Phylogenetic Analysis of Wild Pomegranate (Punica granatum L.) Based on Its Complete Chloroplast Genome from Tibet, China
Abstract
1. Introduction
2. Materials & Methods
2.1. Plant Materials
2.2. Genome Sequencing and Annotation
2.3. Condon Usage Bias Analysis
2.4. Genome Comparison and Sequence Divergence
2.5. Phylogenetic Analysis
3. Results
3.1. Structural Organization and Gene Content of Wild Pomegranate cp Genome
3.2. Codon Usage (RSCU) Analysis
3.3. Analysis of Types via the Number of SSRs in the Chloroplast Genome of Wild Pomegranate
3.4. Structural and Variant Assessment of Chloroplast Genomes
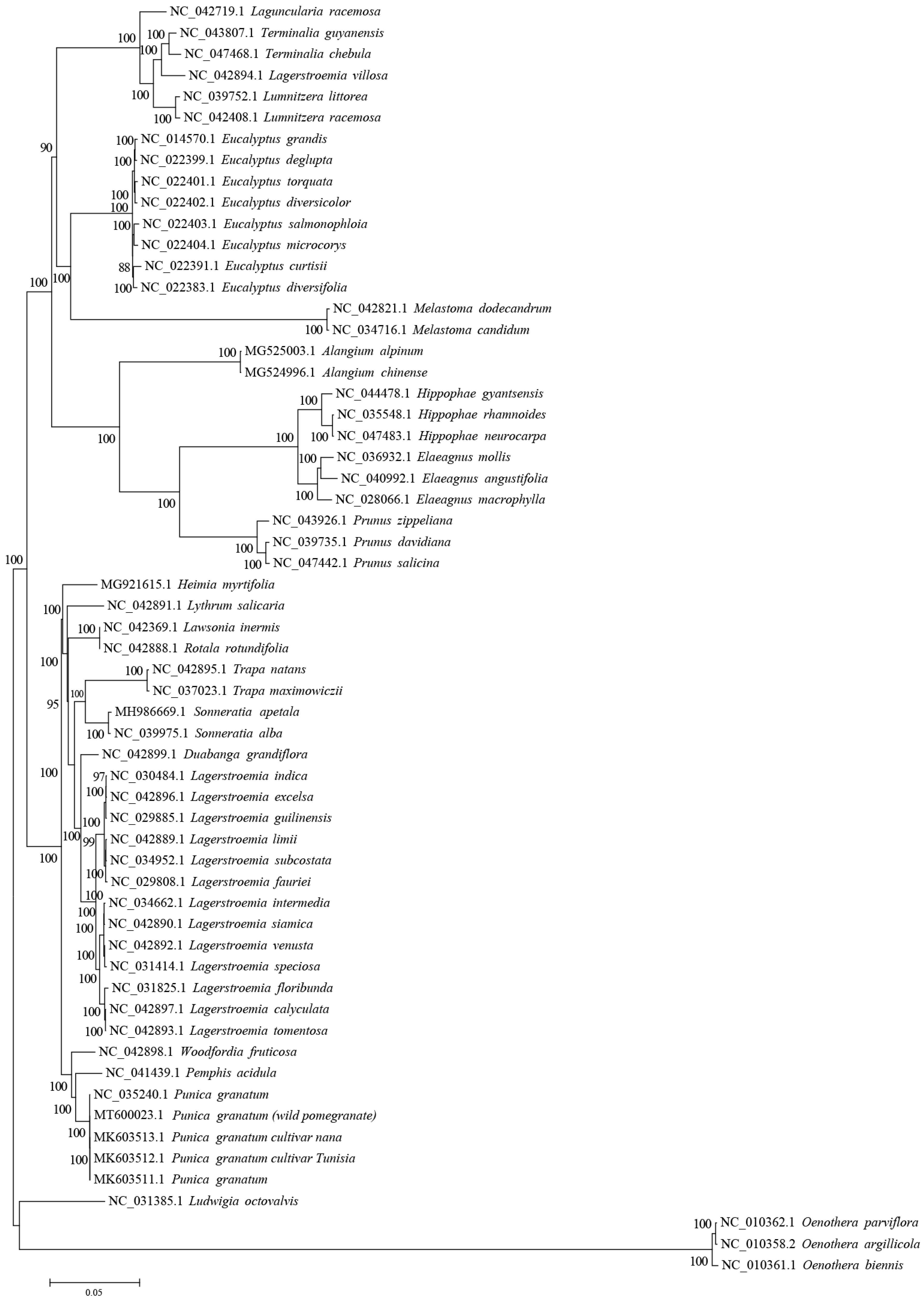
3.5. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Al-Khayri, J.M.; Jain, S.M.; Johnson, D.V. Advances in Plant Breeding Strategies: Fruits; Springer: Berlin/Heidelberg, Germany, 2019; Volume 3. [Google Scholar]
- Yuan, Z.; Fang, Y.; Zhang, T.; Fei, Z.; Han, F.; Liu, C.; Liu, M.; Xiao, W.; Zhang, W.; Wu, S.; et al. The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol. J. 2018, 16, 1363–1374. [Google Scholar] [CrossRef] [PubMed]
- Bonesi, M.; Tundis, R.; Vincenzo, S.; Loizzo, M. The Juice of Pomegranate (Punica granatum L.): Recent Studies on Its Bioactivities Quality Control in the Beverage Industry. In Quality Control in the Beverage Industry (ELSEVIER); Elsevier: Amsterdam, The Netherlands, 2019; Volume 17, pp. 459–489. [Google Scholar] [CrossRef]
- Karimi, M.; Sadeghi, R.; Kokini, J. Pomegranate as a promising opportunity in medicine and nanotechnology. Trends Food Sci. Technol. 2017, 69, 59–73. [Google Scholar] [CrossRef]
- Lansky, E.P.; Newman, R.A. Punica granatum (pomegranate) and its potential for prevention and treatment of inflammation and cancer. J. Ethnopharmacol. 2007, 109, 177–206. [Google Scholar] [CrossRef] [PubMed]
- Mars, M. Pomegranate plant material: Genetic resources and breeding, a review. Options Mediterr. Ser. A 2000, 42, 55–62. [Google Scholar]
- Narzary, D.; Mahar, K.S.; Rana, T.; Ranade, S. Analysis of genetic diversity among wild pomegranates in Western Himalayas, using PCR methods. Sci. Hortic. 2009, 121, 237–242. [Google Scholar] [CrossRef]
- R, M.S.; Vaishali, M.; Ss, B. Molecular Characterization of Cultivated and Wild Genotypes of Punica granatum L. (Pomegranate) by Using SSR Marker. Int. J. Life Sci. Sci. Res. 2018, 4, 1786–1794. [Google Scholar] [CrossRef]
- Aziz, S.; Firdous, S.; Rahman, H.; Awan, S.I.; Michael, V.; Meru, G. Genetic diversity among wild pomegranate (Punica granatum) in Azad Jammu and Kashmir region of Pakistan. Electron. J. Biotechnol. 2020, 46, 50–54. [Google Scholar] [CrossRef]
- Hasnaoui, N.; Buonamici, A.; Sebastiani, F.; Mars, M.; Trifi, M.; Vendramin, G.G. Development and characterization of SSR markers for pomegranate (Punica granatum L.) using an enriched library. Conserv. Genet. Resour. 2010, 2, 283–285. [Google Scholar] [CrossRef]
