Deciphering the Roles of Peanut (Arachis hypogaea L.) Type-One Protein Phosphatase (TOPP) Family in Abiotic Stress Tolerance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Annotation of TOPPs from Peanut
2.2. Multiple Sequence Alignment and Analysis of Phylogenetics
2.3. Gene Structure Analysis and Identification of Conserved Domains
2.4. Chromosomal Location and Gene Duplication
2.5. Prediction of cis-Regulatory Elements
2.6. Expression Analysis of Peanut TOPP Genes
2.7. Prediction of TFs Involved in Controlling Peanut TOPP Expression
2.8. GO and KEGG Enrichment Analysis
3. Results
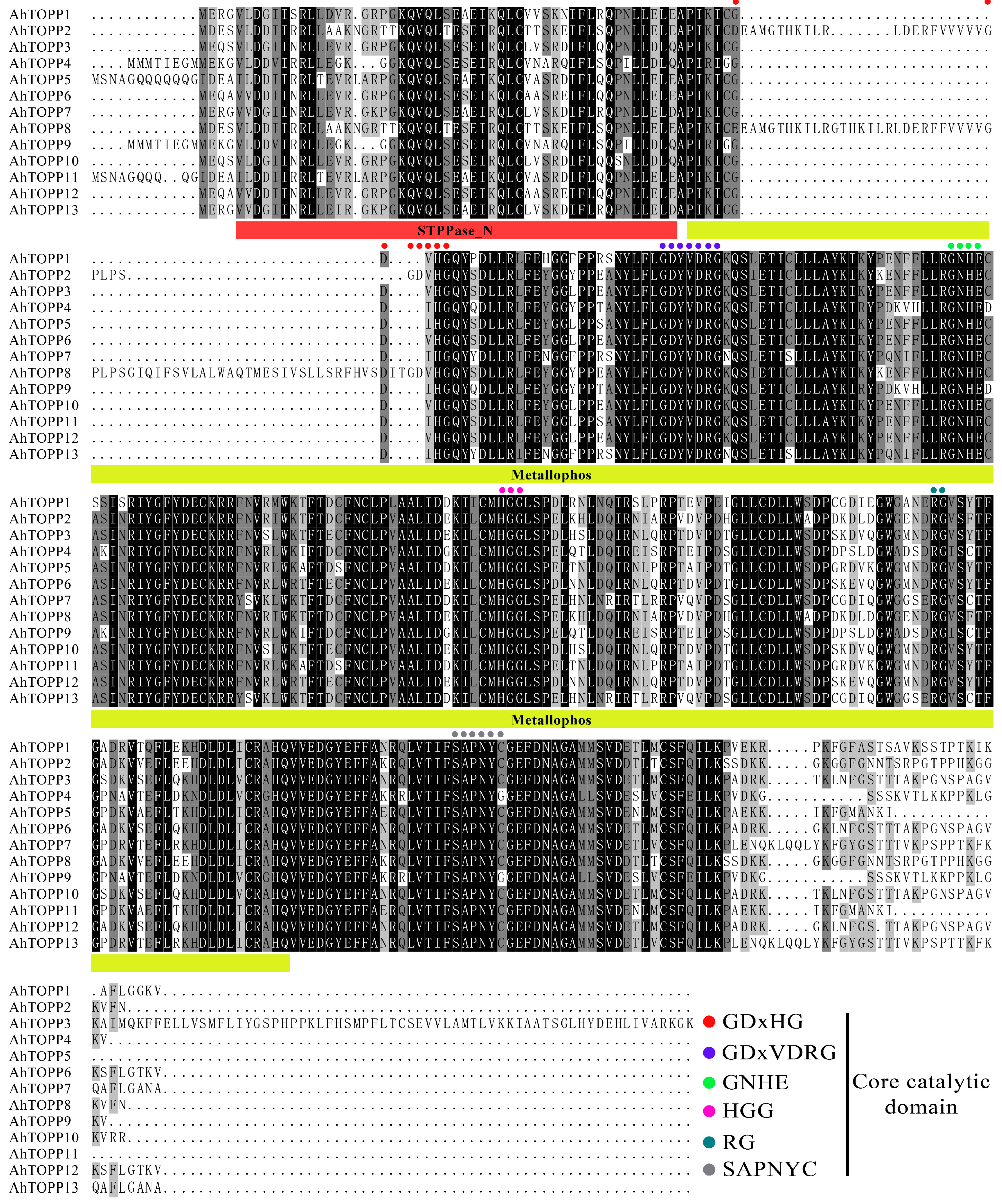
3.1. Identification and Sequence Analysis of the Peanut TOPP Genes
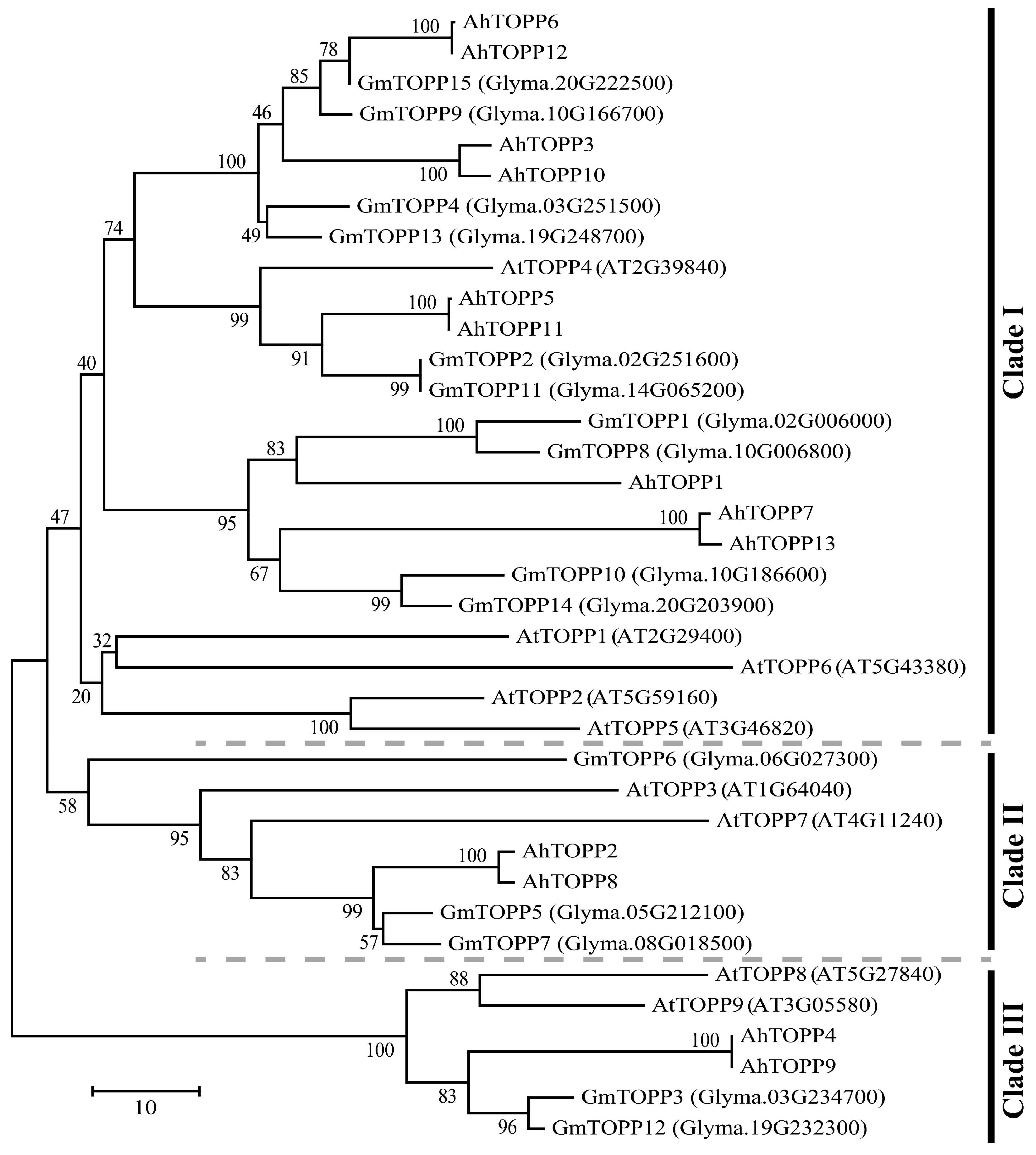
3.2. Phylogenetic Analysis of Peanut TOPP Genes
3.3. Conserved Motifs and Gene Structure
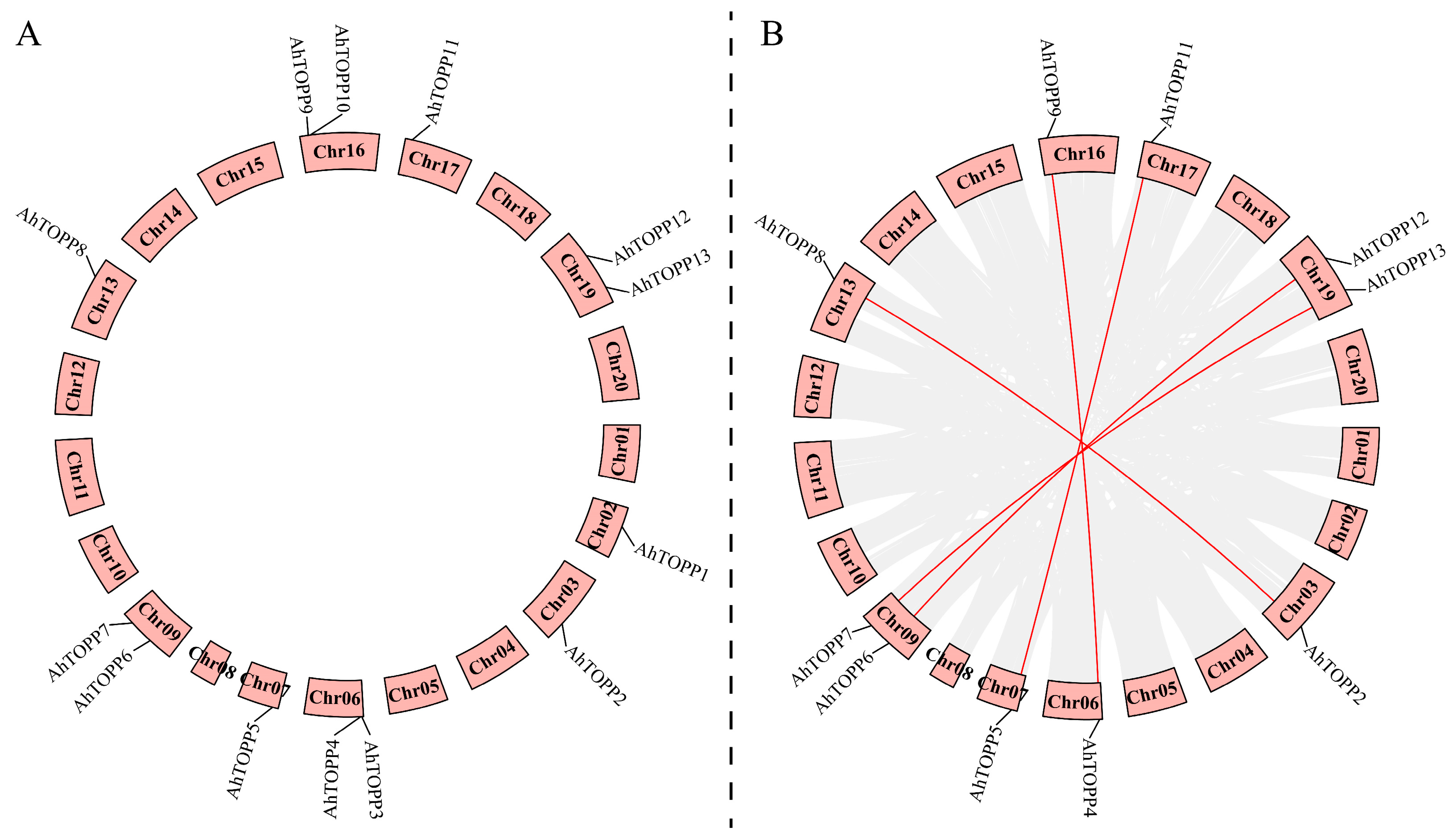
3.4. Chromosomal Localization and Duplication Events
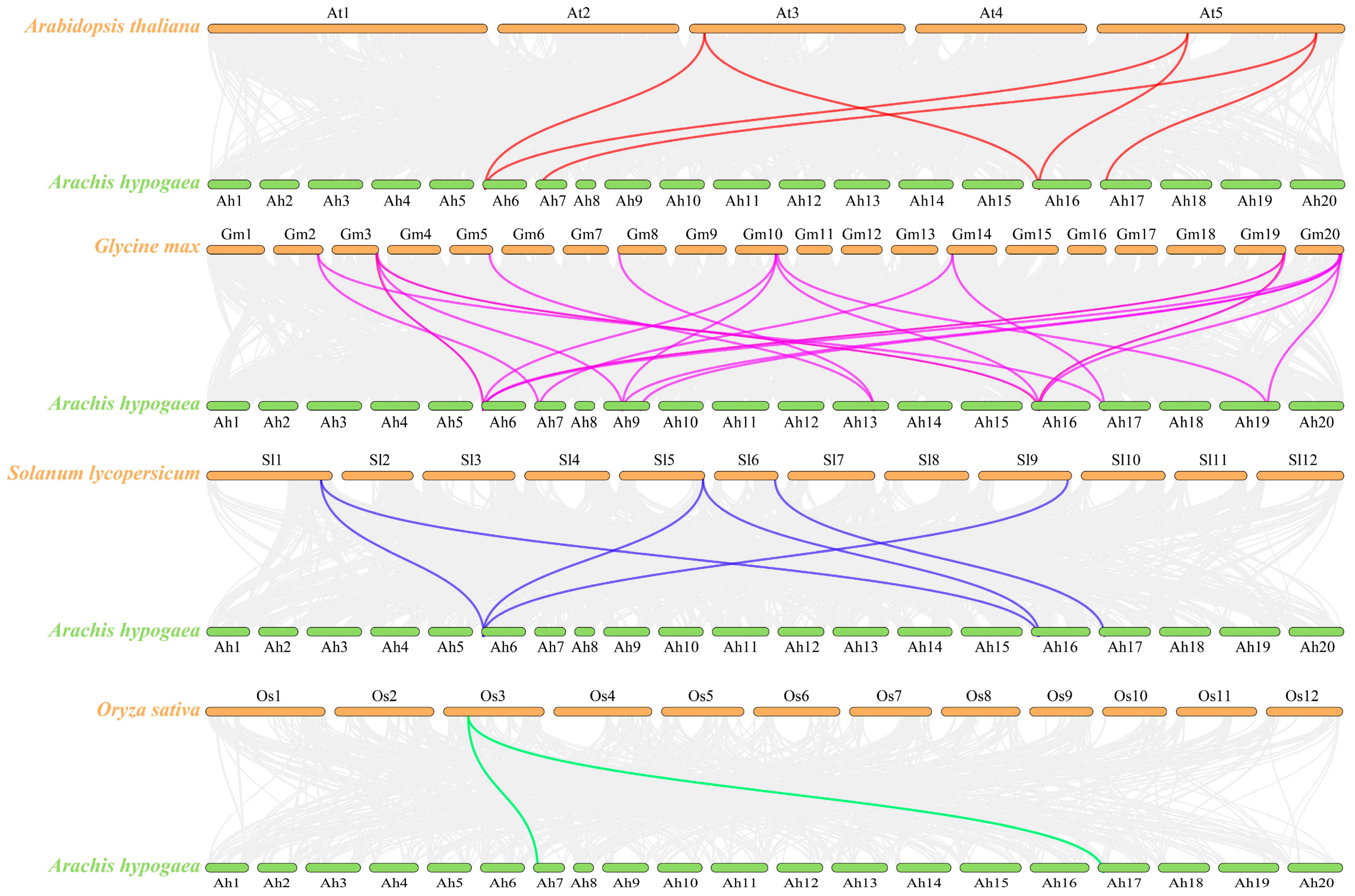
3.5. Analysis of Peanut TOPP Genes Syntenic
3.6. Cis-Regulatory Elements Analysis of Peanut TOPP Genes Promoter
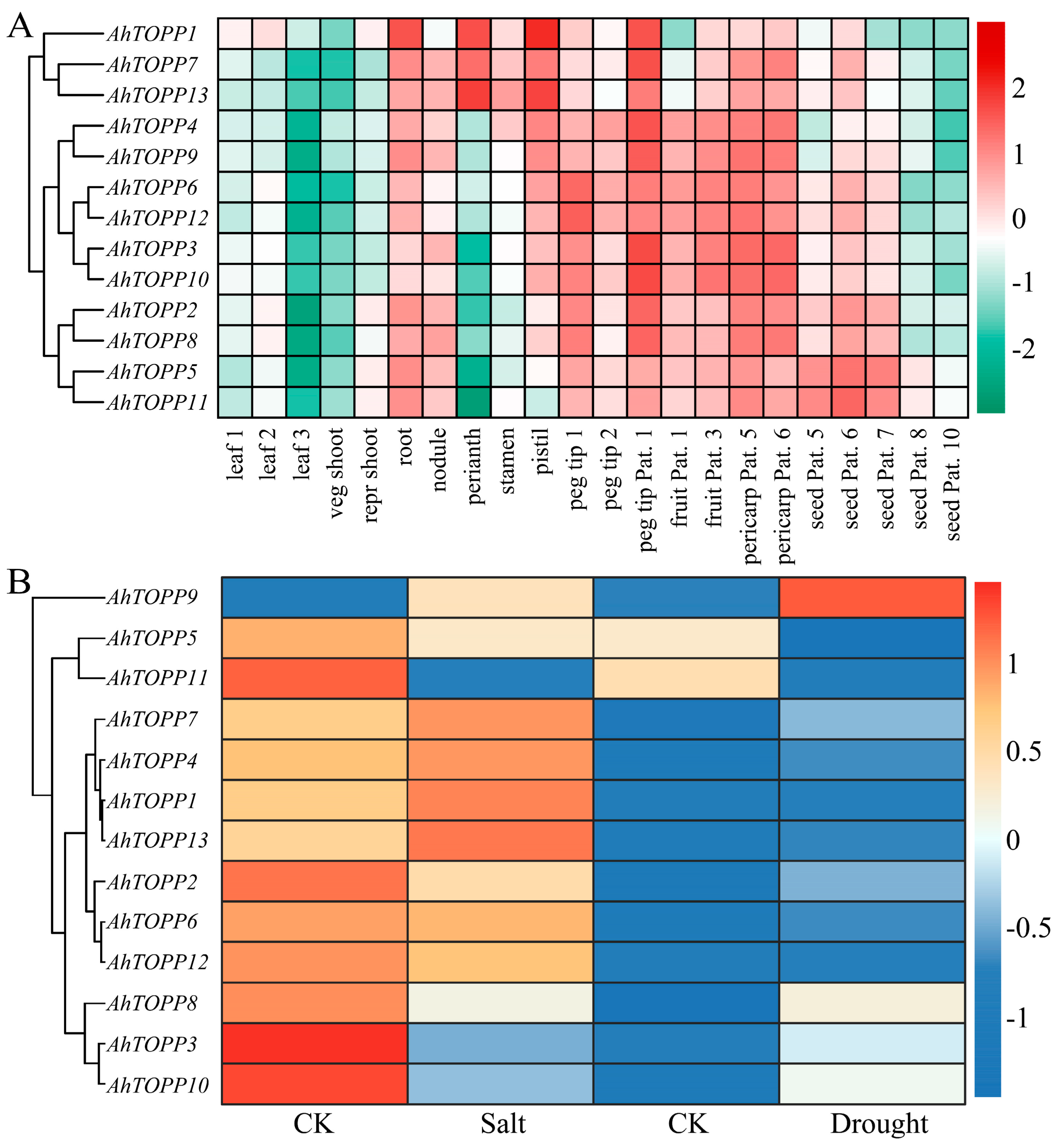
3.7. Transcriptome Profiling of AhTOPPs in Different Tissues and in Response to Stress Treatments
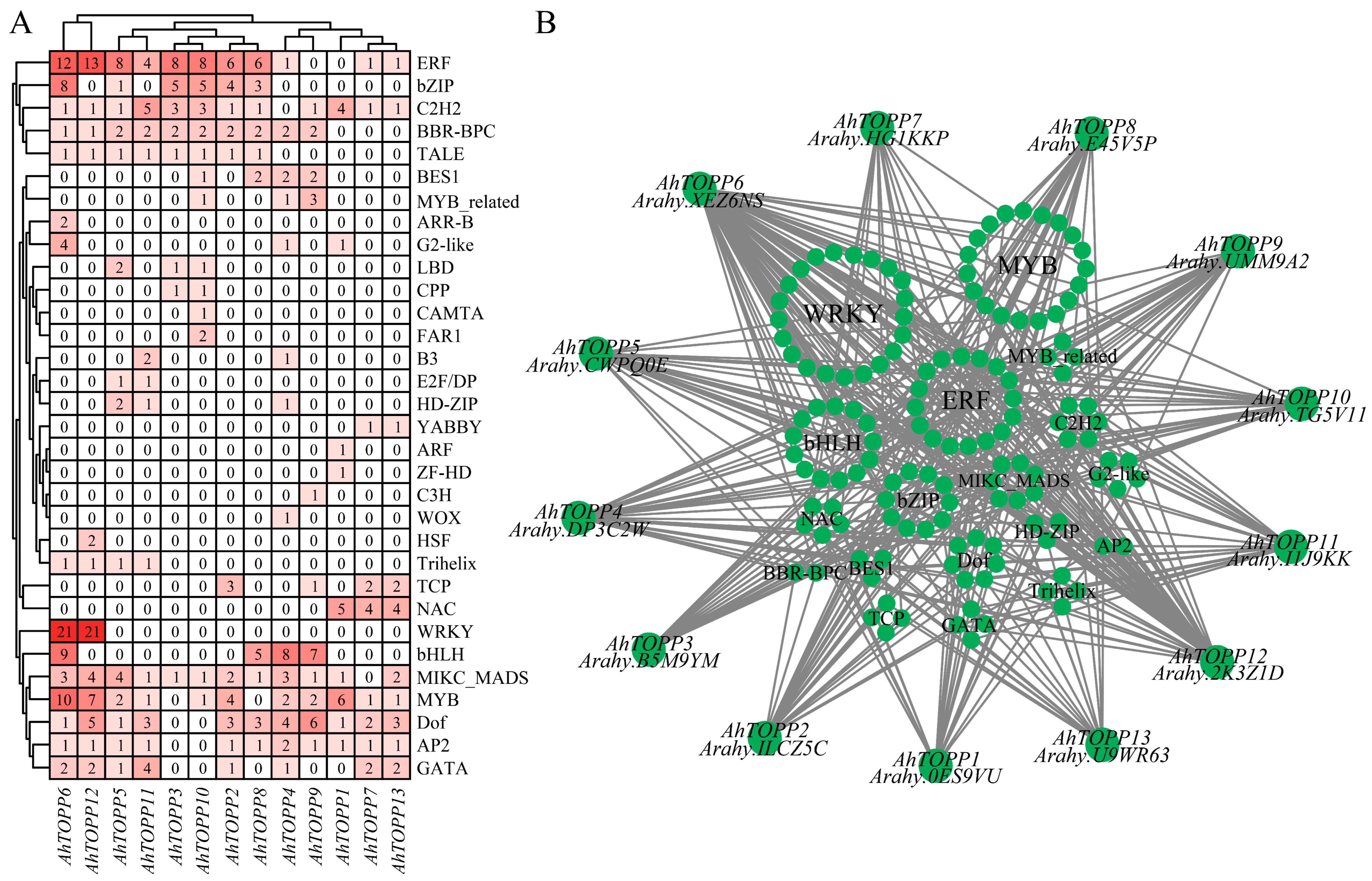
3.8. Prediction of Regulatory Network
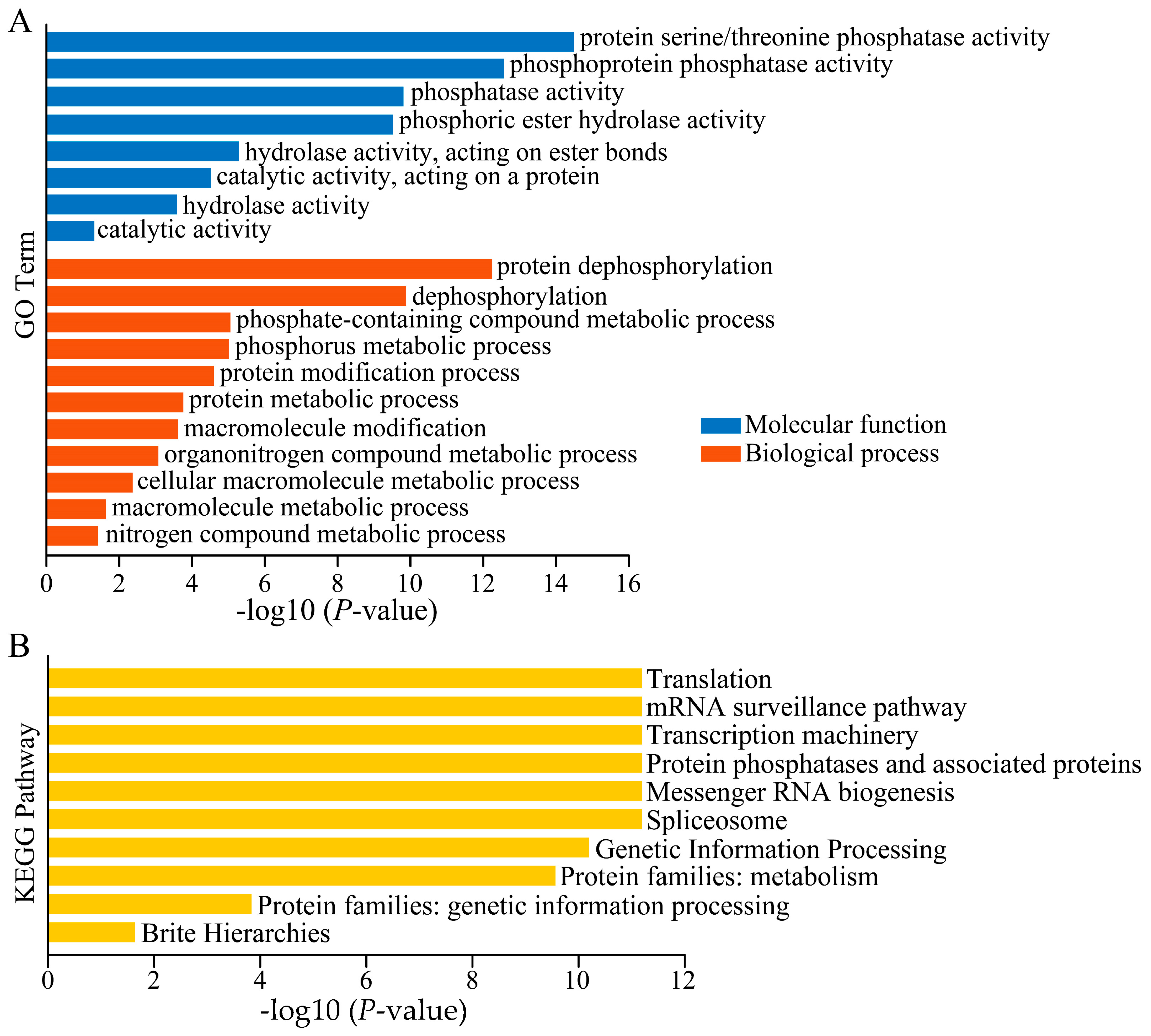
