Abstract
Kiwifruit is an economically important horticultural crop with extremely high values in nutrition and health care. However, the molecular mechanisms underlying fruit quality formation remain largely limited for most kiwifruit varieties. Recently, a new kiwifruit cultivar with a high level of soluble solids, Actinidia arguta cv. Qinziyu (full-red flesh) was discovered through the introduction and propagation test. To provide new insights into fruit quality formation in a typical kiwifruit cultivar, we integrated full-length transcriptome surveys based on PacBio single-molecule real-time (SMRT) sequencing, key enzyme genes expression involved in carbohydrate and amino acids metabolism pathways, and bHLH gene family analysis to enhance the understanding of soluble sugar, organic acid, and anthocyanin biosynthesis in A. arguta cv. Qinziyu. A total of 175,913 CCSs were generated, of which 124,789 were identified as FLNC transcripts. In total, 45,923 (86.99%) transcripts were successfully annotated, and more than 76.05% of the transcripts were longer than 1 Kb. KEGG pathway analysis showed that 630 candidate genes encoding 55 enzymes were mainly involved in carbohydrate and amino acid biosynthesis pathways. Further analysis verified the expression of 12 key enzyme genes (e.g., pyruvate kinase (PK), enolase (ENO), hexokinase (HK), and phosphoglycerate kinase (PGK)) in flowers using quantitative real-time PCR. Furthermore, we also screened 10 AabHLH proteins’ function in anthocyanin biosynthesis and characterized the AabHLH gene family in A. arguta cv. Qinziyu. Overall, our research data generated by SMRT technology provide the first set of gene isoforms from a full-length transcriptome in A. arguta cv. Qinziyu and more comprehensive insights into the molecular mechanism of fruit quality formation.
1. Introduction
Kiwifruit (genus Actinidia, Actinidiaceaeis) is a major liana fruit crop that originated in China and now is planted in temperate areas [1,2,3]. As an indispensable nutrition supplier for daily human consumption, kiwifruit offers abundant nutrients, including high contents of amino acids, minerals, and vitamin C [2,4]. Fruit quality is the most significant breeding goal that highly affects consumer preference and market competitiveness [5]. The most critical agronomic traits of kiwifruit such as taste, color, and size have a major impact on kiwifruit quality [6]. Sweetness is determined by soluble sugars, such as sucrose, glucose, and fructose, and organic acids [2,4], and the ratio of sugars to organic acids has a significant influence on the kiwifruit taste [3], whereas the flesh color is caused by the concentration of chlorophyll, carotenoids, and anthocyanins [7].
In recent years, most research has focused on breeding and evaluations of the horticultural merits of specific germplasms [8]. More than 60 cultivars of Actinidia are grown throughout the world [9]. Commercial cultivars are mainly domesticated or bred from two species: A. chinensis and A. deliciosa due to their various flesh colors (e.g., the green-, yellow-, and red-fleshed kiwifruit), unique flavor, large size, and long storage [10]. Recently, A. arguta, an all-red fleshed kiwifruit, has been domesticated and cultivated successfully in Korea and eastern Russia as well as some cold areas [11]. It is recognized as one of the most popular fruit in the local market, with a distinguishable appearance and flavor and the highest rates of anthocyanin [12,13,14]. At present, a new kiwifruit cultivar, Actinidia arguta cv. Qinziyu (full-red flesh), has been discovered through the introduction and propagation test (Figure 1). A. arguta cv. Qinziyu often has deep purple flesh when ripe with a long shelf life, storing for 2–3 weeks at 3–5 °C, and the fruit size ranges from 16 to 20 g. The advantage properties of A. arguta cv. Qinziyu, such as its unique aroma and its high levels of soluble solids and anthocyanin, are different from other cultivated types. The total soluble solids (TSS) in the fruit of A. arguta cv. Qinziyu are 18~22 Brix higher than that of A. chinensis and A. deliciosa (11.60~19.13 Brix) [3]. In kiwifruit, the TSSs mainly include sugars, acids, vitamins, certain minerals, and other soluble solids [15]. Previous studies have demonstrated that the transport, hydrolysis, and storage courses of sugars from source leaves to fruits are governed by well-understood metabolic pathways, such as central carbon metabolism, sucrose metabolism, glycolysis, and the tricarboxylic acid (TCA) cycle [16]. Moreover, sucrose acts as a signaling molecule and directly mediates amino acid metabolism [17], and other small molecule soluble substances such as organic acids, amino acids, and sugars are synthesized and accumulated [18]. Most studies suggest that carbon metabolism, the TCA cycle, and amino acid metabolism contribute to the accumulation and metabolism of sugars and organic acids, which is an important mechanism for fruit quality, e.g., [2,5,19]. Therefore, studying the soluble sugars and organic acid accumulation and regulation, highly correlated with carbohydrate and amino acid metabolism, of A. arguta cv. Qinziyu has become important for kiwifruit quality.

Figure 1.
The fruit of kiwifruit cultivar A. arguta cv. Qinziyu.
In addition, anthocyanins deriving from the flavonoid branch of the phenylpropanoid metabolic pathway have already been proven to contribute to the red color formation in many fruits [7,20]. Most enzyme genes governing anthocyanin biosynthesis have been extensively studied in kiwifruit [5,21,22]. Many important transcription factors, such as MYBs, bHLHs, and WD40s regulating the tissue-specific expression of typical anthocyanins by forming the MYB-bHLH-WD40 (MBW) complex, have also been isolated and functionally identified [5]. However, previous studies have mainly focused on flavor [2], volatile compounds [23], and flesh coloration [4] in various kiwifruit cultivars in China (e.g., A. chinensis cv. Hongyang, A. deliciosa cv. Xuxiang, A. eriantha, A. deliciosa) [2,4,22,23,24], and key biochemical pathways and regulatory genes that affect the accumulation of associated metabolism during fruit development and ripening have been revealed and identified. In addition, other members of the Actinidia genus, namely, A. chinensis, A. chinensis var. chinensis and A. chinensis cv. Hongyang, have more extensive genomic resources, including sequenced genomes [1,25,26]. Whereas little research was carried out in A. arguta cv. Qinziyu, further mechanism studies underlying its characteristic formation have not been performed. Thus, scientific information governing the TSS regulatory network as well as the color characteristics of kiwifruit cultivars is also limited. Therefore, an in-depth investigation is necessary to identify key candidate genes and transcription factors that regulate the soluble sugar, organic acid, and anthocyanins biosynthesis in A. arguta cv. Qinziyu for enhancing the fruit quality.