- Sarkhosh, A.; Zamani, Z.; Fatahi, R.; Ebadi, A. RAPD markers reveal polymorphism among some Iranian pomegranate (Punica granatum L.) genotypes. Sci. Hortic. 2006, 111, 24–29. [Google Scholar] [CrossRef]
- Yuan, Z.; Yin, Y.; Qu, J.; Zhu, L.; Li, Y. Population Genetic Diversity in Chinese Pomegranate (Punica granatum L.) Cultivars Revealed by Fluorescent-AFLP Markers. J. Genet. Genom. 2007, 34, 1061–1071. [Google Scholar] [CrossRef]
- Ismail, T.; Sestili, P.; Akhtar, S. Pomegranate peel and fruit extracts: A review of potential anti-inflammatory and anti-infective effects. J. Ethnopharmacol. 2012, 143, 397–405. [Google Scholar] [CrossRef]
- Saad, H.; Charrier-El Bouhtoury, F.; Pizzi, A.; Rode, K.; Charrier, B.; Ayed, N. Characterization of pomegranate peels tannin extractives. Ind. Crops Prod. 2012, 40, 239–246. [Google Scholar] [CrossRef]
- Singh, B.; Singh, J.P.; Kaur, A.; Singh, N. Phenolic compounds as beneficial phytochemicals in pomegranate (Punica granatum L.) peel: A review. Food Chem. 2018, 261, 75–86. [Google Scholar] [CrossRef]
- He, L.; Xu, H.; Liu, X.; He, W.; Yuan, F.; Hou, Z.; Gao, Y. Identification of phenolic compounds from pomegranate (Punica granatum L.) seed residues and investigation into their antioxidant capacities by HPLC–ABTS+ assay. Food Res. Int. 2011, 44, 1161–1167. [Google Scholar] [CrossRef]
- Zhang, L.; Fu, Q.; Zhang, Y. Composition of anthocyanins in pomegranate flowers and their antioxidant activity. Food Chem. 2011, 127, 1444–1449. [Google Scholar] [CrossRef]
- Bekir, J.; Mars, M.; Souchard, J.P.; Bouajila, J. Assessment of antioxidant, anti-inflammatory, anti-cholinesterase and cytotoxic activities of pomegranate (Punica granatum) leaves. Food Chem. Toxicol. 2013, 55, 470–475. [Google Scholar] [CrossRef]
- Sreekumar, S.; Sithul, H.; Muraleedharan, P.; Azeez, J.M.; Sreeharshan, S. Pomegranate Fruit as a Rich Source of Biologically Active Compounds. BioMed Res. Int. 2014, 2014, 686921. [Google Scholar] [CrossRef] [PubMed]
- Bassiri-Jahromi, S. Punica granatum (Pomegranate) activity in health promotion and cancer prevention. Oncol. Rev. 2018, 12, 345. [Google Scholar] [CrossRef]
- Malviya, S.; Arvind; Jha, A.; Hettiarachchy, N. Antioxidant and antibacterial potential of pomegranate peel extracts. J. Food Sci. Technol. 2014, 51, 4132–4137. [Google Scholar] [CrossRef]
- Wang, J.-Y.; Zhu, C.; Qian, T.-W.; Guo, H.; Wang, D.-D.; Zhang, F.; Yin, X. Extracts of black bean peel and pomegranate peel ameliorate oxidative stress-induced hyperglycemia in mice. Exp. Ther. Med. 2015, 9, 43–48. [Google Scholar] [CrossRef]