3.9. GO and KEGG Analysis of Peanut TOPP Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Umezawa, T.; Fujita, M.; Fujita, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Engineering drought tolerance in plants: Discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 2006, 17, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Giri, J.; Kapoor, S.; Tyagi, A.K.; Pandey, G.K. Protein phosphatase complement in rice: Genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genom. 2010, 11, 435. [Google Scholar] [CrossRef] [PubMed]
- Hunter, T. Protein kinases and phosphatases: The yin and yang of protein phosphorylation and signaling. Cell 1995, 80, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.; Gack, M.U. Post-translational Control of Intracellular Pathogen Sensing Pathways. Trends Immunol. 2017, 38, 39–52. [Google Scholar] [CrossRef]
- Barford, D.; Das, A.K.; Egloff, M.P. The structure and mechanism of protein phosphatases: Insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct. 1998, 27, 133–164. [Google Scholar] [CrossRef]
- Kerk, D.; Templeton, G.; Moorhead, G.B. Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol. 2008, 146, 351–367. [Google Scholar] [CrossRef]
- Heroes, E.; Lesage, B.; Görnemann, J.; Beullens, M.; Van Meervelt, L.; Bollen, M. The PP1 binding code: A molecular-lego strategy that governs specificity. FEBS J. 2013, 280, 584–595. [Google Scholar] [CrossRef]
- Cohen, P.T. Protein phosphatase 1—Targeted in many directions. J. Cell Sci. 2002, 115, 241–256. [Google Scholar] [CrossRef]
- Bradai, M.; Mahjoubi, H.; Chini, A.; Chabouté, M.E.; Hanin, M.; Ebel, C. Genome wide identification of wheat and Brachypodium type one protein phosphatases and functional characterization of durum wheat TdPP1a. PLoS ONE 2018, 13, e0191272. [Google Scholar] [CrossRef]
- Wang, S.; Guo, J.; Zhang, Y.; Guo, Y.; Ji, W. Genome-wide characterization and expression analysis of TOPP-type protein phosphatases in soybean (Glycine max L.) reveal the role of GmTOPP13 in drought tolerance. Genes Genom. 2021, 43, 783–796. [Google Scholar] [CrossRef]
- Farkas, I.; Dombrádi, V.; Miskei, M.; Szabados, L.; Koncz, C. Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci. 2007, 12, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, D.; Morita, H.; Hattori, T.; Takeda, S. Molecular characterization of the rice protein RSS1 required for meristematic activity under stressful conditions. Plant Physiol. Biochem. 2012, 61, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Takemiya, A.; Kinoshita, T.; Asanuma, M.; Shimazaki, K. Protein phosphatase 1 positively regulates stomatal opening in response to blue light in Vicia faba. Proc. Natl. Acad. Sci. USA 2006, 103, 13549–13554. [Google Scholar] [CrossRef] [PubMed]