Transcriptome sequencing can provide valuable means of underlying the molecular basis of the physiological regulation process [23]. It has been used to identify putative genes for the biosynthesis and accumulation of metabolites such as sugars, amino acids, and anthocyanins in apple [5], plum [27], melon [28], and more. Such studies have successfully captured the important genes involved in the biosynthesis of the metabolites of interest [29], which are indispensable for the development and formation of quality fruit pathways. Single-molecule real-time (SMRT) isoform sequencing (Iso-Seq) using the PacBio platform captures a more accurate reference gene set with a high level of assembly completeness [30]. Thereby, PacBio SMRT sequencing technology has been successfully applied in gene annotation, isoform identification, long non-coding RNA (lncRNA) prediction, and alternative splicing (AS) discovery [31,32]. Hence, to enhance the understanding of the soluble sugar, organic acid, and anthocyanins biosynthesis present in typical kiwifruit variety A. arguta cv. Qinziyu, we integrated full-length transcriptome surveys based on PacBio SMRT sequencing, bHLH gene family analysis, and key enzyme gene expression involved in carbohydrate and amino acid biosynthesis pathways to provide new insights into kiwifruit quality formation. The findings from this study provide more comprehensive insights into the molecular regulatory mechanism of fruit quality formation and will be highly useful for the improvement of kiwifruit molecular breeding.
2. Materials and Methods
2.1. Plant Materials and RNA Preparation
Actinidia arguta cv. Qinziyu used in this study was successfully transplanted from the field to the germplasm nursery of Xi’an Botanical Garden of Shaanxi Province (Shaanxi, China, 34°12′ N, 109°1′ E). The fresh flowers, leaves, stem, petioles, fruit exocarp, and endocarp were sampled and frozen for RNA isolation. Total RNA was isolated from six tissues by using the RNAprep Plant Kit (Tiangen Biotech, Beijing, China). RNA purity (OD260/280) and RNA integrity were measured using Nanodrop2000 (ThermoFisher, Waltham, MA, USA) and the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
2.2. PacBio SMRT Library Preparation and Sequencing
The high-quality RNA was reverse transcribed into cDNA using the SMARTer™ PCR cDNA Synthesis Kit and prepared cDNA libraries as templates. The BluePippin Size Selection System protocol was further screened to generate PacBio SMRT libraries as described by Pacific Biosciences. Full-length transcriptome sequencing was performed on a PacBio Sequel instrument (Pacific Biosciences, Menlo Park, CA, USA). Subsequently, PacBio SMRT-seq data were processed with the SMRTlink v6.0 (http://www.pacb.com/products-and-services/analytical-sofware/smrt-analysis/ (accessed on 1 May 2022)). The CD-HIT v4.6 was used to remove redundancy in the corrected reads to generate the final transcripts for the subsequent analysis [33].
2.3. Structure Analysis
The completeness of transcriptome assembly was estimated by utilizing benchmarking universal single-copy orthologs (BUSCO) [34]. Coding sequences (CDSs) of the unigenes were predicted using Transdecoder v3.0.1 [35]. Alternative splicing analysis was conducted following Liu et al. [36]. Additionally, simple sequence repeats (SSRs) were predicted using MISA v1.0 [37]. The detection parameters were set as follows: repeat numbers of mono-, di-, tri-, tetra-, penta-, and hexanucleotide SSRs were greater than or equal to 10, 6, 5, 5, 5, and 5, respectively.
Long noncoding RNAs (lncRNAs) regulate the transcriptional and post-transcriptional gene expression through numerous and complex molecular mechanisms in plants [38]. Four computational tools, a Coding Potential Calculator (CPC) with an e-value of 1 × 10−10 [39]; a Coding–NonCoding Index (CNCI) [40], a Coding Potential Assessment Tool (CPAT) with a minimum length of 200 [41]; and Pfam (E 0.001 -domE 0.001) [42] were combined to predict the candidate lncRNAs.
2.4. Genes Functional Annotation
The functional annotation of the transcripts was performed by blasting against the following eight databases: NCBI non-redundant protein sequences (NR) [43]; protein family (Pfam) [42]; Swiss-Prot protein sequence database (Swiss-Prot) [44]; Clusters of Orthologous Groups (COG) [45]; euKaryotic Ortholog Groups (KOG) [46]; Clusters of Orthologous Groups of proteins (eggNOG) [47]; Gene Ontology (GO) [48]; and the Kyoto Encyclopedia of Genes and Genomes (KEGG) [49].
2.5. Analysis of bHLH Gene Family in A. arguta cv. Qinziyu
Transcription factors (TFs) were identified by iTAK v1.7a [50]. As one of the largest TF families in A. arguta cv. Qinziyu, according to the TF prediction results, the bHLH gene family was identified for subsequent analysis. In detail, a hidden Markov model of the bHLH domain (PF00010) was downloaded from Pfam [42] and utilized to screen candidate bHLH genes using HMMER [51]. The kiwifruit AabHLH protein sequences were extracted from A. arguta cv. Qinziyu transcriptome using TBtools [52]. Subsequently, these identified AabHLH proteins were further verified using BLASTP with default parameters in the NCBI database.
In order to identify kiwifruit anthocyanin biosynthesis-related bHLH proteins, the identified AabHLH proteins were further mapped to the reported anthocyanin biosynthesis-related bHLHs from Actinidia chinensis [53], Arabidopsis thaliana [54], and Solanum lycopersicum [55]. The basic characteristics of the AabHLH proteins, such as the coding sequence length (CDS), theoretical molecular weights (Mw), and isoelectric point (pI), were determined by ExPASy [56]. The conserved motifs in AabHLH related to anthocyanin synthesis were predicted using MEME [57]. The phylogenetic analysis of anthocyanin-related bHLH proteins was constructed using MEGA v6.0 based on the maximum likelihood (ML) method with 1000 bootstraps [58]. Based on the A. chinensis protein database [53], the gene interaction network was performed using STRING with a required confidence score > 0.7 (https://string-db.org/ (accessed on 10 October 2022)).
2.6. Quantitative RT-PCR Validation
We selected 12 key enzyme genes involved in carbohydrate and amino acid biosynthesis pathways for qRT-PCR and verified the accuracy of the transcriptome results (Table S1). Total RNAs were isolated from Qinziyu flowers using an RNAprep Plant Kit (Tiangen, Beijing, China), and cDNA was synthesized as previously described by Jia et al. [59]. The qRT-PCR was carried out with SuperReal PreMix Plus (Tiangen, Beijing, China) by an ABI Prism 7500 system (Applied Biosystems, Foster City, CA, USA). Each PCR was repeated three times, and the data were analyzed as means ± SDs (n = 3).
3. Results
3.1. Characterization of PacBio SMRT Sequencing Data
A total of 517,297 polymerase reads were obtained from the full-length transcriptome sequencing using the PacBio Sequel platform. Circular consensus sequence (CCS) analysis was performed to further reduce the error rate. A total of 175,913 CCSs were generated with a mean length of 1683 bp (Table 1, Figure S1). In addition, 124,789 full-length non-chimeric (FLNC) sequences were identified, accounting for 70.94%, of which, the number of high-quality consensus isomers was 71,637 with an average length of 1470 bp. After removing the redundant sequences using CD-HIT, 52,793 transcripts were finally obtained with N50 of 2003 bp for subsequent analysis (Table 1). Based on BUSCO analysis, a total of 962 (66.81%) genes from the transcriptome were identified as complete, with 544 (37.78%) single-copy genes and 418 (29.03%) duplicated; 100 (6.94%) were fragmented and 378 (26.65%) were missing BUSCOs (Figure S2).