- McFadden, G.I.; van Dooren, G.G. Evolution: Red algal genome affirms a common origin of all plastids. Curr. Biol. 2004, 14, R514–R516. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Savage, Z.; Duggan, C.; Toufexi, A.; Pandey, P.; Liang, Y.; Segretin, M.E.; Yuen, L.H.; Gaboriau, D.C.A.; Leary, A.Y.; Tumtas, Y.; et al. Chloroplasts alter their morphology and accumulate at the pathogen interface during infection by Phytophthora infestans. Plant J. 2021, 107, 1771–1787. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.L.; Asaf, S.; Lee, I.; Al-Harrasi, A.; Al-Rawahi, A. First reported chloroplast genome sequence of Punica granatum (cultivar Helow) from Jabal Al-Akhdar, Oman: Phylogenetic comparative assortment with Lagerstroemia. Genetica 2018, 146, 461–474. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Shi, T.; Luo, W.; Ni, X.; Iqbal, S.; Ni, Z.; Huang, X.; Yao, D.; Shen, Z.; Gao, Z. Comparative analysis of the complete chloroplast genome among Prunus mume, P. armeniaca, and P. salicina. Hortic. Res.-Engl. 2019, 6, 89. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, X.; Liu, G.; Yin, Y.; Chen, K.; Yun, Q.; Zhao, D.; Al-Mssallem, I.S.; Yu, J. The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.). PLoS ONE 2010, 5, e12762. [Google Scholar] [CrossRef]
- Yao, X.; Tang, P.; Li, Z.; Li, D.; Liu, Y.; Huang, H. The First Complete Chloroplast Genome Sequences in Actinidiaceae: Genome Structure and Comparative Analysis. PLoS ONE 2015, 10, e129347. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Dong, B.; Xu, L.; Tembrock, L.R.; Zheng, S.; Wu, Z. The Complete Chloroplast Genome of Heimia myrtifolia and Comparative Analysis within Myrtales. Molecules 2018, 23, 846. [Google Scholar] [CrossRef] [PubMed]
- Soriano, J.M.; Zuriaga, E.; Rubio, P.; Llmcer, G.; Infante, R.; Badenes, M.L. Development and characterization of microsatellite markers in pomegranate (Punica granatum L.). Mol. Breed. 2011, 27, 119–128. [Google Scholar] [CrossRef]
- Zarei, A.; Sahraroo, A. Molecular characterization of pomegranate (Punica granatum L.) accessions from Fars Province of Iran using microsatellite markers. Hortic. Environ. Biotechnol. 2018, 59, 239–249. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [PubMed]
- Ankenbrand, M.J.; Pfaff, S.; Terhoeven, N.; Qureischi, M.; Gündel, M.; Weiß, C.L.; Hackl, T.; Förster, F. chloroExtractor: Extraction and assembly of the chloroplast genome from whole genome shotgun data. J. Open Source Softw. 2018, 3, 464. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; Yi, T.S.; Li, D.Z. GetOrganelle: A simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. BioRxiv 2018, 4, 256–479. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. A J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; LoCascio, P.F.; Hauser, L.J.; Uberbacher, E.C. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 2012, 28, 2223–2230. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.; Luciani, A.; Eddy, S.; Park, Y.; Lopez, R.; Finn, R. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Molecular Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. BioRxiv 2018, 289660. [Google Scholar]
