2 | ND | ND | 2 | ND | 2 |
| CMGC_MAPK | ND | 1 | 2 | 1 | 2 | ND |
| Group-Pl-4 | 2 | ND | ND | 2 | ND | 2 |
| IRE1 | 1 | ND | ND | 1 | ND | 1 |
| NAK | 3 | ND | ND | 3 | ND | 3 |
| RLK-Pelle_CR4L | 1 | 3 | 1 | 4 | 4 | 1 |
| RLK-Pelle_CrRLK1L-1 | 1 | 1 | 4 | 1 | 4 | 1 |
| RLK-Pelle_DLSV | 47 | 24 | 66 | 76 | 78 | 55 |
| RLK-Pelle_Extensin | ND | 1 | 1 | 1 | 1 | ND |
| RLK-Pelle_L-LEC | 29 | 10 | 16 | 41 | 22 | 32 |
| RLK-Pelle_LRK10L-2 | 8 | ND | 10 | 12 | 10 | 12 |
| RLK-Pelle_LRR-I-1 | 1 | 2 | 4 | 3 | 6 | 1 |
| RLK-Pelle_LRR-II | 2 | ND | ND | 4 | ND | 4 |
| RLK-Pelle_LRR-III | ND | 1 | 1 | 1 | 2 | ND |
| RLK-Pelle_LRR-VI-1 | ND | ND | 1 | ND | 1 | ND |
| RLK-Pelle_LRR-VI-2 | 6 | ND | ND | 6 | ND | 6 |
| RLK-Pelle_LRR-VII-2 | ND | ND | ND | 1 | ND | 15 |
| RLK-Pelle_LRR-VIII-1 | 1 | ND | 2 | 1 | 2 | 1 |
| RLK-Pelle_LRR-Xb-1 | 5 | 1 | ND | 6 | 1 | 26 |
| RLK-Pelle_LRR-Xb-2 | 3 | ND | ND | 3 | ND | 3 |
| RLK-Pelle_LRR-XI-1 | 9 | 3 | 12 | 13 | 15 | 10 |
| RLK-Pelle_LRR-XII-1 | 20 | 7 | 16 | 33 | 19 | 26 |
| RLK-Pelle_LRR-XIIIa | ND | ND | 1 | ND | 1 | ND |
| RLK-Pelle_LRR-XIV | 1 | ND | ND | 1 | ND | 1 |
| RLK-Pelle_LysM | 5 | ND | ND | 5 | ND | 5 |
| RLK-Pelle_PERK-1 | 2 | ND | ND | 5 | ND | 5 |
| RLK-Pelle_RLCK-IV | ND | 2 | 1 | 2 | 3 | ND |
| RLK-Pelle_RLCK-Ixb | 2 | 6 | 2 | 8 | 8 | 2 |
| RLK-Pelle_RLCK-V | ND | ND | 1 | ND | 1 | ND |
| RLK-Pelle_RLCK-VIIa-2 | 12 | 5 | 3 | 20 | 8 | 15 |
| RLK-Pelle_RLCK-VIII | 6 | ND | 1 | 6 | 1 | 6 |
| RLK_Pelle RLCK-XI | ND | ND | ND | 1 | 1 | ND |
| RLK-Pelle_SD-2b | 13 | 8 | 13 | 21 | 18 | 18 |
| RLK_Pelle URK-3 | ND | ND | ND | 1 | 1 | ND |
| RLK-Pelle_WAK | 17 | 7 | 19 | 23 | 24 | 19 |
| RLK-Pelle_WAK_LRK10L-1 | 5 | 14 | 5 | 39 | 19 | 25 |
| STE_STE11 | 4 | ND | 1 | 7 | 1 | 7 |
| STE_STE7 | ND | ND | ND | 2 | 1 | 1 |
| TKL_Gdt | ND | ND | ND | ND | 1 | ND |
| TKL-Pl-4 | ND | ND | ND | 1 | ND | 1 |
| TKL-Pl-6 | 3 | ND | ND | 13 | ND | 13 |
| Motif | Motif Logo | E-Value | Width | Best Match in JASPAR | Significant GO Terms Identified by GOMO |
|---|---|---|---|---|---|
| Motif 1 |  | 6.90 × 10−42 | 29 | MA1723.1 (PRDM9) | MF: transcription factor activity |
| MA1833.1 (Zm00001d049364) | CC: plasma membrane | ||||
| MA2022.1 (LOB) | BP: regulation of transcription | ||||
| Motif 2 | 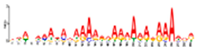 | 5.70 × 10−32 | 29 | MA1267.1 (DOF5.8) | MF: transcription factor activity |
| MA1268.1 (CDF5) | |||||
| MA1274.1 (DOF3.6) | |||||
| Motif 3 |  | 5.20 × 10−18 | 29 | MA1890.1 (Klf15) | MF: translation initiation factor activity |
| MA1833.1 (Zm00001d049364) | CC: CUL4 RING ubiquitin ligase complex | ||||
| MA1821.1 (Zm00001d020595) | BP: translation | ||||
| Motif 4 |  | 1.60 × 10−11 | 27 | MA0752.1 (ZNF410) | MF: structural constituent of ribosome |
