CRISPR/Cas9-Mediated Genome Editing of Soluble Starch Synthesis Enzyme in Rice for Low Glycemic Index
Abstract
:1. Introduction
2. Results
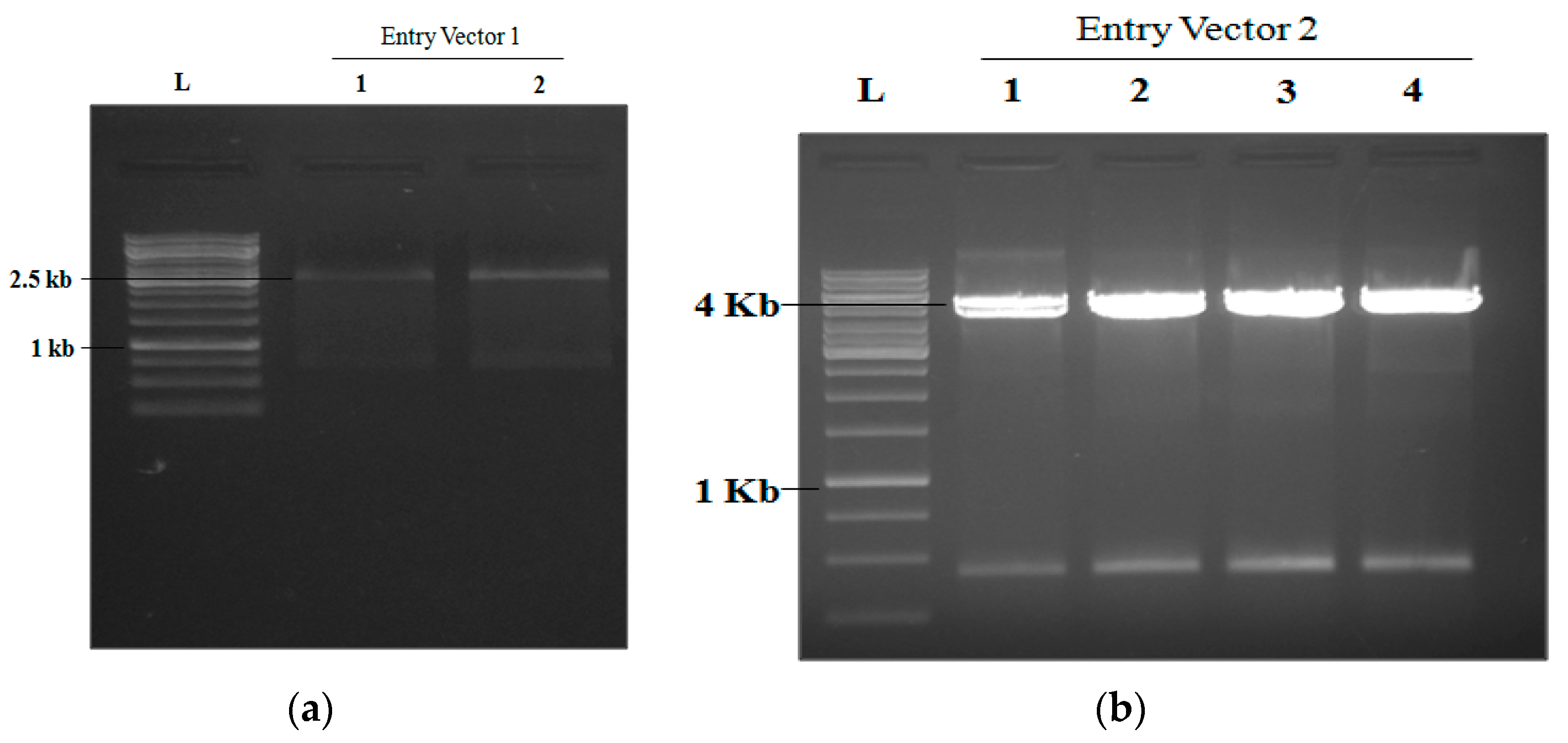
2.1. The Digestion of EV1 and EV2 with Bsa1 Endonuclease
2.2. Confirmation of gRNAs in Recombinant Vectors EV1 and EV2 Using PCR
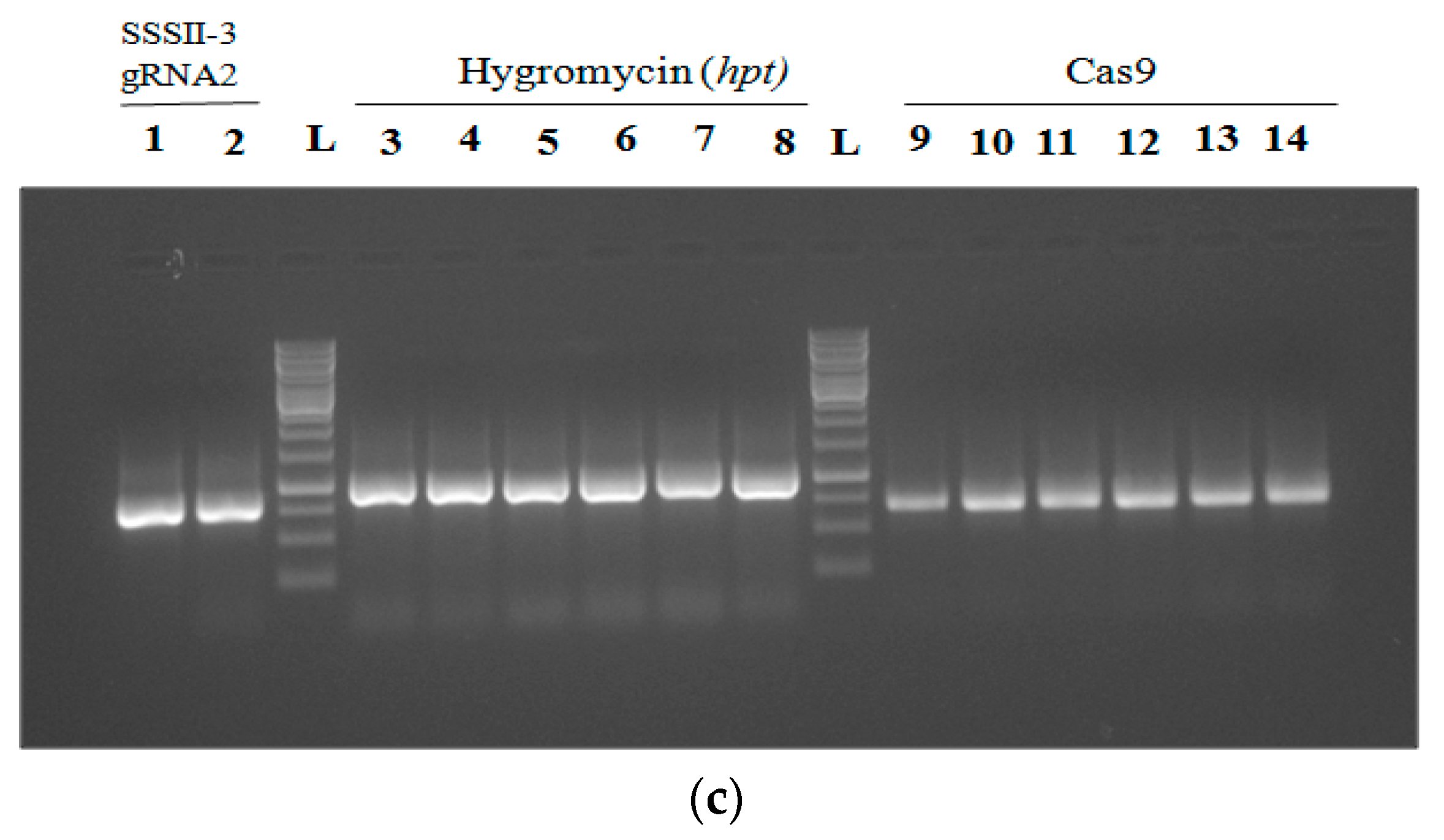
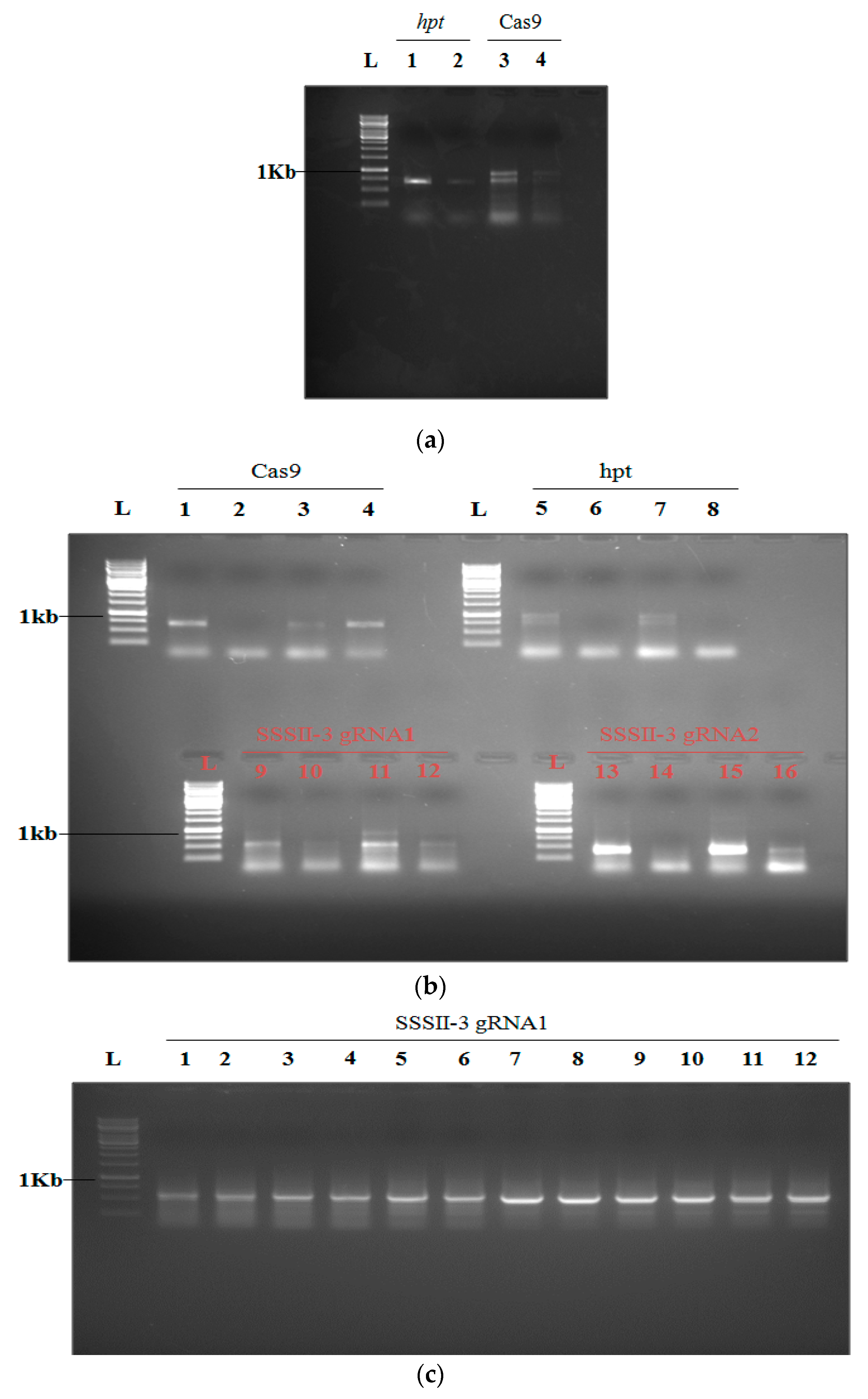
2.3. Confirmation of Ultimate Binary Expression Vector with hpt, Cas9 Gene, and the gRNA1, gRNA2
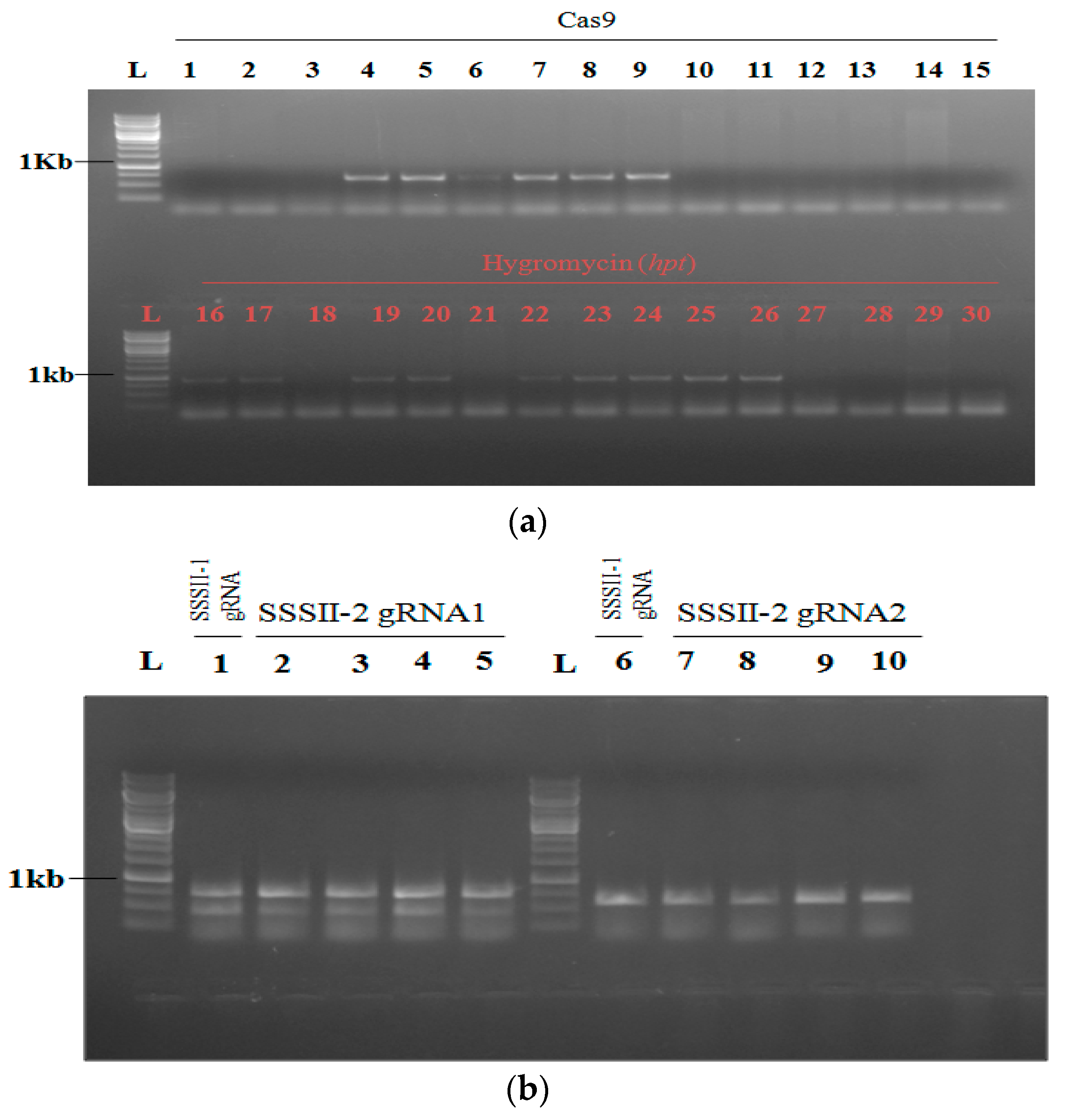
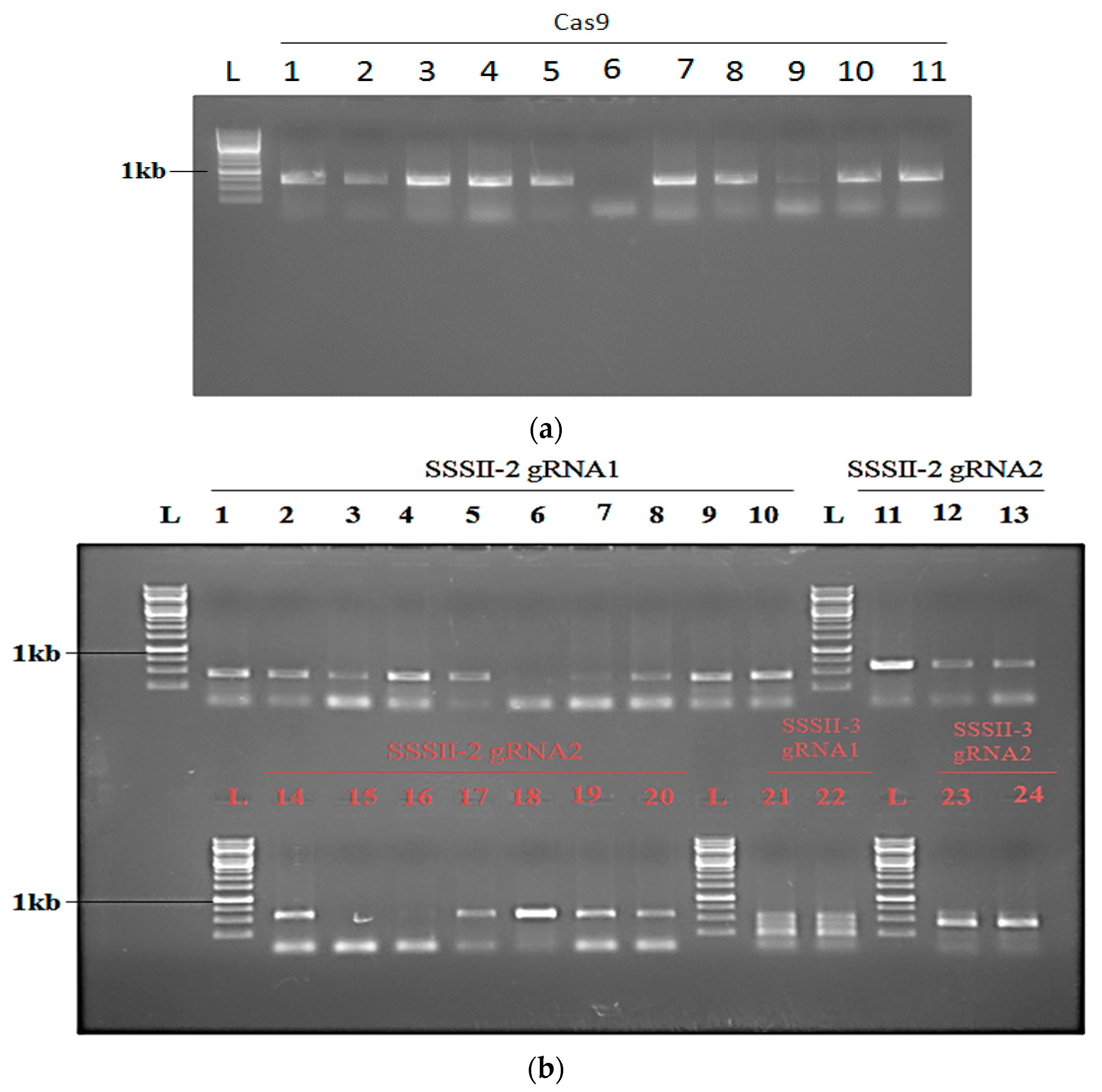
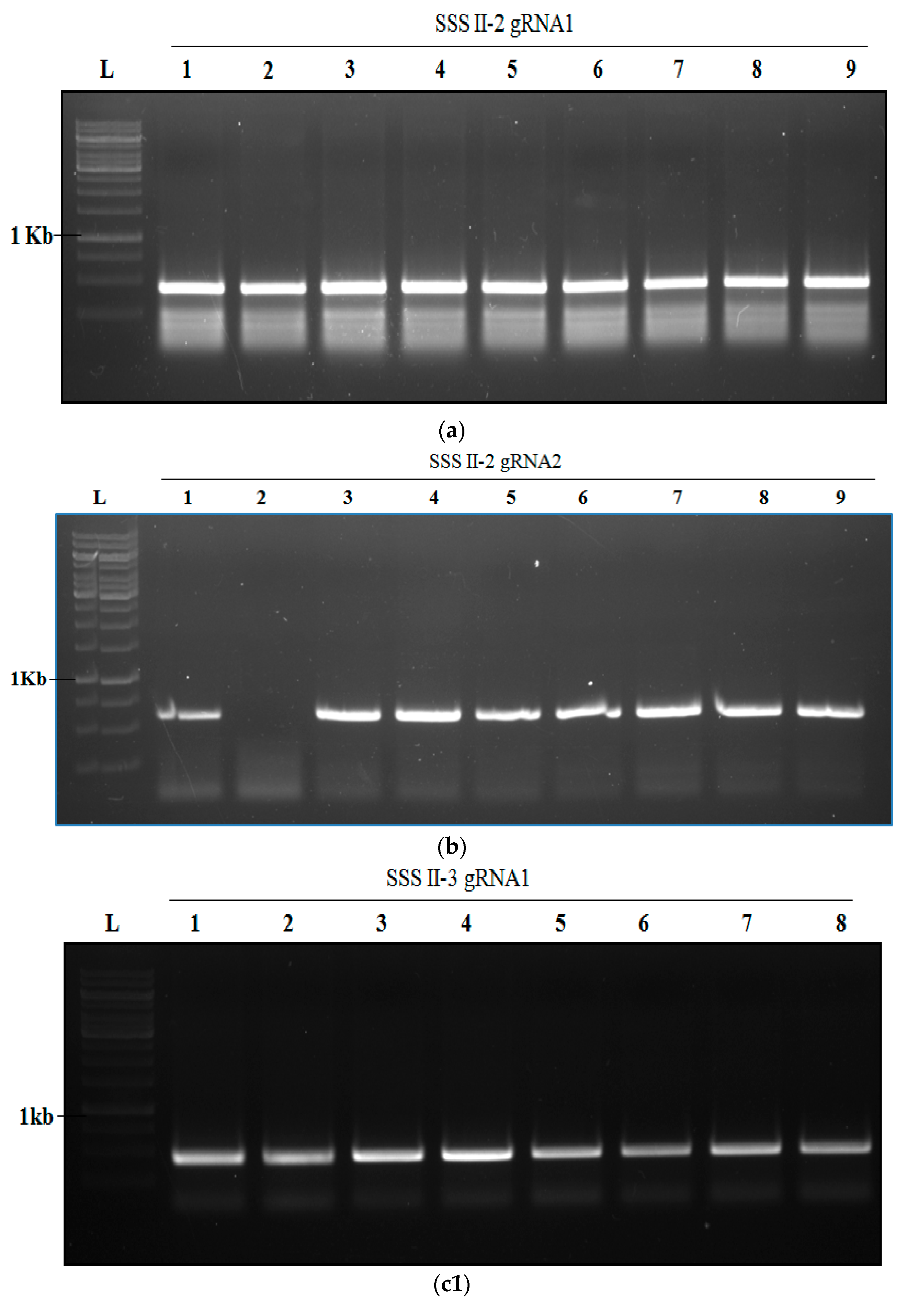
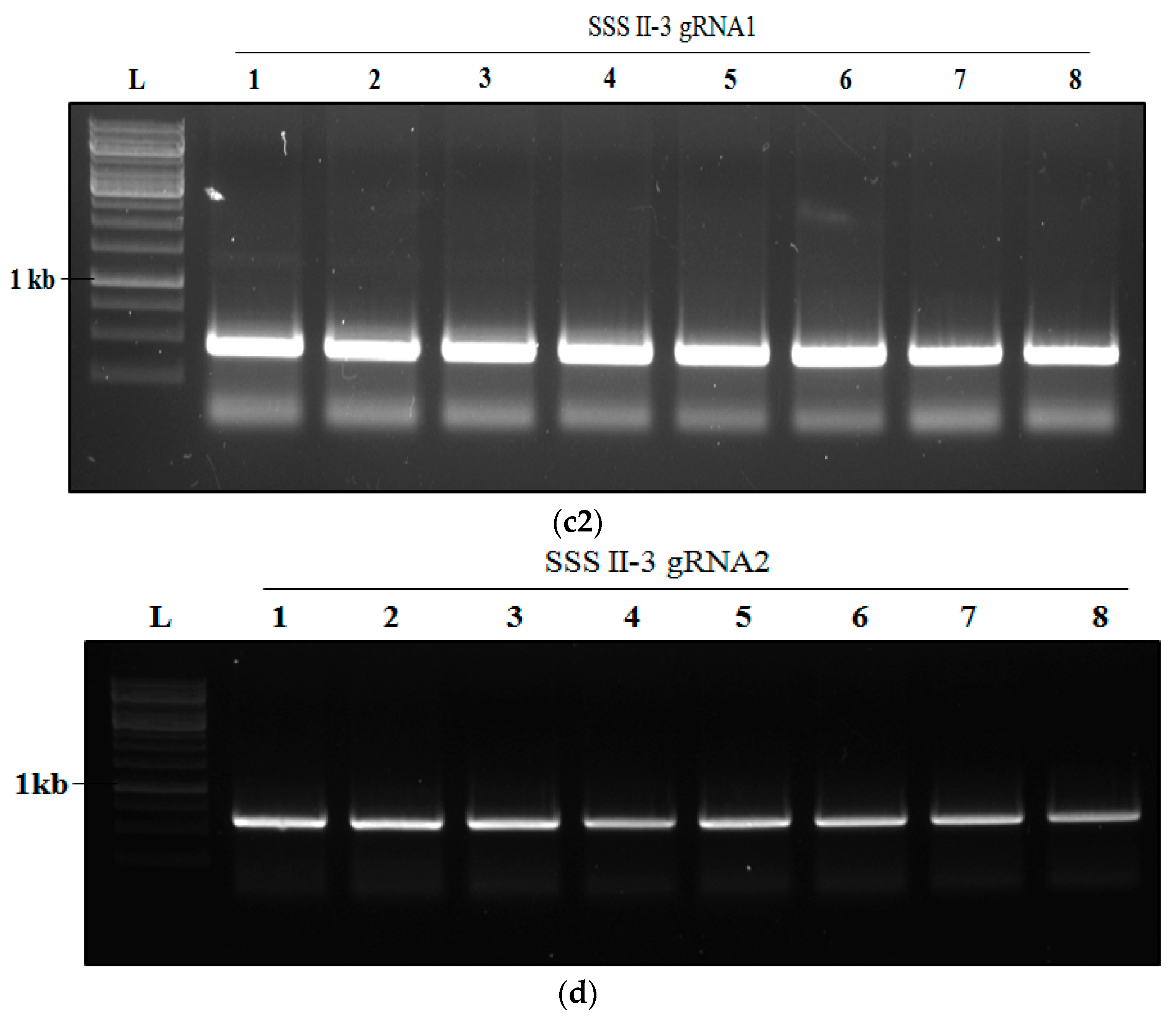
2.4. Transformation of Rice and Generation of SSSII-1, SSSII-2 and SSSII-3 Knockout Lines
2.5. Analysis of Transgenic Rice Lines of T0 Generation
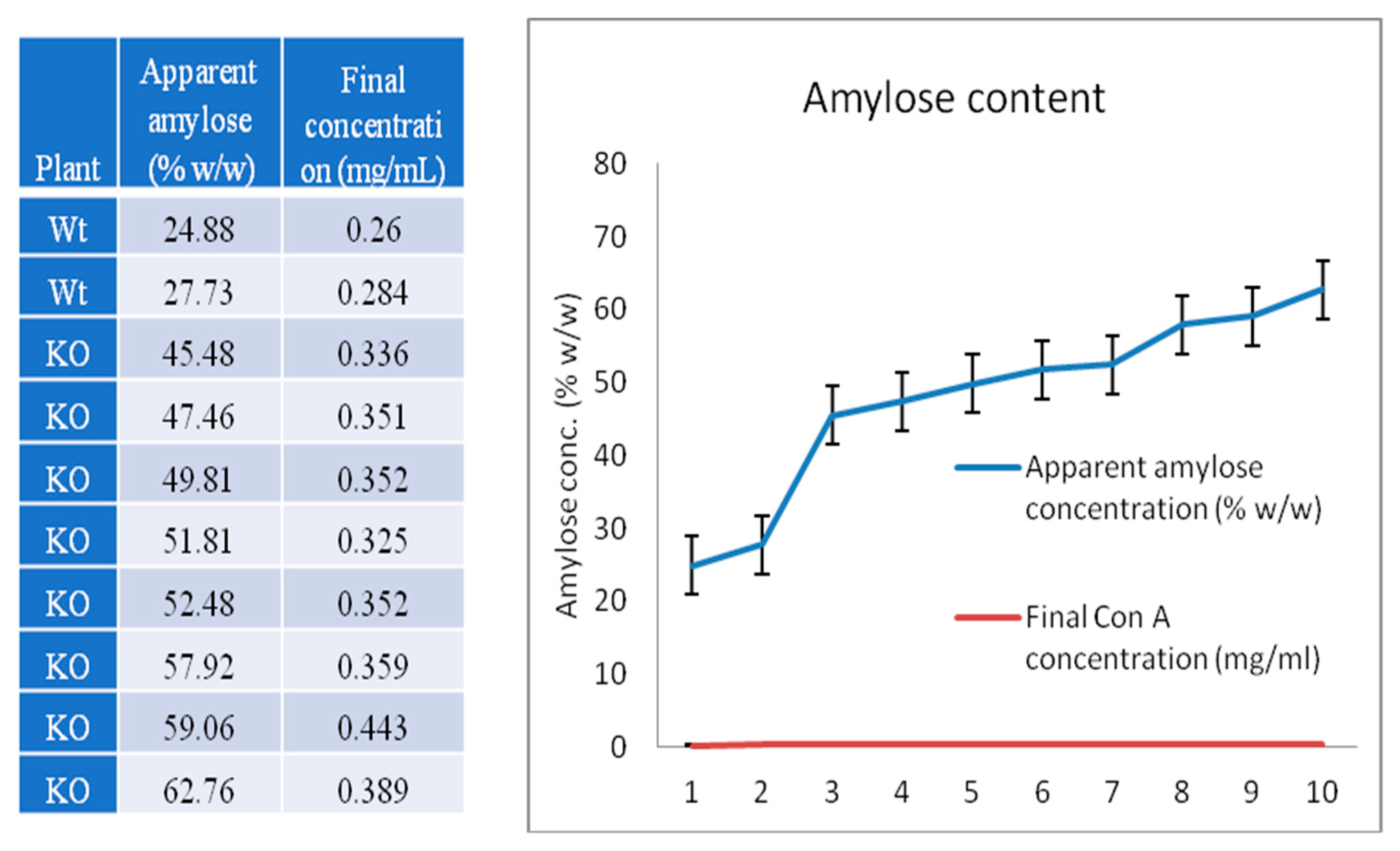
2.6. High Level of RS, High-Amylose, and Gelatinization of Properties of Rice Starch
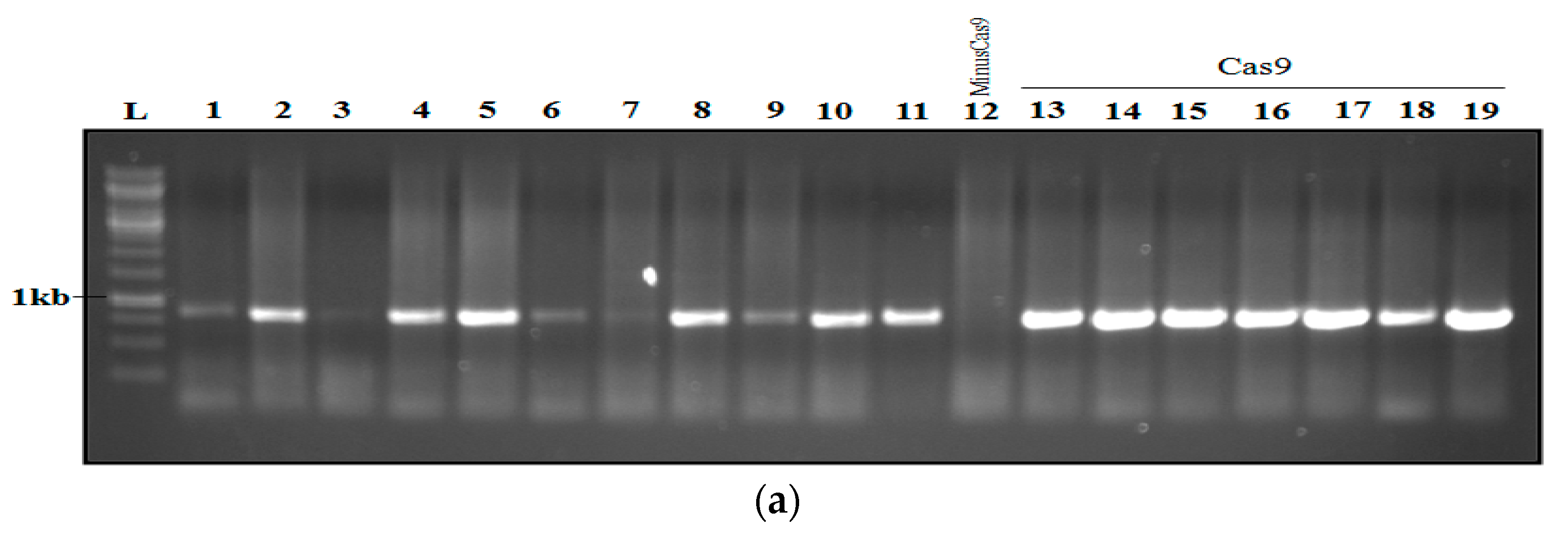
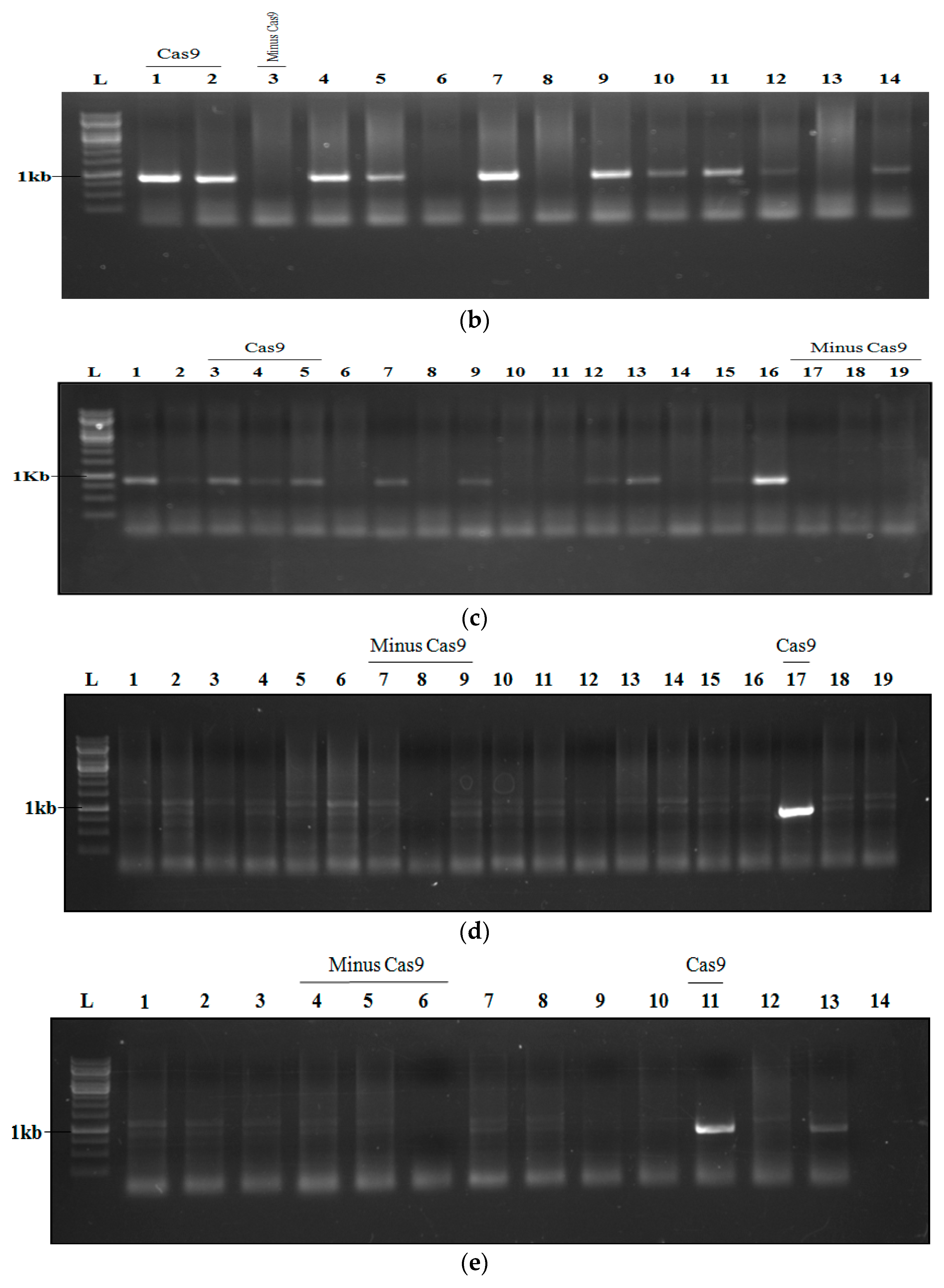
2.7. Analysis and Confirmation of Cas9-Free Edited Lines
2.8. Sequencing Analysis for Cas9-Free Plants
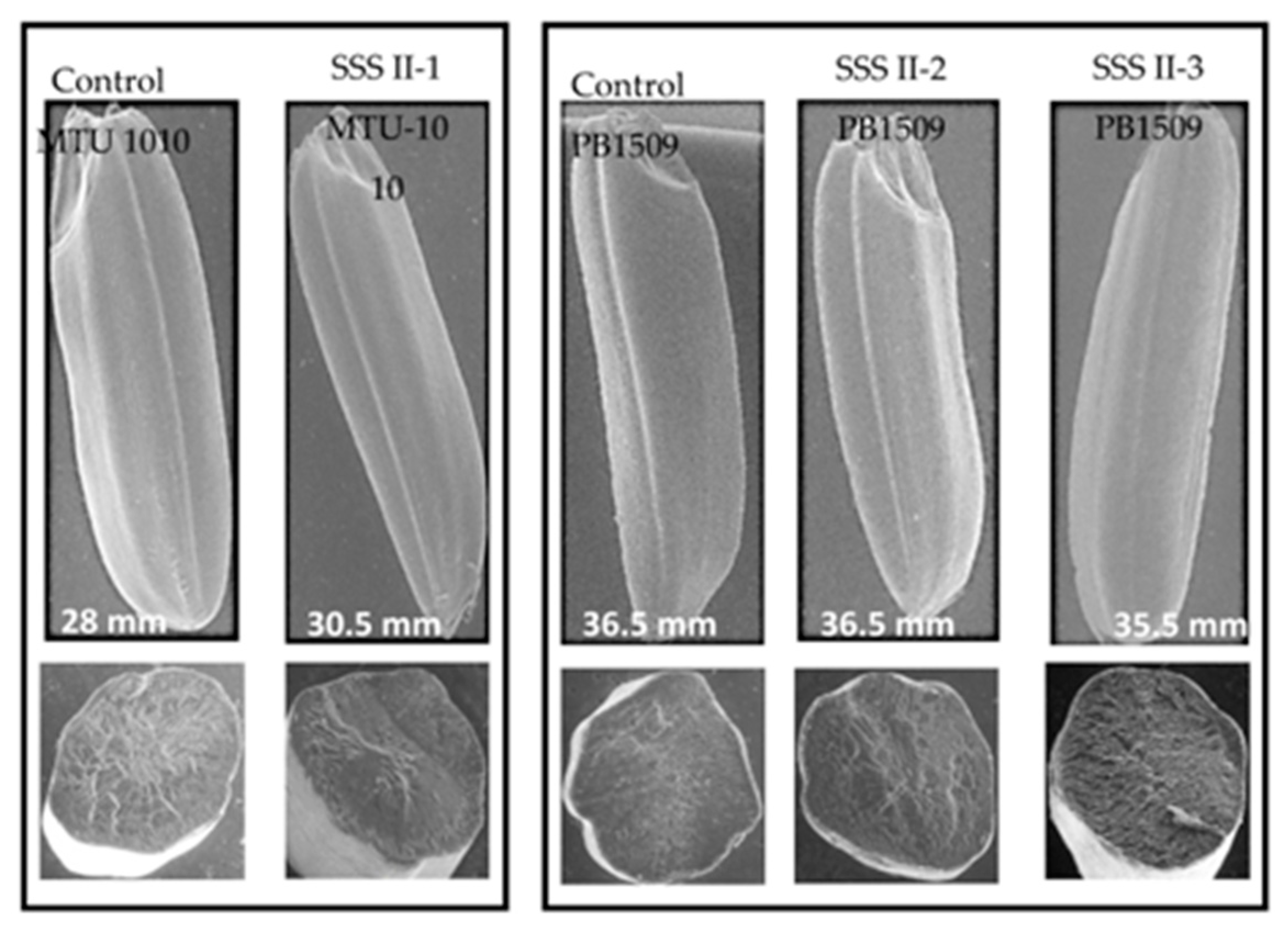
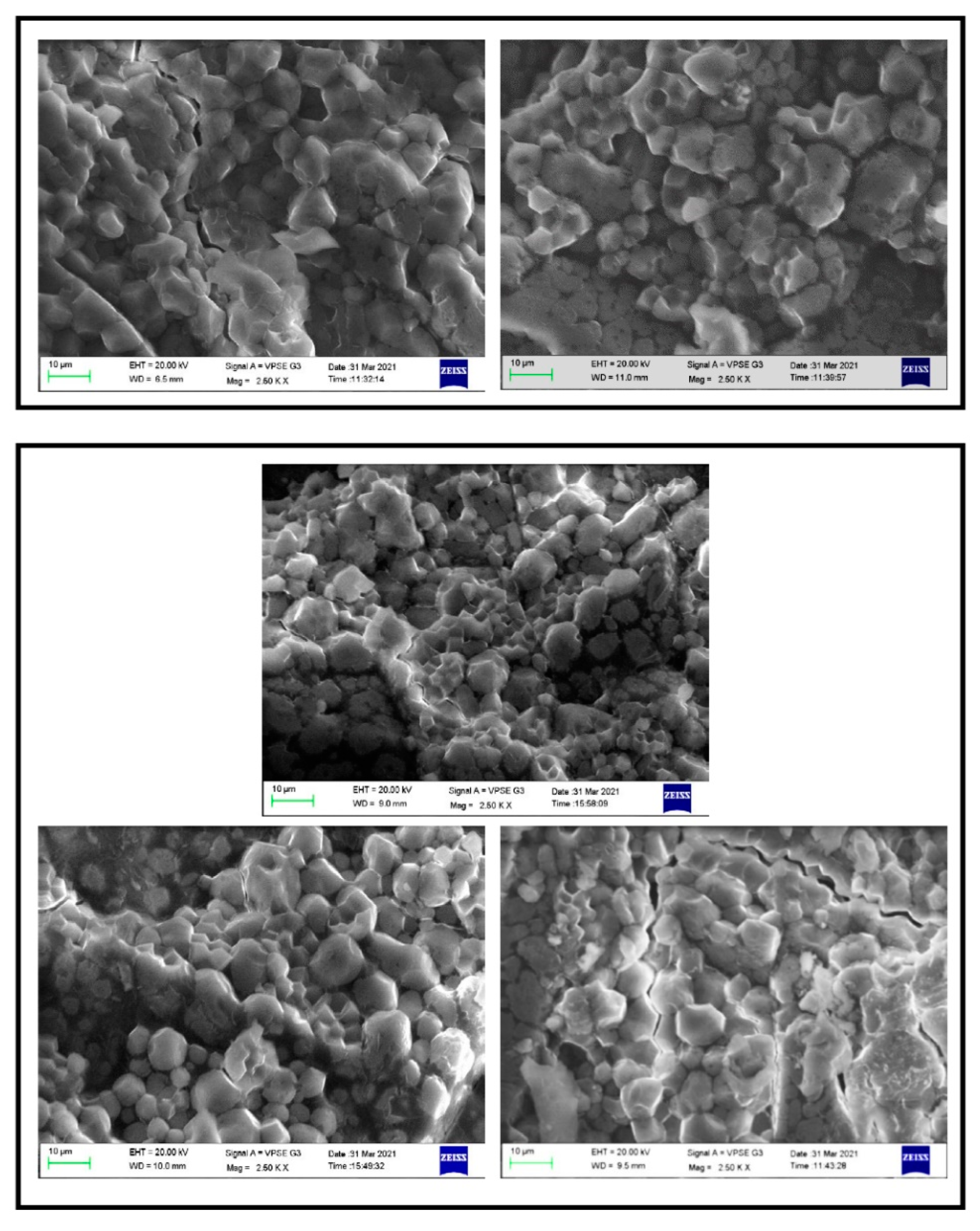
2.9. Size of Rice Grains and Starch Structure
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhu, L.; Gu, M.; Meng, X.; Cheung, S.C.; Yu, H.; Huang, J.; Sun, Y.; Shi, Y.; Liu, Q. High-amylose rice improves indices of animal health in normal and diabetic rats. Plant Biotech. J. 2012, 10, 353–362. [Google Scholar] [CrossRef]