- Kurtz, S. The Vmatch Large Scale Sequence Analysis Software; University of Hamburg: Hamburg, Germany, 2011. [Google Scholar]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple Alignment of Conserved Genomic Sequence with Rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvonen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, J.; Cui, Y.; Wang, Y.; Duan, B.; Yao, H. Identification of Ligularia Herbs Using the Complete Chloroplast Genome as a Super-Barcode. Front. Pharmacol. 2018, 9, 695. [Google Scholar] [CrossRef]
- Yang, J.; Feng, L.; Yue, M.; He, Y.-L.; Zhao, G.F.; Li, Z.H. Species delimitation and interspecific relationships of the endangered herb genus Notopterygium inferred from multilocus variations. Mol. Phylogenetics Evol. 2019, 133, 142–151. [Google Scholar] [CrossRef]
- Yang, J.-B.; Yang, S.-X.; Li, H.T.; Yang, J.; Li, D.-Z. Comparative Chloroplast Genomes of Camellia Species. PLoS ONE 2013, 8, e73053. [Google Scholar] [CrossRef]
- Song, Y.; Yao, X.; Tan, Y.; Gan, Y.; Corlett, R. Complete chloroplast genome sequence of the avocado: Gene organization, comparative analysis, and phylogenetic relationships with other Lauraceae. Can. J. For. Res. 2016, 46, 1293–1301. [Google Scholar] [CrossRef]
- Ivanova, Z.; Sablok, G.; Daskalova, E.; Zahmanova, G.; Apostolova, E.; Yahubyan, G.; Baev, V. Chloroplast Genome Analysis of Resurrection Tertiary Relict Haberlea rhodopensis Highlights Genes Important for Desiccation Stress Response. Front. Plant Sci. 2017, 8, 204. [Google Scholar] [CrossRef]
- Singh, N.V.; Patil, P.G.; Sowjanya, R.P.; Parashuram, S.; Natarajan, P.; Babu, K.D.; Pal, R.K.; Sharma, J.; Reddy, U.K. Chloroplast Genome Sequencing, Comparative Analysis, and Discovery of Unique Cytoplasmic Variants in Pomegranate (Punica granatum L.). Front. Genet. 2021, 12, 704075. [Google Scholar] [CrossRef]
- Nie, X.; Deng, P.; Feng, K.; Liu, P.; Du, X.; You, F.M.; Weining, S. Comparative analysis of codon usage patterns in chloroplast genomes of the Asteraceae family. Plant Mol. Biol. Rep. 2014, 32, 828–840. [Google Scholar] [CrossRef]
- Gu, C.; Tembrock, L.R.; Zheng, S.; Wu, Z. The Complete Chloroplast Genome of Catha edulis: A Comparative Analysis of Genome Features with Related Species. Int. J. Mol. Sci. 2018, 19, 525. [Google Scholar] [CrossRef]
- Park, M.; Park, H.; Lee, H.; Lee, B.-H.; Lee, J. The Complete Plastome Sequence of an Antarctic Bryophyte Sanionia uncinata (Hedw.) Loeske. Int. J. Mol. Sci. 2018, 19, 709. [Google Scholar] [CrossRef]
- Thiel, T.; Michalek, W.; Varshney, R.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar] [CrossRef]
- Mader, M.; Pakull, B.; Blanc-Jolivet, C.; Paulini-Drewes, M.; Bouda, Z.H.-N.; Degen, B.; Small, I.; Kersten, B. Complete Chloroplast Genome Sequences of Four Meliaceae Species and Comparative Analyses. Int. J. Mol. Sci. 2018, 19, 701. [Google Scholar] [CrossRef]