| MA0410.1 (UGA3) | CC: chloroplast thylakoid membrane | ||||
| MA1642.1 (NEUROG2) | BP: trichome branching | ||||
| Motif 5 | 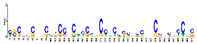 | 4.00 × 10−18 | 41 | MA1819.1 (Zm00001d005892) | MF: structural constituent of ribosome |
| MA1817.1 (Zm00001d020267) | CC: chloroplast stroma | ||||
| MA2022.1 (LOB) | BP: DNA replication initiation | ||||
| Motif 6 | 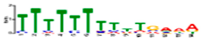 | 3.30 × 10−9 | 14 | MA1125.1 (ZNF384) | |
| MA0277.1 (AZF1) | CC: nucleus | ||||
| MA0548.2 (AGL15) | |||||
| Motif 7 |  | 6.50 × 10−9 | 21 | MA1257.1 (ERF9) | MF: nucleotide binding |
| MA1246.1 (LEP) | CC: chloroplast stroma | ||||
| MA1239.1 (ERF104) | BP: translation | ||||
| Motif 8 |  | 2.20 × 10−5 | 41 | MA1833.1 (Zm00001d049364) | MF: ATP binding |
| MA1246.1 (LEP) | CC: nucleus | ||||
| MA1257.1 (ERF9) | BP: potassium ion transport | ||||
| Motif 9 | 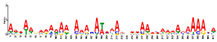 | 4.20 × 10−9 | 41 | MA1267.1 (DOF5.8) | MF: ATP binding |
| MA1871.1 (FoxM) | CC: CUL4 RING ubiquitin ligase complex | ||||
| MA1866.1 (FoxI-a) | BP: DNA replication initiation | ||||
| Motif 10 | 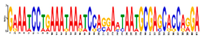 | 1.20 × 10−3 | 41 | MA0762.1 (ETV2) | MF: transcription factor activity |
| MA1367.1 (AT1G76870) | CC: plasma membrane | ||||
| MA0679.2 (ONECUT1) | BP: regulation of transcription | ||||
| Motif 11 |  | 3.80 × 10−3 | 29 | MA1630.2 (ZNF281) | MF: DNA-directed DNA polymerase activity |
| MA1713.1 (ZNF610) | CC: CUL4 RING ubiquitin ligase complex | ||||
| MA0528.2 (ZNF263) | BP: translation |
| Motif | Motif Logo | E-Value | Width | Best Match in JASPAR | Significant GO Terms Identified by GOMO |
|---|---|---|---|---|---|
| Motif 1 |  | 4.90 × 10−88 | 29 | MA1833.1 (Zm00001d049364) | MF: translation initiation factor activity |
| MA1817.1 (Zm00001d020267) | BP: DNA replication initiation | ||||
| MA1820.1 (Zm00001d024324) | CC: CUL4 RING ubiquitin ligase complex | ||||
| Motif 2 | 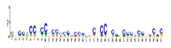 | 2.90 × 10−57 | 41 | MA1890.1 (Klf15) | ND |
| MA1892.1 (Klf5-like) | |||||
| MA1653.1 (ZNF148) | |||||
| Motif 3 |  | 3.20 × 10−35 | 15 | MA1274.1 (DOF3.6) | MF: structural constituent of ribosome |
| MA1268.1 (CDF5) | BP: megagametogenesis | ||||
| MA1267.1 (DOF5.8) | CC: CUL4 RING ubiquitin ligase complex | ||||
| Motif 4 |  | 4.80 × 10−25 | 41 | MA1818.1 (Zm00001d052229) | MF: ATP binding |
| MA1833.1 (Zm00001d049364) | BP: xanthophyll biosynthetic process | ||||
| MA1817.1 (Zm00001d020267) | CC: chloroplast stroma | ||||
| Motif 5 |  | 6.90 × 10−19 | 29 | MA1267.1 (DOF5.8) | MF: structural constituent of ribosome |
| MA1274.1 (DOF3.6) | BP: translation | ||||