- Miao, J.; Guo, D.; Zhang, J.; Huang, Q.; Qin, G.; Zhang, X.; Wan, J.; Gu, H.; Qu, L.J. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013, 23, 1233–1236. [Google Scholar] [CrossRef]
- Atwell, S.; Huang, Y.S.; Vilhjálmsson, B.J.; Willems, G.; Horton, M.; Li, Y.; Meng, D.; Platt, A.; Tarone, A.M.; Hu, T.T.; et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 2010, 465, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Umemoto, T.; Horibata, T.; Aoki, N.; Hiratsuka, M.; Yano, M.; Inouchi, N. Effects of variations in starch synthase on starch properties and eating quality of rice. Plant Prod. Sci. 2008, 11, 472–480. [Google Scholar] [CrossRef]
- Liu, W.C.; Hung, C.C.; Chen, S.C.; Yeh, S.M.; Lin, M.Y.; Chiu, Y.W.; Kuo, M.C.; Chang, J.M.; Hwang, S.J.; Chen, H.C. Association of hyperuricemia with renal outcomes, cardiovascular disease, and mortality. Clin. J. Am. Soc. Nephrol. 2012, 7, 541–548. [Google Scholar] [CrossRef]
- Sun, Y.; Jiao, G.; Liu, Z.; Zhang, X.; Li, J.; Guo, X.; Du, W.; Du, J.; Francis, F.; Zhao, Y.; et al. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 2017, 8, 298. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Bird, A.; Regina, A.; Li, Z.; Ral, J.P.; McMaugh, S.; Topping, D.; Morell, M. Resistant starch in cereals: Exploiting genetic engineering and genetic variation. J. Cereal Sci. 2007, 46, 251–260. [Google Scholar] [CrossRef]