Table 1.
PacBio SMRT sequencing data statistics.
3.2. Prediction of AS, LncRNA, and SSRs
In total, 46,252 coding sequences (CDSs) were predicted using TransDecoder sequencing data, of which, 27,753 coding sequences had initial and termination codons, and these sequences were defined as a complete open reading frame (ORF). The number of candidate alternatively spliced (AS) events that met all the criteria in Qinziyu was 657. Additionally, four computational approaches (CPC, CNCI, CPAT, and Pfam) were combined and predicted 52,793, 12,977, 13,921, and 9541 as lncRNAs, respectively. There were 9037 common sequences identified in four tools at the same time (Figure S3).
In addition, 27,717 SSR candidates were detected from 17,932 transcripts; the di-nucleotide repeat motifs (17,509, 63.17%) were the most abundant, followed by mono-nucleotide repeats (5948, 21.46%) and tri-nucleotide repeats (3299, 11.90%) (Table S2).
3.3. Functional Annotation
To generate a comprehensive annotation of A. arguta cv. Qinziyu, there were 45,762, 33,927, 36,258, 28,850, 18,257, 45,049, 34,444, and 22,522 transcripts annotated by searching against the NR, Swissprot, Pfma, KOG, COG, eggNOG, GO, and KEGG databases, respectively (Figure 2A, Table S3). Among the eight databases, 45,923 transcripts were functionally annotated, interactively generating 71,646 annotated transcripts (64.11%) (Table S3). In addition, the NR alignment indicated that 8524 transcripts had the highest homology with Vitis vinifera, accounting for 18.67% of the total transcripts. Quercus suber, Juglans regia, and Coffea canephora accounted for 4.51%, 3.56%, and 3.53% of the total, respectively (Figure 2B).
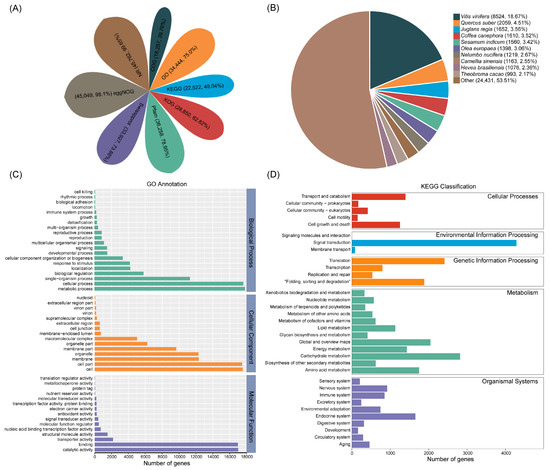
Figure 2.
(A) Function annotation of the assembled full-length transcripts in the Nr, COG, eggNOG, GO, KEGG, KOG, Pfam, and Swissprot databases. (B) The distribution of homologous species annotated in NCBI nonredundant protein sequences. (C) GO annotation of the assembled full-length transcripts in A. arguta cv. Qinziyu. (D) KEGG classification of the assembled full-length transcripts in A. arguta cv. Qinziyu.
GO terms were classified into three main functional categories: biological processes, cellular components, and molecular functions (Figure 2C). In the biological processes, ‘metabolic process’ (17,794, 51.66%), ‘cellular process’ (17,584, 51.05%), and ‘single-organism process’ (11,257, 32.68%) were the most prominent subcategories. Within the cellular components, the first three components were ‘cell’ (17,527, 50.89%), ‘cell part’ (17,452, 50.67%), and ‘membrane’ (12,318, 35.76%). For the molecular functions, the largest subcategories were ‘catalytic activity’ (16,989, 49.32%), ‘binding’ (16,960, 49.24%), and ‘transporter activity’ (2148, 6.24%) (Figure 2C).
To further predict and classify the transcripts’ functions, the annotated transcripts were analyzed according to KEGG pathway assignments. There were 34 primary pathways assigned to five categories including cellular processes, environmental information processing, genetic information processing, metabolism, and organismal systems (Figure 2D). The most common pathways were ‘signal transduction’ (4281, 19.01%), ‘carbohydrate metabolism’ (2818, 12.51%), ‘translation’ (2413, 10.71%), and ‘global and overview maps’ (2046, 9.08%) (Figure 2D). In addition, eggNOG domains could be divided into 25 groups according to eggNOG functional classification. Among them, the transcripts belonging to ‘posttranslational modification, protein turnover, chaperones’ were the most common with a total of 3986, accounting for 8.66%. There were 2988 ‘Transcription’ and 2758 ‘Signal transduction mechanisms’ accounting for 6.49% and 5.99%, respectively (Figure S4).
3.4. Candidate Genes Related to Soluble Sugar and Organic Acid Biosynthesis in A. arguta cv. Qinziyu
To address the regulatory roles of the key enzymes and the encoding genes potentially controlling soluble sugar and organic acid biosynthesis, we selected the regulatory network of carbohydrate metabolism and amino acid metabolism pathways considering their major contribution to quality formation in A. arguta cv. Qinziyu, e.g., [2,5,19]. In this study, KEGG pathway analysis indicated that 630 candidate genes encoding 55 enzymes were mainly involved in the carbohydrate and amino acid synthesis pathways (Table 2, Table S4 and Figure 3). The important screened enzyme gene families from all major steps of the carbohydrate and amino acid biosynthesis pathways were mainly distributed as follows: pyruvate kinase (PK, 43 members), malate dehydrogenase (MDH, 32 members), glyceraldehyde 3-phosphate dehydrogenase (GAPD, 27 members), enolase (ENO, 19 members), transketolase (TKT, 18 members), hexokinase (HK, 17 members), 6-phosphofructokinase 1 (PFK1 16 members), aconitate hydratase (ACON, 15 members), phosphoglycerate kinase (PGK, 14 members), citrate synthase (CS, 11 members), and alanine transaminase (ALT, 9 members) (Table 2).

Table 2.
List of candidate enzymes and genes comprising carbohydrate metabolism and amino acid biosynthesis pathways involved in A. arguta cv. Qinziyu.
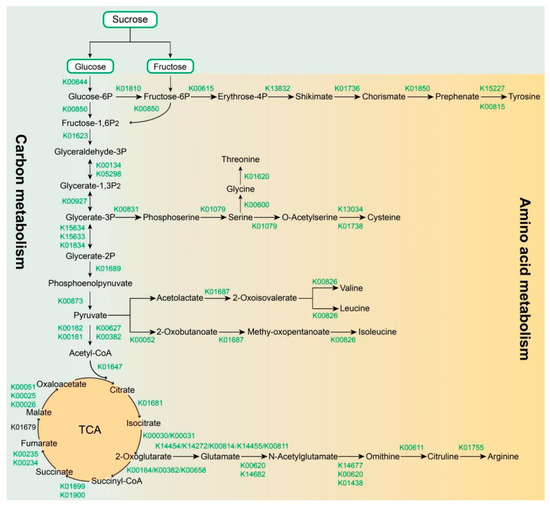
Figure 3.