- Coulibaly, D.; Huang, X.; Ting, S.; Iqbal, S.; Ni, Z.; Ouma, K.O.; Hayat, F.; Tan, W.; Hu, G.; Ma, C.; et al. Comparative Analysis of Complete Chloroplast Genome and Phenotypic Characteristics of Japanese Apricot Accessions. Horticulturae 2022, 8, 794. [Google Scholar] [CrossRef]
- Sobreiro, M.B.; Vieira, L.D.; Nunes, R.; Novaes, E.; Coissac, E.; Silva-Junior, O.B.; Grattapaglia, D.; Collevatti, R.G. Chloroplast genome assembly of Handroanthus impetiginosus: Comparative analysis and molecular evolution in Bignoniaceae. Planta 2020, 252, 91. [Google Scholar] [CrossRef]
- Ni, L.; Zhao, Z.; Dorje, G.; Ma, M. The Complete Chloroplast Genome of Ye-Xing-Ba (Scrophularia dentata; Scrophulariaceae), an Alpine Tibetan Herb. PLoS ONE 2016, 11, e0158488. [Google Scholar] [CrossRef]
- Li, C.; Zheng, Y.; Huang, P. Molecular markers from the chloroplast genome of rose provide a complementary tool for variety discrimination and profiling. Sci. Rep. 2020, 10, 12188. [Google Scholar] [CrossRef]
- Luo, C.; Huang, W.; Yer, H.; Kamuda, T.; Li, X.; Li, Y.; Rong, Y.; Yan, B.; Wen, Y.; Wang, Q.; et al. Complete Chloroplast Genomes and Comparative Analyses of Three Ornamental Impatiens Species. Front. Genet. 2022, 13, 816123. [Google Scholar] [CrossRef]
- Gu, C.; Tembrock, L.R.; Johnson, N.G.; Simmons, M.P.; Wu, Z. The Complete Plastid Genome of Lagerstroemia fauriei and Loss of rpl2 Intron from Lagerstroemia (Lythraceae). PLoS ONE 2016, 11, e0150752. [Google Scholar] [CrossRef]
- Luo, C.; Huang, W.; Sun, H.; Yer, H.; Li, X.; Li, Y.; Yan, B.; Wang, Q.; Wen, Y.; Huang, M.; et al. Comparative chloroplast genome analysis of Impatiens species (Balsaminaceae) in the karst area of China: Insights into genome evolution and phylogenomic implications. BMC Genom. 2021, 22, 571. [Google Scholar] [CrossRef]
- Yan, M.; Zhao, X.; Zhou, J.; Huo, Y.; Ding, Y.; Yuan, Z. The Complete Chloroplast Genomes of Punica granatum and a Comparison with Other Species in Lythraceae. Int. J. Mol. Sci. 2019, 20, 2886. [Google Scholar] [CrossRef]
- Chen, Y.; Hu, N.; Wu, H. Analyzing and Characterizing the Chloroplast Genome of Salix wilsonii. BioMed Res. Int. 2019, 2019, 5190425. [Google Scholar] [CrossRef]
- Zhou, J.; Cui, Y.; Chen, X.; Li, Y.; Xu, Z.; Duan, B.; Li, Y.; Song, J.; Yao, H. Complete Chloroplast Genomes of Papaver rhoeas and Papaver orientale: Molecular Structures, Comparative Analysis, and Phylogenetic Analysis. Molecules 2018, 23, 437. [Google Scholar] [CrossRef]
- Shen, X.; Wu, M.; Liao, B.; Liu, Z.; Bai, R.; Xiao, S.; Li, X.; Zhang, B.; Xu, J.; Chen, S. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of the Medicinal Plant Artemisia annua. Molecules 2017, 22, 1330. [Google Scholar] [CrossRef]
- Guo, S.; Guo, L.; Zhao, W.; Xu, J.; Li, Y.; Zhang, X.; Shen, X.; Wu, M.; Hou, X. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Paeonia ostia. Molecules 2018, 23, 246. [Google Scholar] [CrossRef]