- Butardo, V.M.; Fitzgerald, M.A.; Bird, A.R.; Gidley, M.J.; Flanagan, B.M.; Larroque, O.; Resurreccion, A.P.; Laidlaw, H.K.; Jobling, S.A.; Morell, M.K.; et al. Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA-and hairpin RNA-mediated RNA silencing. J. Exp. Bot. 2011, 62, 4927–4941. [Google Scholar] [CrossRef]
- Denyer, K.; Sidebottom, C.; Hylton, C.M.; Smith, A.M. Soluble isoforms of starch synthase and starch-branching enzyme also occur within starch granules in developing pea embryos. Plant J. 1993, 4, 191–198. [Google Scholar] [CrossRef]
- Edwards, R. Characterisation of glutathione transferases and glutathione peroxidases in pea (Pisum sativum). Physiol Plant. 1996, 98, 594–604. [Google Scholar] [CrossRef]
- Forster-Mu, C.; Huang, R.; Powers, J.R.; Harriman, R.W.; Knight, M.; Singletary, G.W.; Keeling, P.L.; Wasserman, B.P. Physical association of starch biosynthetic enzymes with starch granules of maize endosperm (granule-associated forms of starch synthase I and starch branching enzyme II). Plant Physiol. 1996, 111, 821829. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, J.; Xia, L. Precise genome modification via sequence-specific nucleases-mediated gene targeting for crop improvement. Front. Plant Sci. 2016, 7, 1928. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, X.; Wu, C.; He, Y.; Ma, Y.; Hou, H.; Guo, X.; Du, W.; Zhao, Y.; Xia, L. Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol. Plant 2016, 9, 628–631. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, V.; Underwood, J.L.; Zhao, S. Dual-targeting by CRISPR/Cas9 for precise excision of transgenes from rice genome. Plant Cell Tissue Organ Cult. PCTOC 2017, 129, 153–160. [Google Scholar] [CrossRef]
- Hu, X.; Cui, Y.; Dong, G.; Feng, A.; Wang, D.; Zhao, C.; Zhang, Y.; Hu, J.; Zeng, D.; Guo, L.; et al. Using CRISPR-Cas9 to generate semi-dwarf rice lines in elite landraces. Sci. Rep. 2019, 9, 19096. [Google Scholar] [CrossRef]
- Kim, Y.A.; Moon, H.; Park, C.J. CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. oryzae. Rice 2019, 12, 67. [Google Scholar] [CrossRef]
- Nekrasov, V.; Staskawicz, B.; Weigel, D.; Jones, J.D.; Kamoun, S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nature Biotech. 2013, 31, 691–693. [Google Scholar] [CrossRef]
- Jameel, M.R.; Ansari, Z.; Qureshi, M.I. From design to validation of CRISPR/gRNA primers towards genome editing. Bioinformation 2022, 18, 471–477. [Google Scholar] [CrossRef]
- Buntru, M.; Gärtner, S.; Staib, L.; Kreuzaler, F.; Schlaich, N. Delivery of multiple transgenes to plant cells by an improved version of MultiRound Gateway technology. Transgenic Res. 2013, 22, 153–167. [Google Scholar] [CrossRef]