Carbohydrate metabolism (Ko01200) and amino acid metabolism pathways (Ko01230) in A. arguta cv. Qinziyu were adapted from the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, available online (http://www.genome.jp/kegg/pathway.html, accessed on 10 October 2022).
Furthermore, the expression patterns of 12 representative key genes, AaPK (dazi_transcript_3491), AaMDH (dazi_transcript_14608), AaGAPD (dazi_transcript_62211), AaENO (dazi_transcript_4206), AaTKT (dazi_transcript_1017), AaHK (dazi_transcript_37294), AaPFK1 (dazi_transcript_38033), AaACON (dazi_transcript_46018), AaPGK (dazi_transcript_36237), AaCS (dazi_transcript_3928), AaALT (dazi_transcript_4056), and AaGPI (dazi_transcript_45611) were performed via qRT-PCR to verify the accuracy of the transcriptome results (Table S1, Figure 4).
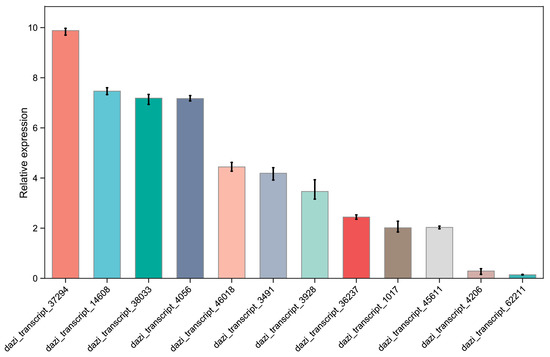
Figure 4.
Expression profiles of key genes involved in carbohydrate metabolism and amino acid metabolism in A. arguta cv. Qinziyu.
3.5. Identification of bHLH Gene Family
A total of 4657 TFs were confirmed belonging to 56 TF families based on the iTAK. The largest group of TFs was the bHLH family (1782), followed by the MYB-related (1326) and NAC (1069), whereas other TFs belonged to the WRKY (1063) and FAR1 (1021) families (Figure 5). To identify bHLH proteins for A. arguta cv. Qinziyu, 107 candidate bHLH members were originally generated using HMMER and BLASTP with default parameters. After removing redundant sequences, 64 AabHLH proteins were identified in A. arguta cv. Qinziyu (Table S5).
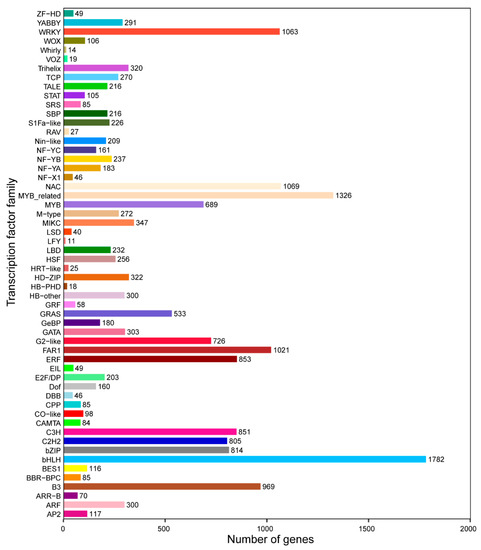
Figure 5.
Distribution of transcription factor types in A. arguta cv. Qinziyu.
3.6. Screening Potential Anthocyanin Biosynthesis-Related AabHLH Proteins
In total, 10 AabHLH proteins that function as anthocyanin regulators have been identified in A. arguta cv. Qinziyu using homologous protein sequence alignment (Table 3). The similarity between AabHLH15 and AcbHLH42 (A. chinensis) was higher than 93.68%, whereas other AabHLH protein similarities with AtEGL3, AtGL3, AtTT8, and SlAH were all more than 50%. These AabHLH proteins range in size from 138 (AabHLH39) to 520 (AabHLH15) amino acids (aa), with molecular weights ranging from 14,914.02 Da to 57,493.60 Da and the isoelectric point ranging from 5.06 to 9.89 (Table 3). All the AabHLH proteins were localized in the nucleus (Table 3).

Table 3.
Details of candidate anthocyanin biosynthesis-related AabHLH gene family in A. arguta cv. Qinziyu.
In addition, conserved motif analysis showed that six putative conserved protein motifs were predicted in the AabHLH proteins by MEME (Motifs 1–6, Figure 6A). Among them, Motif1 contains most of the AabHLH domain, and adjacent Motif 2 only co-existed in AabHLH 4, 13, 25, and 31. Whereas Motif 5 was specifically distributed in AabHLH9 and AabHLH10, Motif 1 and Motif 3 were only contained in AabHLH15 and AabHLH39, respectively (Figure 6A).
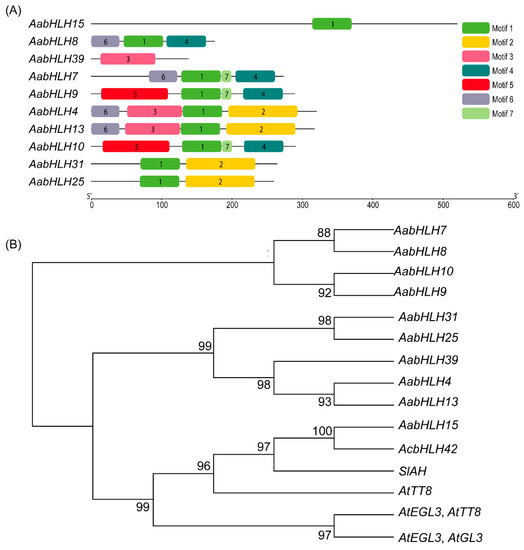
Figure 6.
(A) The amino acid motifs of AabHLH family genes in A. arguta cv. Qinziyu. The motifs, numbers 1–6, are displayed in different colored boxes. (B) The phylogenetic tree was constructed among anthocyanin biosynthesis-related bHLHs.
3.7. Phylogenetic and Protein Interaction Network Analysis
Phylogenetic analysis indicated that the 10 anthocyanin biosynthesis-related AabHLHs could be divided into two groups, which was consistent with the results of protein similarity and conserved motif analysis (Figure 6A). AabHLH15, AcbHLH42, AtEGL3, AtGL3, AtTT8, and SlAH were clustered into one group; this group of AabHLH15 showed the highest similarity to well-known anthocyanin biosynthesis-related kiwifruit AcbHLH42 (A. chinensis), and the remaining AabHLHs were clustered into another group (Figure 6B).