- Bheri, M.; Mahiwal, S.; Sanyal, S.K.; Pandey, G.K. Plant protein phosphatases: What do we know about their mechanism of action? FEBS J. 2021, 288, 756–785. [Google Scholar] [CrossRef]
- Hou, Y.J.; Zhu, Y.; Wang, P.; Zhao, Y.; Xie, S.; Batelli, G.; Wang, B.; Duan, C.G.; Wang, X.; Xing, L.; et al. Type One Protein Phosphatase 1 and Its Regulatory Protein Inhibitor 2 Negatively Regulate ABA Signaling. PLoS Genet. 2016, 12, e1005835. [Google Scholar] [CrossRef]
- Templeton, G.W.; Nimick, M.; Morrice, N.; Campbell, D.; Goudreault, M.; Gingras, A.C.; Takemiya, A.; Shimazaki, K.; Moorhead, G.B. Identification and characterization of AtI-2, an Arabidopsis homologue of an ancient protein phosphatase 1 (PP1) regulatory subunit. Biochem. J. 2011, 435, 73–83. [Google Scholar] [CrossRef]
- Qin, Q.; Wang, W.; Guo, X.; Yue, J.; Huang, Y.; Xu, X.; Li, J.; Hou, S. Arabidopsis DELLA protein degradation is controlled by a type-one protein phosphatase, TOPP4. PLoS Genet. 2014, 10, e1004464. [Google Scholar] [CrossRef]
- Yue, J.; Qin, Q.; Meng, S.; Jing, H.; Gou, X.; Li, J.; Hou, S. TOPP4 Regulates the Stability of Phytochrome Interacting FACTOR5 During Photomorphogenesis in Arabidopsis. Plant Physiol. 2016, 170, 1381–1397. [Google Scholar] [CrossRef]