- Hiei, Y.; Komari, T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nature Prot. 2008, 3, 824–834. [Google Scholar] [CrossRef]
- Manna, M.; Achary, V.; Islam, T.; Agrawal, P.K.; Reddy, M.K. The development of a phosphite-mediated fertilization and weed control system for rice. Sci. Rep. 2016, 6, 24941. [Google Scholar] [CrossRef] [PubMed]
- Nishi, A.; Nakamura, Y.; Tanaka, N.; Satoh, H. Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol. 2001, 127, 459–472. [Google Scholar] [CrossRef] [PubMed]


| Soluble Starch Synthase-II Isoform-1 |
|---|
| Target 1 |
| Wt-SSII-1: CGCAGGCGGAGCAGGGTGAGCGGGGTTTGGTGGCACTTGTATGGTGGCA |
| KO-SSII-1.1: CGCAGGCGGAGCAGGGTGAGCGGGGTTTGGTGGCACTT--------TGGTGGCA |
| Target 2 |
| Wt-SSII-1: AAGGATCTAGGTGTCCGCAAACGTTACAGGGTAGCTGGACAGGTAAGAAA |
| KO-SSII-1.2: AAGGATCTAGGTGTCCGCAA--------------------GGGTAGCTGGACAGGTAAGAAA |
| Soluble Starch Synthase-II Isoform-2 |
| Target 1 |
| Wt-SSII-2: GTGAGCGGGGTTTGGTGGCACTTGTA----------------------TGGTGGCACGGGGCT |
| KO-SSII-2.1: GTGAGCGGGGTTTGGTGGCACTTGTAGTTGCAGTCACTGGTGGCACGGGGCT |
| Target 2 |
| Wt-SSII-2: CAGAAGCCAAGGATCTAGGTGTCCGCAAACGT-TACAGGGTAGCTGGACAGG |
| KO-SSII-2.2: CAGAAGCCAAGGATCTAGGTGTCCGCAAACGTTTACAGGGTAGCTGGACAGG |
| Soluble Starch Synthase-II Isoform-3 |
| Wt-SSII-3: AGGCGACTAGATCTTCCCCTATTCCTGCGGTAGAAGAGGAGACGTGGGATTTCAAGAAAT |