In order to further confirm the function of 10 anthocyanin biosynthesis-related AabHLH, protein interaction network analysis was performed to reconstruct the interaction network of the AabHLH using STRING. Only two AabHLHs (AabHLH39 and AabHLH13) were proved to be able to predict the interacting proteins (Figure 7). AabHLH39 was identified to be a homologous protein of AtTT8 (A. thaliana), while AabHLH13 was defined as a homologous protein of SlAH (S. lycopersicum). In addition, four anthocyanin-related proteins, including dazi_transcript_66562, dazi_transcript_5715, dazi_transcript_5480, dazi_transcript_17686, and dazi_transcript_59720, were also presumed to interact with bHLH proteins (Figure 7).
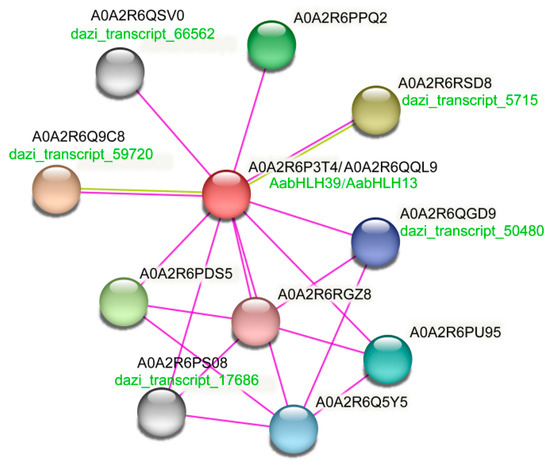
Figure 7.
Protein interaction network for the candidate anthocyanin biosynthesis-related AabHLHs according to bHLHs orthologs in A. chinensis.
4. Discussion
The regulatory mechanisms underlying fruit quality are complicated, and the genes and regulatory factors that control agronomic traits are distributed genome-wide [60]. Transcriptomic analyses have been widely employed to identify genes that affect various fruit quality traits, such as flavor, size, and color [61,62,63]. SMRT technology can produce more accurate full-length transcripts and improve the annotation information of the genome [64,65,66]. Here, 45,923 (86.99%) transcripts were successfully annotated to the Nr, COG, eggNOG, KOG, Pfam, Swiss-Prot, KEGG, and GO databases (Table S3). Moreover, 657 AS events, 9037 lncRNAs, and 27,717 SSR were identified (Table S2). Those new findings provided important information for improving the draft genome annotation and full characterization of the A. arguta cv. Qinziyu transcriptome.
The total soluble solids of fruit crops are important criteria for sensory quality and consumer recognition [3]. In general, the total soluble solids of kiwifruit vary from 11.60 to 19.13 Brix, which is mostly composed of soluble sugar and organic acid [3]. Thus, sugar content and organic acid accumulation are essential indicators of the buildup of the flavor quality of kiwifruit. To understand the regulation of soluble sugar and organic acid biosynthesis, 630 candidate genes encoding 55 enzymes were observed in A. arguta cv. Qinziyu based on the full-length transcriptome data. All of the candidate genes (e.g., PK, ENO, HK, PGK) were completely mapped to the carbohydrate and amino acid biosynthesis pathways and covered almost all of the key genes of these pathways (Table 2, Table S4 and Figure 3). In addition to the known candidate genes, a similar synthesis pattern of carbohydrate and amino acid metabolism has been found in other kiwifruit cultivars [2]. For example, glucose and fructose converted by sucrose are phosphorylated into glucose 6-phosphate (G6P) and fructose 6-phosphate (F6P) by HK and FK [67,68]. HK has a significant role in plant metabolism because its substrate, glucose, regulates the expression of different classes of genes [69,70]. Additionally, HK and FK have been proven to be important factors for regulating the glycolytic pathway in fruit development, especially under low oxygen conditions [71]. Pyruvate is produced by phosphoenol-pyruvate through PK and then converted into acetyl-CoA by pyruvate dehydrogenase (PDH) and followed by further oxidation in the TCA cycle [72]. Organic acids (e.g., malate, ketoglutarate, and succinate) are produced via the TCA cycle and provide a carbon skeleton for amino acid biosynthesis [73,74,75]. The complexity of these biosynthesis pathways clearly demonstrated the relationship between metabolites and genes and also laid a potential molecular foundation for further research of soluble sugar and organic acid biosynthesis in A. arguta cv. Qinziyu.
Furthermore, anthocyanins are plant secondary metabolites that act as visual color pigments in plant tissues [7,76]. TFs are well known to play essential roles in plant anthocyanin biosynthesis by triggering structural genes [7]. The bHLH family is one of the largest TFs in plants, and many members have been inferred to regulate anthocyanin biosynthesis [77,78,79]. The flesh of kiwifruit has a variety of colors, and the coloring mechanisms of the green-, yellow-, and red-fleshed kiwifruit have become a special focus for increasing numbers of researchers [4,7]. In this study, we screened 10 anthocyanin biosynthesis-related AabHLH proteins from 64 identified AabHLH TFs in A. arguta cv. Qinziyu based on the full-length transcriptome data (Table 3 and Table S5). These anthocyanin biosynthesis-related AabHLH proteins were highly conserved by homologous alignment, motif structure, and phylogenetics (Figure 6). Additionally, according to the protein interaction network, two AabHLHs members, AabHLH39 and AabHLH13, were confirmed to be homologous proteins of AtTT8 and SlAH, respectively (Figure 7). Arabidopsis TT8 is required for the expression of anthocyanin biosynthesis by forming a stabilized MBW complex with TT2 and TTG1 [80]. Moreover, SlAH controls anthocyanin accumulation by up-regulating the transcript levels of anthocyanin biosynthetic genes in S. lycopersicum [55]. Besides that, the gene sequence of AabHLH15 is highly similar to that of AcbHLH42 (A. chinensis), which has been reported to be involved in anthocyanin biosynthesis [53]. However, other TFs besides AabHLH proteins may participate in regulating anthocyanin accumulation in kiwifruit, and the detailed mechanisms require further investigation.
5. Conclusions
We first report the full-length transcriptome data for the characterization of carbohydrate and amino acid metabolism pathways and the key transcription factors regulating anthocyanin biosynthesis in A. arguta cv. Qinziyu. This knowledge is critical for understanding soluble sugar, organic acid, and anthocyanin biosynthesis to provide more comprehensive insights into the molecular mechanism of fruit quality formation in kiwifruit and will be highly useful for the breeding of horticultural crops.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy13010143/s1, Figure S1: Length distributions of circular consensus (CCS) reads. Figure S2: Statistics of BUSCO integrity assessment. Figure S3: Venn diagram of long non-coding RNAs (lncRNAs). CPC: coding potential calculator; CNCI: coding-noncoding index; CPAT: coding potential assessment tool; Pfam: protein families. Figure S4: eggNOG classification of the assembled full-length transcripts in A. arguta cv. Qinziyu. Table S1: Characteristics of 12 primers used for qRT-PCR analysis. Table S2: Summary of SSR results in A. arguta cv. Qinziyu. Table S3: Annotated transcripts numbers based on eight databases in A. arguta cv. Qinziyu. Table S4: Annotated transcripts in the carbohydrate metabolism and amino acid metabolism pathways. Table S5: Details of AabHLH gene family in A. arguta cv. Qinziyu.