- Wang, W.; Yu, H.; Wang, J.; Lei, W.; Gao, J.; Qiu, X.; Wang, J. The Complete Chloroplast Genome Sequences of the Medicinal Plant Forsythia suspensa (Oleaceae). Int. J. Mol. Sci. 2017, 18, 2288. [Google Scholar] [CrossRef]
- Maréchal, A.; Brisson, N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010, 186, 299–317. [Google Scholar] [CrossRef]
- Deng, Y.; Luo, Y.; He, Y.; Qin, X.; Li, C.; Deng, X. Complete Chloroplast Genome of Michelia Shiluensis and a Comparative Analysis with Four Magnoliaceae Species. Forests 2020, 11, 267. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, T.; Duan, D.; Yang, J.; Feng, L.; Zhao, G. Comparative Analysis of the Complete Chloroplast Genomes of Five Quercus Species. Front. Plant Sci. 2016, 7, 959. [Google Scholar] [CrossRef]
- Ebert, D.; Peakall, R. Chloroplast simple sequence repeats (cpSSRs): Technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant species. Mol. Ecol. Resour. 2009, 9, 673–690. [Google Scholar] [CrossRef]
- Mohammad-Panah, N.; Shabanian, N.; Khadivi, A.; Rahmani, M.-S.; Emami, A. Genetic structure of gall oak (Quercus infectoria) characterized by nuclear and chloroplast SSR markers. Tree Genet. Genomes 2017, 13, 70. [Google Scholar] [CrossRef]
- Provan, J.; Powell, W.; Hollingsworth, P.M. Chloroplast microsatellites: New tools for studies in plant ecology and evolution. Trends Ecol. Evol. 2001, 16, 142–147. [Google Scholar] [CrossRef]
- Kim, K.; Lee, H.L. Complete Chloroplast Genome Sequences from Korean Ginseng (Panax schinseng Nees) and Comparative Analysis of Sequence Evolution among 17 Vascular Plants. DNA Res. 2004, 11, 247–261. [Google Scholar] [CrossRef]
- Liu, L.; Li, R.; Worth, J.R.P.; Li, X.; Li, P.; Cameron, K.M.; Fu, C. The Complete Chloroplast Genome of Chinese Bayberry (Morella rubra, Myricaceae): Implications for Understanding the Evolution of Fagales. Front. Plant Sci. 2017, 8, 968. [Google Scholar] [CrossRef]
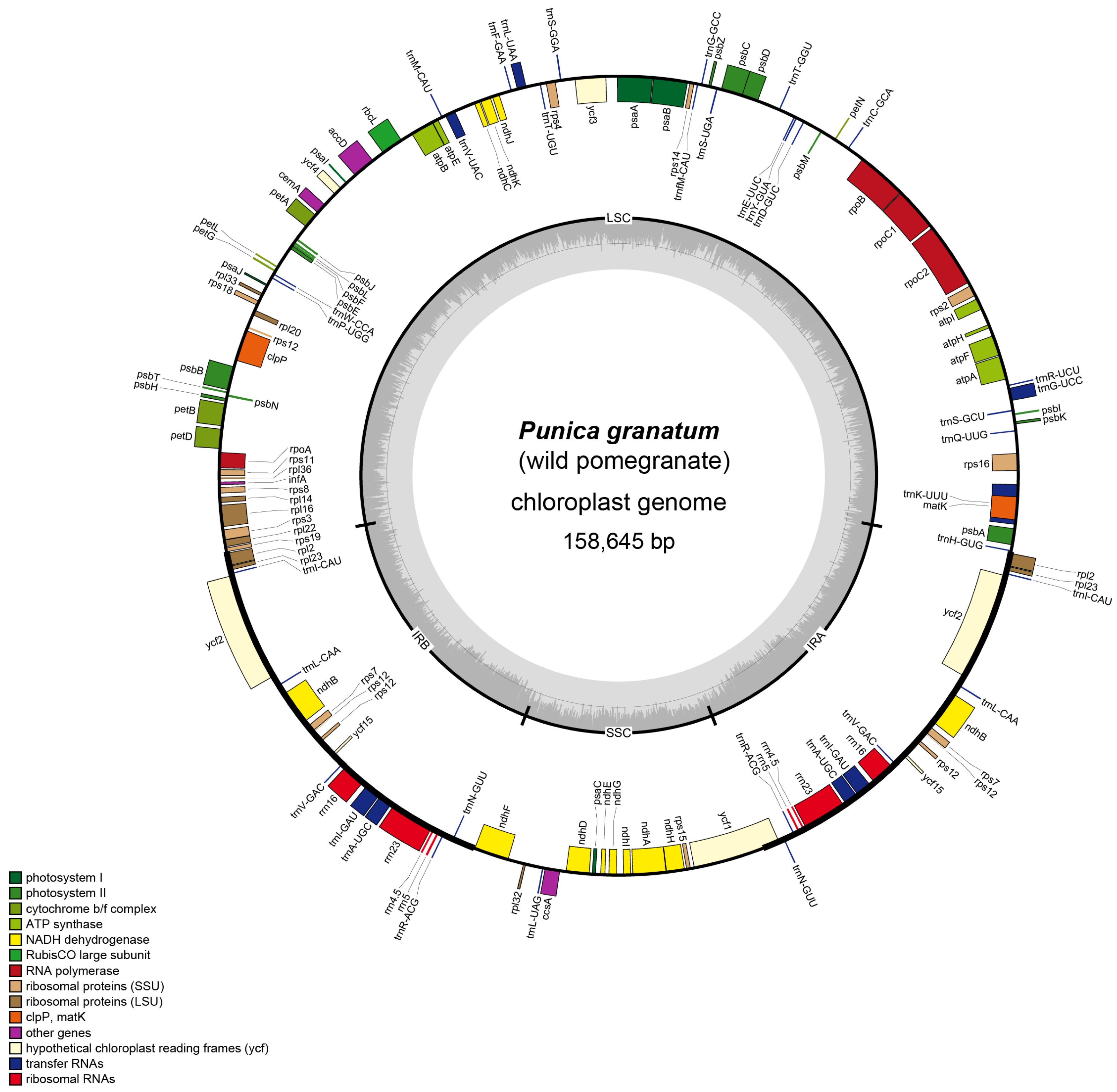
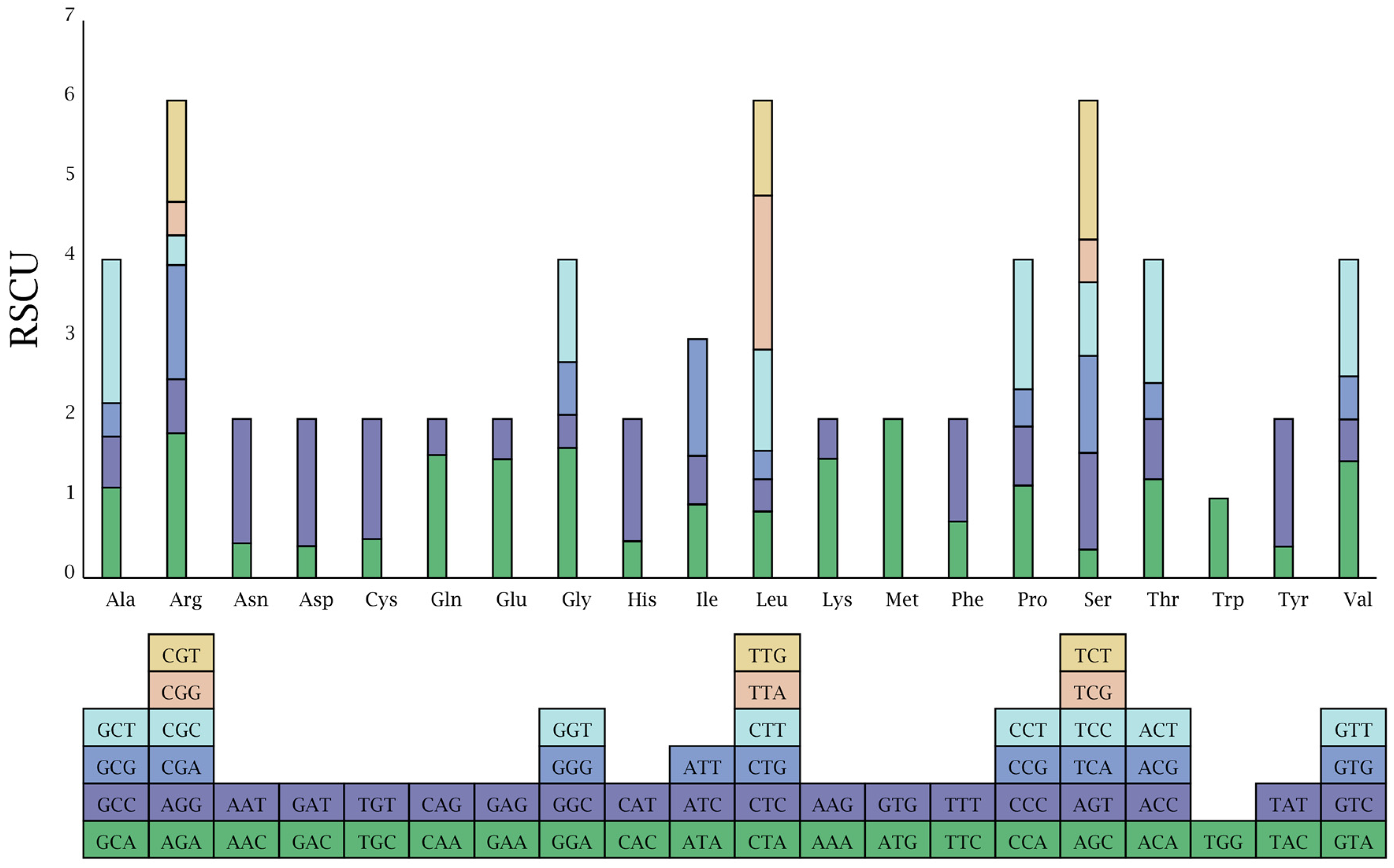
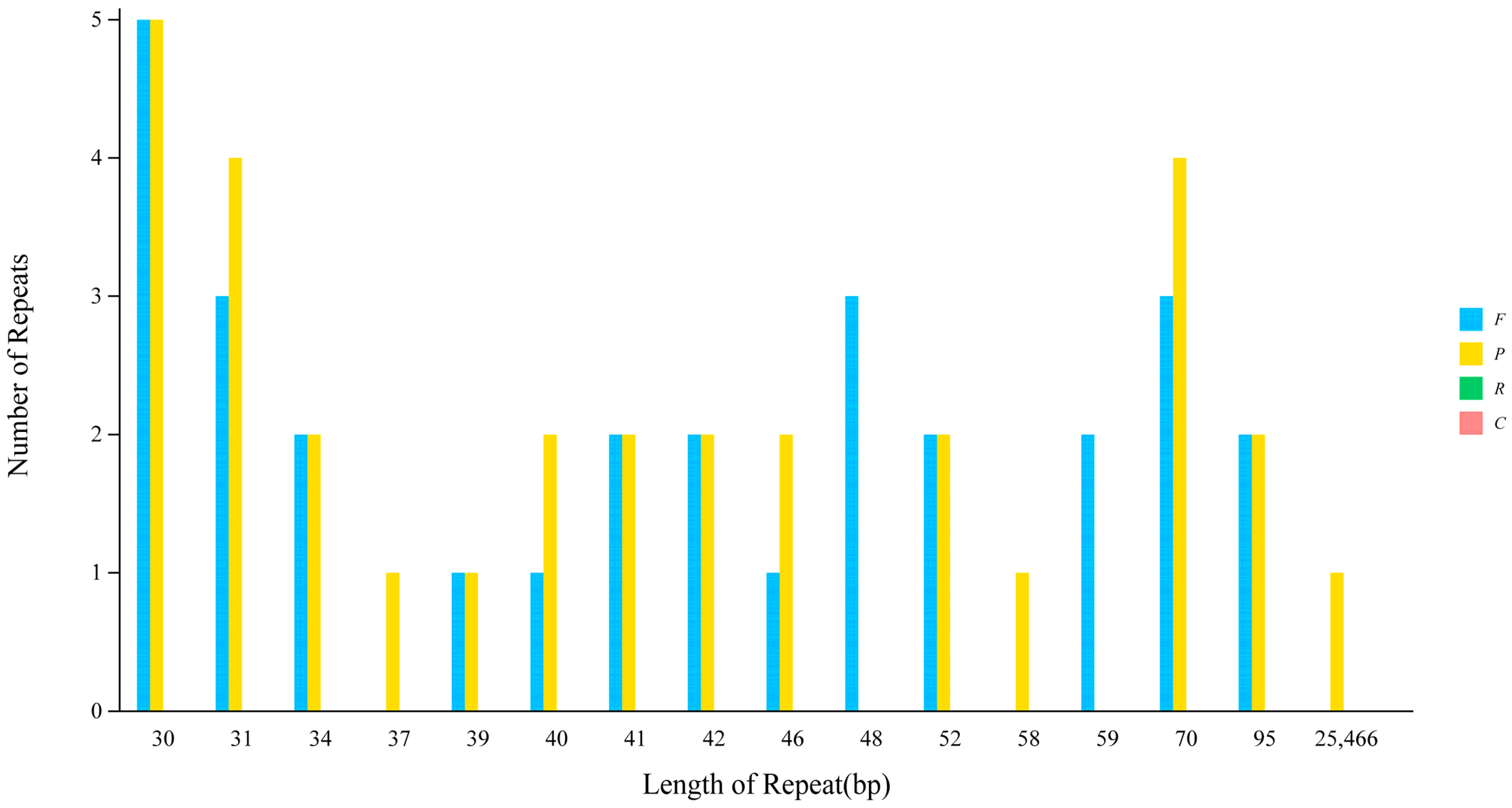
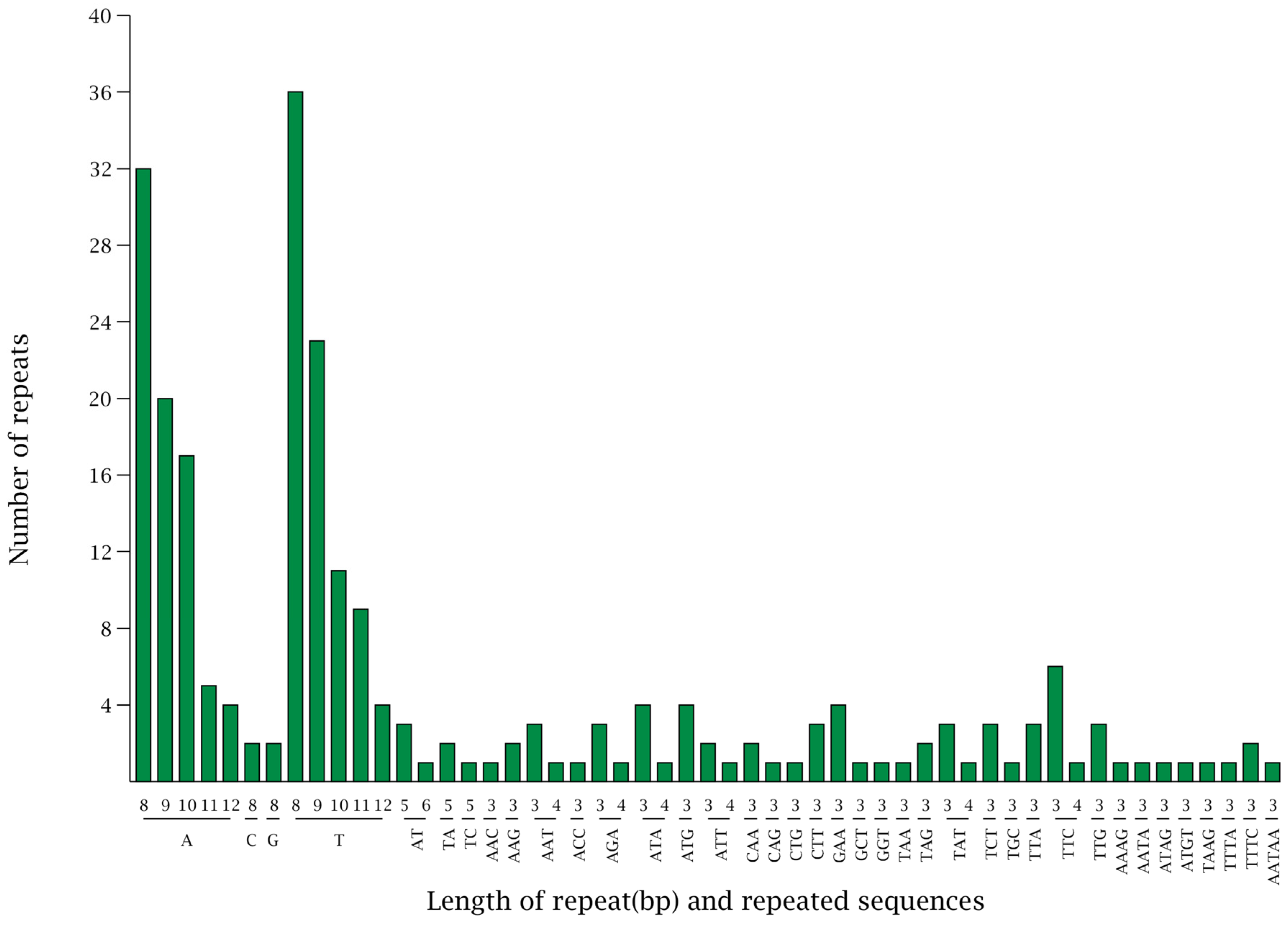


| Parameters | Wild Pomegranate | Punica granatum cultivar ‘Nana’ | Punica granatum cultivar ‘Tunisia’ | Heimia myrtifolia | Lagerstroemia indica | Lagerstroemia intermedia | Lagerstroemia speciose | Lagerstroemia subcostata | Sonneratia alba | Trapa maximowicizz |
|---|---|---|---|---|---|---|---|---|---|---|
| Genome size (bp) | 158,645 | 158,638 | 158,639 | 159,219 | 152,025 | 152,330 | 152,476 | 152,049 | 153,061 | 155,577 |
| LSC size (bp) | 89,028 | 89,021 | 89,022 | 88,571 | 84,046 | 83,987 | 84,051 | 83,890 | 78,226 | 88,528 |
| SSC size(bp) | 18,686 | 18,684 | 18,684 | 18,821 | 16,914 | 16,873 | 16,979 | 16,909 | 18,032 | 18,272 |
| IR size (bp) | 25,465 | 25,467 | 25,467 | 25,914 | 25,623 | 25,736 | 25,723 | 25,625 | 23,902 | 24,389 |
| Number of genes | 132 | 113 | 113 | 112 | 113 | 130 | 129 | 129 | 107 | 110 |
| Protein genes | 84 | 79 | 79 | 78 | 79 | 85 | 85 | 84 | 29 | 77 |