| MA1268.1 (CDF5) | CC: CUL4 RING ubiquitin ligase complex | ||||
| Motif 6 | 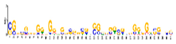 | 5.60 × 10−15 | 41 | MA1833.1 (Zm00001d049364) | MF: structural constituent of ribosome |
| MA1819.1 (Zm00001d005892) | BP: translation | ||||
| MA1832.1 (Zm00001d002364) | CC: respiratory chain complex I | ||||
| Motif 7 |  | 1.30 × 10−15 | 21 | MA0528.2 (ZNF263) | ND |
| MA1226.1 (DREB2G) | |||||
| MA1890.1 (Klf15) | |||||
| Motif 8 |  | 2.00 × 10−8 | 41 | MA1354.1 (AT4G12670) | MF: transcription factor activity |
| MA1210.2 (HAT22) | BP: regulation of transcription | ||||
| MA0389.1 (SRD1) | CC: nucleus | ||||
| Motif 9 | 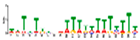 | 2.90 × 10−8 | 21 | MA1267.1 (DOF5.8) | MF: transcription factor activity |
| MA1281.1 (DOF5.1) | BP: response to brassinosteroid stimulus | ||||
| MA1277.1 (DOF1.7) | CC: nucleus | ||||
| Motif 10 |  | 3.00 × 10−6 | 50 | MA1007.1 (PHYPADRAFT_173530) | MF: transcription factor activity |
| MA1023.1 (PHYPADRAFT_28324) | BP: response to water deprivation | ||||
| MA0986.1 (DREB2C) | CC: nucleus | ||||
| Motif 11 | 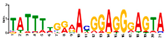 | 2.10 × 10−5 | 21 | MA0687.1 (SPIC) | MF: transcription factor activity |
| MA0752.1 (ZNF410) | BP: regulation of transcription | ||||
| MA0410.1 (UGA3) | CC: nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahriari, A.G.; Majláth, I.; Aliakbari, M.; Ghodoum Parizipour, M.H.; Tahmasebi, A.; Nami, F.; Tahmasebi, A.; Taherishirazi, M. Identifying Critical Regulators in the Viral Stress Response of Wheat (Triticum aestivum L.) Using Large-Scale Transcriptomics Data. Agronomy 2023, 13, 2610. https://doi.org/10.3390/agronomy13102610
Shahriari AG, Majláth I, Aliakbari M, Ghodoum Parizipour MH, Tahmasebi A, Nami F, Tahmasebi A, Taherishirazi M. Identifying Critical Regulators in the Viral Stress Response of Wheat (Triticum aestivum L.) Using Large-Scale Transcriptomics Data. Agronomy. 2023; 13(10):2610. https://doi.org/10.3390/agronomy13102610
Chicago/Turabian StyleShahriari, Amir Ghaffar, Imre Majláth, Massume Aliakbari, Mohamad Hamed Ghodoum Parizipour, Aminallah Tahmasebi, Fatemeh Nami, Ahmad Tahmasebi, and Mohsen Taherishirazi. 2023. "Identifying Critical Regulators in the Viral Stress Response of Wheat (Triticum aestivum L.) Using Large-Scale Transcriptomics Data" Agronomy 13, no. 10: 2610. https://doi.org/10.3390/agronomy13102610
APA StyleShahriari, A. G., Majláth, I., Aliakbari, M., Ghodoum Parizipour, M. H., Tahmasebi, A., Nami, F., Tahmasebi, A., & Taherishirazi, M. (2023). Identifying Critical Regulators in the Viral Stress Response of Wheat (Triticum aestivum L.) Using Large-Scale Transcriptomics Data. Agronomy, 13(10), 2610. https://doi.org/10.3390/agronomy13102610