Author Contributions
Conceptualization, Y.W. (Yongpeng Wu), Y.Z. and Y.J.; methodology, Y.J. and Y.Z.; software, Y.J.; formal analysis, Y.J.; investigation, Y.W. (Yaling Wang), Y.Z, L.Z., F.W., G.Y., Y.W. (Yongpeng Wu) and X.K.; data curation, Y.J.; writing—original draft preparation, Y.J.; writing—review and editing, Y.J., Y.Z. and Y.W. (Yaling Wang); project administration, Y.W. (Yongpeng Wu) and Y.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the Xi’an Bureau of Science and Technology under Grants (NO. 2017050NC/NY008 (5), 20NYYF0003, 22NYYF002); the Department of Science and Technology of Shaanxi Province (NO. 2018ZDXM-NY-012, 2020NY-044, 2019TG-005, 2021NY-058, 2023-JC-QN-0253); and the Shaanxi Academy of Sciences (NO. 2019k-01(2021), 2022K-09).
Data Availability Statement
The PacBio SMRT-seq raw reads of this study were deposited NCBI under the following accession number PRJNA909198.
Acknowledgments
We are grateful to Da-Feng Xu for helpful suggestions and comments on our text.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Huang, S.X.; Ding, J.; Deng, D.; Tang, W.; Sun, H.H.; Liu, D.Y.; Zhang, L.; Niu, X.; Zhang, X.L.; Meng, M.; et al. Draft genome of the kiwifruit Actinidia chinensis. Nat. Commun. 2013, 4, 2640. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.C.; Shu, P.; Zhang, C.; Zhang, J.L.; Chen, Y.; Zhang, Y.X.; Du, K.; Xie, Y.; Li, M.Z.; Ma, T.; et al. Integrative analyses of metabolome and genome-wide transcriptome reveal the regulatory network governing flavor formation in kiwifruit (Actinidia chinensis). New Phytol. 2022, 233, 373–389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, Q.; Lan, T.; Geng, T.; Gao, C.; Yuan, Q.; Zhang, Q.; Xu, P.; Sun, X.; Liu, X.; et al. Comparative analysis of physicochemical characteristics, nutritional and functional components and antioxidant capacity of fifteen kiwifruit (Actinidia) cultivars-comparative analysis of fifteen kiwifruit (Actinidia) cultivars. Foods 2020, 9, 1267. [Google Scholar] [CrossRef]
- Liao, G.L.; He, Y.Q.; Li, X.S.; Zhong, M.; Huang, C.H.; Yi, S.Y.; Liu, Q.; Xu, X.B. Effects of bagging on fruit flavor quality and related gene expression of AsA synthesis in Actinidia eriantha. Sci. Hortic. 2019, 256, 108511. [Google Scholar] [CrossRef]
- Wang, F.; Ge, S.F.; Xu, X.X.; Xing, Y.; Du, X.; Zhang, X.; Lv, M.X.; Liu, J.Q.; Zhu, Z.L.; Jiang, Y.M. Multiomics analysis reveals new insights into the apple fruit quality decline under high nitrogen conditions. J. Agric. Food Chem. 2021, 69, 5559–5572. [Google Scholar] [CrossRef]
- Lu, X.M.; Man, Y.P.; Lei, R.; Liu, Y.B.; Wu, J.H.; Wang, Y.C. Structural analysis of Actinidia arguta natural populations and preliminary application in association mapping of fruit traits. Sci. Hortic. 2022, 304, 111306. [Google Scholar] [CrossRef]
- Liu, Y.F.; Zhou, B.; Qi, Y.W.; Chen, X.; Liu, C.H.; Liu, Z.D.; Ren, X.L. Expression Differences of pigment structural genes and transcription factors explain flesh coloration in three contrasting kiwifruit cultivars. Front. Plant Sci. 2017, 8, 1507. [Google Scholar] [CrossRef]
- Huang, H.; Ferguson, A.R. Genetic resources of kiwifruit: Domestication and breeding. Hort. Rev. 2007, 33, 1–121. [Google Scholar]
- Liu, Y.; Li, D.; Zhang, Q.; Song, C.; Zhong, C.; Zhang, X.; Wang, Y.; Yao, X.; Wang, Z.; Zeng, S.; et al. Rapid radiations of both kiwifruit hybrid lineages and their parents shed light on a two-layer mode of species diversification. New Phytol. 2017, 215, 877–890. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Z.Z.; Wang, Y.C.; Jiang, Z.W.; Wang, S.M.; Huang, H.W. Vitamin C, flower color and ploidy variation of hybrids from a ploidy-unbalanced Actinidia interspecific cross and SSR characterization. Euphytica 2010, 175, 133–143. [Google Scholar] [CrossRef]
- Li, Y.; Fang, J.; Qi, X.; Lin, M.; Zhong, Y.; Sun, L.; Cui, W. Combined analysis of the fruit metabolome and transcriptome reveals candidate genes involved in flavonoid biosynthesis in Actinidia arguta. Int. J. Mol. Sci. 2018, 19, 1471. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.; Pinto, D.; Santos, J.; Vinha, A.F.; Palmeira, J.; Ferreira, H.N.; Rodrigues, F.; Oliveira, M.B.P. Hardy kiwifruit leaves (Actinidia arguta): An extraordinary source of value-added compounds for food industry. Food Chem. 2018, 259, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fang, J.; Qi, X.; Lin, M.; Zhong, Y.; Sun, L. A key structural gene, AaLDOX, is involved in anthocyanin biosynthesis in all red-fleshed kiwifruit (Actinidia arguta) based on transcriptome analysis. Gene 2018, 648, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cui, W.; Wang, R.; Lin, M.; Fang, J. Microrna858-mediated regulation of anthocyanin biosynthesis in kiwifruit (Actinidia arguta) based on small RNA sequencing. PLoS ONE 2019, 14, e0217480. [Google Scholar] [CrossRef] [PubMed]
- Wojdyło, A.; Nowicka, P.; Oszmia’nski, J.; Golis, T. Phytochemical compounds and biological effects of Actinidia fruits. J. Funct. Foods 2017, 30, 194–202. [Google Scholar] [CrossRef]
- Pang, Q.; Zhang, A.; Zang, W.; Wei, L.; Yan, X. Integrated proteomics and metabolomics for dissecting the mechanism of global responses to salt and alkali stress in Suaeda corniculata. Plant Soil. 2016, 402, 379–394. [Google Scholar] [CrossRef]
- Jia, X.; Zhu, Y.; Hu, Y.; Zhang, R.; Cheng, L.; Zhu, Z.L.; Zhao, T.; Zhang, X.; Wang, Y.X. Integrated physiologic, proteomic, and metabolomic analyses of Malus halliana adaptation to saline-alkali stress. Hortic. Res. 2019, 6, 91–109. [Google Scholar] [CrossRef]
- Guo, J.; Lu, X.; Tao, Y.; Guo, H.; Min, W. Comparative ionomics and metabolic responses and adaptive strategies of cotton to salt and alkali stress. Front. Plant Sci. 2022, 13, 871387. [Google Scholar] [CrossRef]