- Guo, X.; Qin, Q.; Yan, J.; Niu, Y.; Huang, B.; Guan, L.; Li, Y.; Ren, D.; Li, J.; Hou, S. Type-One Protein PHOSPHATASE4 Regulates Pavement Cell Interdigitation by Modulating PIN-FORMED1 Polarity and Trafficking in Arabidopsis. Plant Physiol. 2015, 167, 1058–1075. [Google Scholar] [CrossRef]
- Liao, Y.D.; Lin, K.H.; Chen, C.C.; Chiang, C.M. Oryza sativa protein phosphatase 1a (OsPP1a) involved in salt stress tolerance in transgenic rice. Mol. Breed. 2016, 36, 22. [Google Scholar] [CrossRef]
- Mou, Y.; Sun, Q.; Yuan, C.; Zhao, X.; Wang, J.; Yan, C.; Li, C.; Shan, S. Identification of the LOX Gene Family in Peanut and Functional Characterization of AhLOX29 in Drought Tolerance. Front. Plant Sci. 2022, 13, 832785. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Peng, Z.; Xu, P.; Tang, G.; Ma, C.; Zhu, J.; Shan, L.; Wan, S. Genome-Wide Identification of NAC Transcription Factors and Their Functional Prediction of Abiotic Stress Response in Peanut. Front. Genet. 2021, 12, 630292. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Xiao, D.; Wang, X.; Zhan, J.; Wang, A.; He, L. Genome-wide identification, evolutionary and expression analyses of LEA gene family in peanut (Arachis hypogaea L.). BMC Plant Biol. 2022, 22, 155. [Google Scholar] [CrossRef]