| tRNA genes | 37 | 30 | 30 | 30 | 30 | 37 | 36 | 37 | 24 | 29 |
| rRNA genes | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| GC content (%) | 36.92 | 36.92 | 36.92 | 37.59 | 36.95 | 37.6 | 37.6 | 37.6 | 37.29 | 36.4 |
| GenBank accession | MT600023 | MK603513 | MK603512 | MG921615 | NC_030484 | NC_034662 | NC_031414 | NC_034952 | NC_039975 | NC_037023 |
| Category | Gene Group | Gene Name |
|---|---|---|
| Photosynthesis | Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of NADH dehydrogenase | ndhA *, ndhB * (2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Subunits of cytochrome b/f complex | petA, petB *, petD *, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF *, atpH, atpI | |
| Large subunit of rubisco | rbcL | |
| Self-replication | Proteins of large ribosomal subunit | rpl14, rpl16 *, rpl2 (2), rpl20, rpl22, rpl23 (2), rpl32, rpl33, rpl36 |
| Proteins of small ribosomal subunit | rps11, rps12 ** (2), rps14, rps15, rps16 *, rps18, rps19, rps2, rps3, rps4, rps7 (2), rps8 | |
| Subunits of RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | |
| Ribosomal RNAs | rrn16 (2), rrn23 (2), rrn4.5 (2), rrn5 (2) | |
| Transfer RNAs | trnA-UGC * (2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC *, trnH-GUG, trnI-CAU (2), trnI-GAU * (2), trnK-UUU *, trnL-CAA (2), trnL-UAA *, trnL-UAG, trnM-CAU, trnN-GUU (2), trnP-UGG, trnQ-UUG, trnR-ACG (2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC (2), trnV-UAC *, trnW-CCA, trnY-GUA, trnfM-CAU | |
| Other genes | Maturase | matK |
| Protease | clpP ** | |
| Envelope membrane protein | cemA | |
| Acetyl-CoA carboxylase | accD | |
| c-type cytochrome synthesis gene | ccsA | |
| Translation initiation factor | # infA | |
| Genes of unknown function | Conserved hypothetical chloroplast ORF | # ycf15 (2), ycf1, ycf2 (2), ycf3 **, ycf4 |
| Gene | Location | Exon I | Intron I | Exon II | Intron II | Exon II |
|---|---|---|---|---|---|---|
| trnK-UUU | LSC | 37 | 2489 | 35 | ||
| rps16 | LSC | 40 | 855 | 227 | ||
| trnG-UCC | LSC | 23 | 738 | 48 | ||
| atpF | LSC | 145 | 759 | 410 | ||
| rpoC1 | LSC | 432 | 749 | 1608 | ||
| ycf3 | LSC | 124 | 727 | 230 | 767 | 153 |
| trnL-UAA | LSC | 35 | 517 | 50 | ||
| trnV-UAC | LSC | 38 | 603 | 35 | ||
| rps12 | IRa | 114 | - | 232 | 548 | 26 |
| clpP | LSC | 71 | 607 | 292 | 811 | 228 |
| petB | LSC | 6 | 779 | 642 | ||
| petD | LSC | 8 | 766 | 475 | ||
| rpl16 | LSC | 9 | 994 | 399 | ||
| ndhB | IRb | 777 | 683 | 756 | ||
| rps12 | IRb | 232 | - | 26 | 548 | 114 |
| trnI-GAU | IRb | 37 | 949 | 35 | ||
| trnA-UGC | IRb | 38 | 803 | 35 | ||
| ndhA | SSC | 553 | 1049 | 539 | ||
| trnA-UGC | IRa | 38 | 803 | 35 | ||
| trnI-GAU | IRa | 37 | 949 | 35 | ||
| ndhB | IRa | 777 | 683 | 756 |
| Amino Acid | Codon | Count | RSCU | Amino Acid | Codon | Count | RSCU |
|---|---|---|---|---|---|---|---|