- Cheng, H.; Kong, W.; Tang, T.; Ren, K.; Zhang, K.; Wei, H.; Lin, T. Identification of key gene networks controlling soluble sugar and organic acid metabolism during oriental melon fruit development by integrated analysis of metabolic and transcriptomic analyses. Front. Plant Sci. 2022, 13, 830517. [Google Scholar] [CrossRef]
- Cho, K.; Cho, K.S.; Sohn, H.B.; Ha, I.J.; Hong, S.Y.; Lee, H.; Kim, Y.M.; Nam, M.H. Network analysis of the metabolome and transcriptome reveals novel regulation of potato pigmentation. J. Exp. Bot. 2016, 67, 1519–1533. [Google Scholar] [CrossRef]
- Zhang, L.; Man, Y.P.; Jiang, Z.W.; Wang, Y.C. Cloning and expression of anthocyanin pathway genes, AcCHS and AcLDOX, in Actinidia chinensis. Acta Hortic. Sin. 2012, 39, 2124–2132. [Google Scholar]
- Li, W.B.; Liu, Y.F.; Zeng, S.H.; Xiao, G.; Wang, G.; Wang, Y.; Peng, M.; Huang, H.W. Gene expression profiling of development and anthocyanin accumulation in Kiwifruit (Actinidia chinensis) based on transcriptome sequencing. PloS ONE 2015, 10, e0138743. [Google Scholar]
- Luo, J.; Guo, L.L.; Huang, Y.N.; Wang, C.; Qiao, C.K.; Pang, R.L.; Li, J.; Pang, T.; Wang, R.P.; Xie, H.Z.; et al. Transcriptome analysis reveals the effect of pre-harvest CPPU treatment on the volatile compounds emitted by kiwifruit stored at room temperature. Food Res. Int. 2017, 102, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Huan, C.; Zhang, J.; Jia, Y.; Li, S.E.; Jiang, T.J.; Shen, S.L.; Zheng, X.L. Effect of 1-methylcyclopropene treatment on quality, volatile production and ethanol metabolism in kiwifruit during storage at room temperature. Sci. Hortic. 2020, 265, 109266. [Google Scholar] [CrossRef]
- Pilkington, S.M.; Crowhurst, R.; Hilario, E.; Nardozza, S.; Fraser, L.; Peng, Y.; Gunaseelan, K.; Simpson, R.; Tahir, J.; Deroles, S.C.; et al. A manually annotated Actinidia chinensis var. chinensis (kiwifruit) genome highlights the challenges associated with draft genomes and gene prediction in plants. BMC Genomics 2018, 19, 257. [Google Scholar] [CrossRef]
- Wu, H.; Ma, T.; Kang, M.; Ai, F.; Zhang, J.; Dong, G.; Liu, J.Q. A high-quality Actinidia chinensis (kiwifruit) genome. Hortic Res. 2019, 6, 117. [Google Scholar] [CrossRef] [PubMed]
- Li, X.L.; Cheng, Y.D.; Wang, M.; Cui, S.J.; Guan, J.F. Weighted gene coexpression correlation network analysis reveals a potential molecular regulatory mechanism of anthocyanin accumulation under different storage temperatures in ‘Friar’ plum. BMC Plant Biol. 2021, 21, 576. [Google Scholar] [CrossRef]
- Xin, M.; Li, C.; Khoo, H.E.; Li, L.; He, X.; Yi, P.; Tang, Y.; Sun, J. Dynamic analyses of transcriptome and metabolic profiling: Revealing molecular insight of aroma synthesis of mango (Mangifera indica L. var. Tainong). Front. Plant Sci. 2012, 12, 666805. [Google Scholar] [CrossRef]
- Abbas, H.M.K.; Huang, H.X.; Wang, A.J.; Wu, T.Q.; Xue, S.D.; Ahmad, A.; Xie, D.S.; Li, J.X.; Zhong, Y.J. Metabolic and transcriptomic analysis of two Cucurbita moschata germplasms throughout fruit development. BMC Genom. 2020, 21, 365. [Google Scholar] [CrossRef]
- Hong, C.P.; Kim, C.K.; Lee, D.J.; Jeong, H.J.; Lee, Y.; Park, S.G.; Kim, H.J.; Kang, J.N.; Ryu, H.; Kwon, S.J.; et al. Long-read transcriptome sequencing provides insight into lignan biosynthesis during fruit development in Schisandra chinensis. BMC Genom. 2022, 23, 17. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Hamilton, M.; Jacobi, J.L.; Ngam, P.; Devitt, N.; Schilkey, F.; Ben-Hur, A.; Reddy, A.S. A survey of the sorghum transcriptome using single-molecule long reads. Nat. Commun. 2016, 7, 11706. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, H.; Kohnen, M.V.; Prasad, K.V.S.K.; Gu, L.; Reddy, A.S.N. Analysis of Transcriptome and Epitranscriptome in Plants Using PacBio Iso-Seq and Nanopore-Based Direct RNA Sequencing. Front. Genet. 2019, 10, 253. [Google Scholar] [CrossRef]
- Fu, L.M.; Niu, B.F.; Zhu, Z.W.; Wu, S.T.; Li, W.Z. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Simao, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Mei, W.; Soltis, P.S.; Soltis, D.E.; Barbazuk, W.B. Detecting alternatively spliced transcript isoforms from single-molecule long-read sequences without a reference genome. Mol. Ecol. Resour. 2017, 17, 1243–1256. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Fonouni-Farde, C.; Ariel, F.; Crespi, M. Plant long noncoding RNAs: New players in the field of post-transcriptional regulations. Non-Coding RNA 2021, 7, 12. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Wang, L.G.; Park, H.J.; Dasari, S.; Wang, S.Q.; Kocher, J.P.; Li, W. CPAT: Coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Jaroszewski, L.; Godzik, A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 2002, 18, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Bairoch, A.; Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL. Nucleic Acids Res. 2000, 28, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S.; et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, R7. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Harris, M.A. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Zheng, Y.; Jiao, C.; Sun, H.H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.F.; Banf, M.; Dai, X.B.; Martin, G.B.; Giovannoni, L.J.; et al. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tang, W.; Hu, Y.; Zhang, Y.; Sun, J.; Guo, X.; Lu, H.; Yang, Y.; Fang, C.; Niu, X.; et al. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 2019, 99, 359–378. [Google Scholar] [CrossRef]
- Bernhardt, C.; Lee, M.M.; Gonzalez, A.; Zhang, F.; Lloyd, A.; Schiefelbein, J. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 2003, 130, 6431–6439. [Google Scholar] [CrossRef]