- Yu, S.; Wang, C.; Wang, Q.; Sun, Q.; Zhang, Y.; Dong, J.; Yin, Y.; Zhang, S.; Yu, G. Identification and Analysis of SOD Family Genes in Peanut (Arachis hypogaea L.) and Their Potential Roles in Stress Responses. Agronomy 2023, 13, 1959. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Goldberg, T.; Hecht, M.; Hamp, T.; Karl, T.; Yachdav, G.; Ahmed, N.; Altermann, U.; Angerer, P.; Ansorge, S.; Balasz, K.; et al. LocTree3 prediction of localization. Nucleic Acids Res. 2014, 42, W350–W355. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Clevenger, J.; Chu, Y.; Scheffler, B.; Ozias-Akins, P. A Developmental Transcriptome Map for Allotetraploid Arachis hypogaea. Front. Plant Sci. 2016, 7, 1446. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, X.; Sun, Q.; Yan, C.; Wang, J.; Yuan, C.; Li, C.; Shan, S.; Liu, F. Comparative Transcriptome Analysis Reveals Molecular Defensive Mechanism of Arachis hypogaea in Response to Salt Stress. Int. J. Genom. 2020, 2020, 6524093. [Google Scholar] [CrossRef]
- Zhao, X.; Li, C.; Wan, S.; Zhang, T.; Yan, C.; Shan, S. Transcriptomic analysis and discovery of genes in the response of Arachis hypogaea to drought stress. Mol. Biol. Rep. 2018, 45, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef] [PubMed]