| KO-SSII-3.1: AGGGGAGTGGATCTTCCCGTATTC----------------------------------------------------------------AAGAAAT |
| Soluble Starch Synthase II-1 |
|---|
| Ct-SSS-II-1:ACTTGGGCAGTGCCGAGGAGGAGCAGGTTGGAGTGGGGGC-GTGTGGAGGCTCAGAA-TT |
| KO-SSS-II-1.1:ACTTGGGCAGTGCCGAGGAGGAGCAGGTTGGAGTGGGGGCGGTGTGGAGGCTCAGAATT |
| Soluble Starch Synthase II-2 |
| Target 1 |
| Ct-SSS-II-2:GAGCGGGGTTTGGTGGCACTTGTATGGTGGCACGGGGCTGCGGTTGCATTGGGAGCGGCG |
| KO-SSS-II-2.1: GAGCGGGGTTTGGTGGCAC-------ATGGTGGCACGGGGCTGCGGTTGCATTGGGAGCGGCG |
| Target 2 |
| Ct-SSS-II-2:TTATGGTTGTGATACCAAGATACGGAGAATATGCAGAAGCCAAGGATCTAGGTGTCCGCA |
| KO-SSS-II-2.1: TTA-GGTTGTGATACCAAGATATGGAGAATATGCAGAAGCCAAGGATCTAGGTGTCCGCA |
| Target 1 |
| Ct-SSS-II-2: GAGCGGGGTTTGGTGGCACTTGTATGGTGGCACGGGGCTGCGGTTGCATTGGGAGCGGCG |
| KO-SSS-II-2.2:GAGCGGGGTTTGGTGGCACTTGGATGGCGGC------GGGCTGCTGCTGCTTTGTTGGCAGCG |
| Target 2 |
| Ct-SSS-II-2: TTATGGTTGTGATACCAAGATACGGAGAATATGCAGAAGCCAAGGATCTAGGTGTCCGCA |
| KO-SSS-II-2.2:TTA-GGTTGTGATACCAAGATACGGAGAATATGCAGAAGCCAAGGATCTAGGTGTCCGCA |
| Target 1 |
| Ct-SSS-II-2: GGGTTTGGTTCGGGACGGAGCCGTCGTGTGCTCGGCGTCGGCCGCCGGTGGTGAGGATGG |
| KO-SSS-II-2.3:GGG-----GGTTGTGCACGAAGCCGCCGTGTGGTCCGCGGCGGCCGCCGGTGGTGAGGATGG |
| Target 2 |
| Ct-SSS-II-2: TTATGGTTGTGATACCAAGATACGGAGAATATGCAGAAGCCAAGGATCTAGGTGTCCGCA |
| KO-SSS-II-2.3:TTA-GGTTGTGATACCAAGATACGGAGAATATGCAGAAGCCAAGGATCTAGGTGTCCGCA |
| Target 1 |