- Qiu, Z.; Wang, X.; Gao, J.; Guo, Y.; Huang, Z.; Du, Y. The tomato Hoffman’s anthocyaninless gene encodes a bHLH transcription factor involved in anthocyanin biosynthesis that is developmentally regulated and induced by low temperatures. PLoS ONE 2016, 11, e0151067. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; De Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Wu, Y.P.; Wang, F.W.; Zhang, L.; Yu, G.; Wang, Y.L.; Zhang, Y. Full-length transcriptome sequencing analysis and characterization of gene isoforms involved in flavonoid biosynthesis in the seedless kiwifruit cultivar ‘Chengxiang’ (Actinidia arguta). Diversity 2022, 14, 424. [Google Scholar] [CrossRef]
- Zhang, C.M.; Hao, Y.J. Advances in genomic, transcriptomic, and metabolomic analyses of fruit quality in fruit Crops. Hortic. Plant J. 2020, 6, 361–371. [Google Scholar] [CrossRef]
- Bai, Y.; Dougherty, L.; Cheng, L.L.; Zhong, G.Y.; Xu, K.N. Uncovering co-expression gene network modules regulating fruit acidity in diverse apples. BMC Genom. 2015, 16, 612. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.Q.; Liao, L.; Fang, T.; Peng, Q.; Ogutu, C.; Zhou, H.; Ma, F.W.; Han, Y.P. A Ma10 gene encoding P-type ATPase is involved in fruit organic acid accumulation in apple. Plant Biotechnol. J. 2019, 17, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.Q.; Liang, B.W.; Chang, B.; Yan, J.Y.; Liu, L.; Wang, Y.; Yang, Y.Z.; Zhao, Z.Y. Transcriptome profiling of anthocyanin biosynthesis in the peel of ‘Granny Smith’ apples (Malus domestica) after bag removal. BMC Genom. 2019, 20, 353. [Google Scholar] [CrossRef]
- He, Z.; Su, Y.; Wang, T.; Wu, H.W.; Zhu, J.F.; Wang, F.Z.; Cao, P.P.; Yang, X.Y.; Zhang, H.X. Full-Length Transcriptome Analysis of Four Different Tissues of Cephalotaxus oliveri. Int. J. Mol. Sci. 2021, 22, 787. [Google Scholar] [CrossRef]
- Chao, Y.; Yuan, J.; Guo, T.; Xu, L.; Mu, Z.; Han, L. Analysis of transcripts and splice isoforms in Medicago sativa L. by single molecule long-read sequencing. Plant Mol. Biol. 2019, 99, 219–235. [Google Scholar] [CrossRef]
- Liu, L.Y.; Teng, K.; Fan, X.F.; Han, C.; Zhang, H.; Wu, J.Y.; Chang, Z.H. Combination analysis of single-molecule long-read and Illumina sequencing provides insights into the anthocyanin accumulation mechanism in an ornamental grass, Pennisetum setaceum cv. Rubrum. Plant Mol. Biol. 2022, 109, 159–175. [Google Scholar] [CrossRef]
- Koch, K. Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 2004, 7, 235–246. [Google Scholar] [CrossRef]
- Xiong, Y.; Yan, P.; Du, K.; Li, M.Z.; Xie, Y.; Gao, P. Nutritional component analyses of kiwifruit in different development stages by metabolomic and transcriptomic approaches. J. Sci. Food Agr. 2020, 100, 2399–2409. [Google Scholar] [CrossRef] [PubMed]
- Claeyssen, E.; Rivoal, J. Plant hexokinase isozymes: Occurrence, properties, and functions. Phytochemistry 2007, 68, 709–731. [Google Scholar] [CrossRef]
- Menu, T.; Saglio, P.; Granot, D.; Dai, N.; Raymond, P.; Ricard, B. High hexokinase activity in tomato fruit perturbs carbon and energy metabolism and reduces fruit and seed size. Plant Cell Environ. 2004, 27, 89–98. [Google Scholar] [CrossRef]
- Fox, T.C.; Green, B.J.; Kennedy, R.A.; Rumpho, M.E. Changes in hexokinase activity in Echinochloa phyllopogon and Echinochloa crus-pavonis in response to abiotic stress. Plant Physiol. 1998, 118, 1403–1409. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.S.; Roche, T.E. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. Faseb J. 1990, 4, 3224–3233. [Google Scholar] [CrossRef] [PubMed]
- Katz, E.; Boo, K.H.; Kim, H.Y.; Eigenheer, R.A.; Phinney, B.S.; Shulaev, V.; Negre-Zakharov, F.; Sadka, A.; Blumwald, E. Label-free shotgun proteomics and metabolite analysis reveal a significant metabolic shift during citrus fruit development. J. Exp. Bot. 2011, 62, 5367–5384. [Google Scholar] [CrossRef] [PubMed]
- Sadka, A.; Dahan, E.; Or, E.; Roose, M.L.; Marsh, K.B.; Cohen, L. Comparative analysis of mitochondrial citrate synthase gene structure, transcript level and enzymatic activity in acidless and acid-containing Citrus varieties. Funct. Plant. Biol. 2001, 28, 383–390. [Google Scholar] [CrossRef]
- Amiour, N.; Imbaud, S.; Clment, G. The use of metabolomics integrated with transcriptomic and proteomic studies for identifying key steps involved in the control of nitrogen metabolism incrops such as maize. Exp. Bot. 2012, 63, 5017–5033. [Google Scholar] [CrossRef]
- Meng, J.X.; Gao, Y.; Han, M.L.; Liu, P.Y.; Yang, C.; Shen, T.; Li, H.H. In vitro anthocyanin induction and metabolite analysis in Malus spectabilis leaves under low nitrogen conditions. Hortic. Plant J. 2020, 6, 284–292. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, F.; Wang, B.; Wu, H.; Wu, J.; Liu, J.; Sun, Y.; Cheng, C.; Qiu, D. Identification, characterization and expression analysis of anthocyanin biosynthesis-related bHLH genes in blueberry (Vaccinium corymbosum L.). Int. J. Mol. Sci. 2021, 22, 13274. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, Y.; Li, L.; Meng, H.; Yang, Y.; Dong, Z.; Wang, L.; Wu, G. Genome-wide identification and characterization of bHLH transcription factors related to anthocyanin biosynthesis in red walnut (Juglans regia L.). Front. Genet. 2021, 12, 632509. [Google Scholar] [CrossRef]
- Wang, X.J.; Peng, X.Q.; Shu, X.C.; Li, Y.H.; Wang, Z.; Zhuang, W.B. Genome-wide identifcation and characterization of PdbHLH transcription factors related to anthocyanin biosynthesis in colored-leaf poplar (Populus deltoids). BMC Genom. 2022, 23, 244. [Google Scholar]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).