| Ter | UAA | 50 | 1.7856 | Met | GUG | 1 | 0.0032 |
| Ter | UAG | 19 | 0.6786 | Asn | AAC | 277 | 0.4362 |
| Ter | UGA | 15 | 0.5358 | Asn | AAU | 993 | 1.5638 |
| Ala | GCA | 391 | 1.1348 | Pro | CCA | 313 | 1.1648 |
| Ala | GCC | 222 | 0.6444 | Pro | CCC | 199 | 0.7404 |
| Ala | GCG | 144 | 0.418 | Pro | CCG | 126 | 0.4688 |
| Ala | GCU | 621 | 1.8028 | Pro | CCU | 437 | 1.626 |
| Cys | UGC | 73 | 0.4882 | Gln | CAA | 717 | 1.547 |
| Cys | UGU | 226 | 1.5118 | Gln | CAG | 210 | 0.453 |
| Asp | GAC | 222 | 0.403 | Arg | AGA | 479 | 1.818 |
| Asp | GAU | 880 | 1.597 | Arg | AGG | 179 | 0.6792 |
| Glu | GAA | 1041 | 1.4946 | Arg | CGA | 378 | 1.4346 |
| Glu | GAG | 352 | 0.5054 | Arg | CGC | 99 | 0.3756 |
| Phe | UUC | 521 | 0.7108 | Arg | CGG | 110 | 0.4176 |
| Phe | UUU | 945 | 1.2892 | Arg | CGU | 336 | 1.275 |
| Gly | GGA | 731 | 1.6364 | Ser | AGC | 122 | 0.3594 |
| Gly | GGC | 186 | 0.4164 | Ser | AGU | 413 | 1.2156 |
| Gly | GGG | 296 | 0.6624 | Ser | UCA | 413 | 1.2156 |
| Gly | GGU | 574 | 1.2848 | Ser | UCC | 315 | 0.9276 |
| His | CAC | 145 | 0.4632 | Ser | UCG | 181 | 0.5328 |
| His | CAU | 481 | 1.5368 | Ser | UCU | 594 | 1.749 |
| Ile | AUA | 703 | 0.9267 | Thr | ACA | 410 | 1.2396 |
| Ile | AUC | 463 | 0.6102 | Thr | ACC | 251 | 0.7588 |
| Ile | AUU | 1110 | 1.4631 | Thr | ACG | 149 | 0.4504 |
| Lys | AAA | 1065 | 1.501 | Thr | ACU | 513 | 1.5512 |
| Lys | AAG | 354 | 0.499 | Val | GUA | 521 | 1.4676 |
| Leu | CUA | 387 | 0.8376 | Val | GUC | 186 | 0.524 |
| Leu | CUC | 187 | 0.4044 | Val | GUG | 193 | 0.5436 |
| Leu | CUG | 164 | 0.3546 | Val | GUU | 520 | 1.4648 |
| Leu | CUU | 590 | 1.2768 | Trp | UGG | 459 | 1 |
| Leu | UUA | 892 | 1.9302 | Tyr | UAC | 192 | 0.3966 |
| Leu | UUG | 553 | 1.1964 | Tyr | UAU | 776 | 1.6034 |
| Met | AUG | 607 | 1.9968 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Ren, Y.; Zhao, J.; Wang, Y.; Liu, X.; Zhao, X.; Yuan, Z. Phylogenetic Analysis of Wild Pomegranate (Punica granatum L.) Based on Its Complete Chloroplast Genome from Tibet, China. Agronomy 2023, 13, 126. https://doi.org/10.3390/agronomy13010126
Chen L, Ren Y, Zhao J, Wang Y, Liu X, Zhao X, Yuan Z. Phylogenetic Analysis of Wild Pomegranate (Punica granatum L.) Based on Its Complete Chloroplast Genome from Tibet, China. Agronomy. 2023; 13(1):126. https://doi.org/10.3390/agronomy13010126
Chicago/Turabian StyleChen, Lide, Yuan Ren, Jun Zhao, Yuting Wang, Xueqing Liu, Xueqing Zhao, and Zhaohe Yuan. 2023. "Phylogenetic Analysis of Wild Pomegranate (Punica granatum L.) Based on Its Complete Chloroplast Genome from Tibet, China" Agronomy 13, no. 1: 126. https://doi.org/10.3390/agronomy13010126
APA StyleChen, L., Ren, Y., Zhao, J., Wang, Y., Liu, X., Zhao, X., & Yuan, Z. (2023). Phylogenetic Analysis of Wild Pomegranate (Punica granatum L.) Based on Its Complete Chloroplast Genome from Tibet, China. Agronomy, 13(1), 126. https://doi.org/10.3390/agronomy13010126