| Ct-SSS-II-2: GGATGGCGTCGCGAAGGCGAAGACGAAGTCAGCGGGGAGCTC-GAAGGCGGTCGC-TGTG |
| KO-SSS-II-2.4: GGATGCCTCCCCGAAGGCAAAGGCAAA-TCCGCGGGGAACTCCGAGGGCG-TCGCCTGTG |
| Target 2 |
| Ct-SSS-II-2: TTATGGTTGTGATACCAAGATACGGAGAATATGCAGAAGCCAAGGATCTAGGTGTCCGCA |
| KO-SSS-II-2.4: TTA-GGTTGTGATACCAAGATATGGAGAATATGCAGAAGCCAAGGATCTAGGTGTCCGCA |
| Target 1 |
| Ct-SSS-II-2: GAGCGGGGTTTGGTGGCACTTGTATGGTGGCACGGGGCTGCGGTTGCATTGGGAGCGGCG |
| KO-SSS-II-2.5: GAGCGGGGTTTGGTGGCACTTGTATGGGGGC—GGGGCTCTGGTTGTGCTGTGAGCGCCG |
| Target 2 |
| Ct-SSS-II-2: TTATGGTTGTGATACCAAGATACGGAGAATATGCAGAAGCCAAGGATCTAGGTGTCCGCA |
| KO-SSS-II-2.5: TTA-GGTTGTGATACCAAGATATGGAGAATATGCAGAAGCCAAGGATCTAGGTGTCCGCA |
| Soluble Starch Synthase II-3 |
| Target 1 |
| Ct-SSS-II-3: TCCGCTCCTCTCCCCAAGCCTGACAATTCGGAATTTGCAGAGGATAAGAGCGCAAAAGTT |
| KO-SSS-II-3.1: TCCGCTCCTCTCCCCT--GCCTGAC--ATTCGAAATTTGCAAAGGATAAGAGCGCAG—AGTT |
| Target 2 |
| Ct-SSS-II-3: CTCCGAAGCCAAAGGCGACTAGATCTTCCCCTATTC-CTGCGGTAGAAGAGGAGACGTGG |
| KO-SSS-II-3.1: CTCCGAAGCCAAAGGCGACTAGATCTTCCCCTATTCTCTGCGGTAGAAGAGGAGACGTGG |
| Target 1 |
| Ct-SSS-II-3: GAAGCTCCCAGCGCCGGACTCCCCCGTGATCCTTCCATCCGTAGACAAGCCGCAGCCGGA |
| KO-SSS-II-3.2: GAAGCTCCCAGCGCCGGACTCCCCCGTGATCCTTCCATCCATA--AC--AGCCGCAGCCTGA |
| Target 2 |
| Ct-SSS-II-3: CAAAGGCGACTAGATCTTCCCCTATTC-CTGCGGTAGAAGAGGAGACGTGGGATTTCAAG |
| Ct-SSS-II-3.2: CAAAGGCGACTAGATCTTCCCCTATTCTCTGCGGTAGAAGAGGAGACGTGGGATTTCAAG |