- Powell, S.; Forslund, K.; Szklarczyk, D.; Trachana, K.; Roth, A.; Huerta-Cepas, J.; Gabaldón, T.; Rattei, T.; Creevey, C.; Kuhn, M.; et al. eggNOG v4.0: Nested orthology inference across 3686 organisms. Nucleic Acids Res. 2014, 42, D231–D239. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Uhrig, R.G.; Labandera, A.M.; Moorhead, G.B. Arabidopsis PPP family of serine/threonine protein phosphatases: Many targets but few engines. Trends Plant Sci. 2013, 18, 505–513. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, X.; Wang, X.; Zhou, M.; Zhou, X.; Ye, X.; Wei, X. An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New Phytol. 2012, 196, 1155–1170. [Google Scholar] [CrossRef]
- Olive, M.R.; Walker, J.C.; Singh, K.; Dennis, E.S.; Peacock, W.J. Functional properties of the anaerobic responsive element of the maize Adh1 gene. Plant Mol. Biol. 1990, 15, 593–604. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, J.; Qin, Q.; Zhang, J.; Chen, Y.; Zhao, L.; He, K.; Hou, S. Type one protein phosphatases (TOPPs) contribute to the plant defense response in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 360–377. [Google Scholar] [CrossRef]
- Wang, Q.; Li, X.; Guo, C.; Wen, L.; Deng, Z.; Zhang, Z.; Li, W.; Liu, T.; Guo, Y. Senescence-related receptor kinase 1 functions downstream of WRKY53 in regulating leaf senescence in Arabidopsis. J. Exp. Bot. 2023, 74, 5140–5152. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, X.; Ren, Z.; Abou-Elwafa, S.F.; Pu, X.; Zhu, Y.; Dou, D.; Su, H.; Cheng, H.; Liu, Z.; et al. ZmERF21 directly regulates hormone signaling and stress-responsive gene expression to influence drought tolerance in maize seedlings. Plant Cell Environ. 2022, 45, 312–328. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Shan, S. Deciphering the Roles of Peanut (Arachis hypogaea L.) Type-One Protein Phosphatase (TOPP) Family in Abiotic Stress Tolerance. Agronomy 2023, 13, 2444. https://doi.org/10.3390/agronomy13102444
Wang Q, Shan S. Deciphering the Roles of Peanut (Arachis hypogaea L.) Type-One Protein Phosphatase (TOPP) Family in Abiotic Stress Tolerance. Agronomy. 2023; 13(10):2444. https://doi.org/10.3390/agronomy13102444
Chicago/Turabian StyleWang, Qi, and Shihua Shan. 2023. "Deciphering the Roles of Peanut (Arachis hypogaea L.) Type-One Protein Phosphatase (TOPP) Family in Abiotic Stress Tolerance" Agronomy 13, no. 10: 2444. https://doi.org/10.3390/agronomy13102444
APA StyleWang, Q., & Shan, S. (2023). Deciphering the Roles of Peanut (Arachis hypogaea L.) Type-One Protein Phosphatase (TOPP) Family in Abiotic Stress Tolerance. Agronomy, 13(10), 2444. https://doi.org/10.3390/agronomy13102444