| Target Genes Encoding for Various Isoforms | Gene | Localization on Chromosome | |
|---|---|---|---|
| Soluble Starch Synthase (SSS) | SSSI | SSSI | Chr 6 |
| SSSII | SSSII-1 | Chr 10 | |
| SSSII-2 | Chr 2 | ||
| SSSII-3 | Chr 6 | ||
| SSSIII | SSSIII-1 | Chr 4 | |
| SSSIII-2 | Chr 8 | ||
| SSSIV-1 | Chr 1 | ||
| SSSIV-2 | Chr 5 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jameel, M.R.; Ansari, Z.; Al-Huqail, A.A.; Naaz, S.; Qureshi, M.I. CRISPR/Cas9-Mediated Genome Editing of Soluble Starch Synthesis Enzyme in Rice for Low Glycemic Index. Agronomy 2022, 12, 2206. https://doi.org/10.3390/agronomy12092206
Jameel MR, Ansari Z, Al-Huqail AA, Naaz S, Qureshi MI. CRISPR/Cas9-Mediated Genome Editing of Soluble Starch Synthesis Enzyme in Rice for Low Glycemic Index. Agronomy. 2022; 12(9):2206. https://doi.org/10.3390/agronomy12092206
Chicago/Turabian StyleJameel, Mohd Rizwan, Zubaida Ansari, Asma A. Al-Huqail, Sheeba Naaz, and Mohammad Irfan Qureshi. 2022. "CRISPR/Cas9-Mediated Genome Editing of Soluble Starch Synthesis Enzyme in Rice for Low Glycemic Index" Agronomy 12, no. 9: 2206. https://doi.org/10.3390/agronomy12092206
APA StyleJameel, M. R., Ansari, Z., Al-Huqail, A. A., Naaz, S., & Qureshi, M. I. (2022). CRISPR/Cas9-Mediated Genome Editing of Soluble Starch Synthesis Enzyme in Rice for Low Glycemic Index. Agronomy, 12(9), 2206. https://doi.org/10.3390/agronomy12092206







