Intraspecific Variability and Distribution Difference within the Ribosomal Introns of the Discrete Plasmodiophora brassicae Group in Japan: A Case Study for Complex Dynamics of Intron Evolution
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clubroot Samples Collected
2.2. DNA Extraction
2.3. Primer Designation and Sequencing Analysis
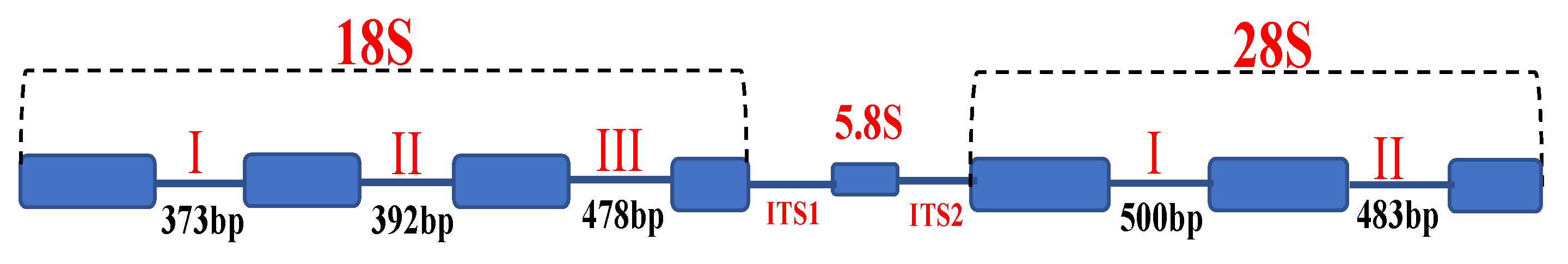
2.4. Sequence Annotation
3. Results
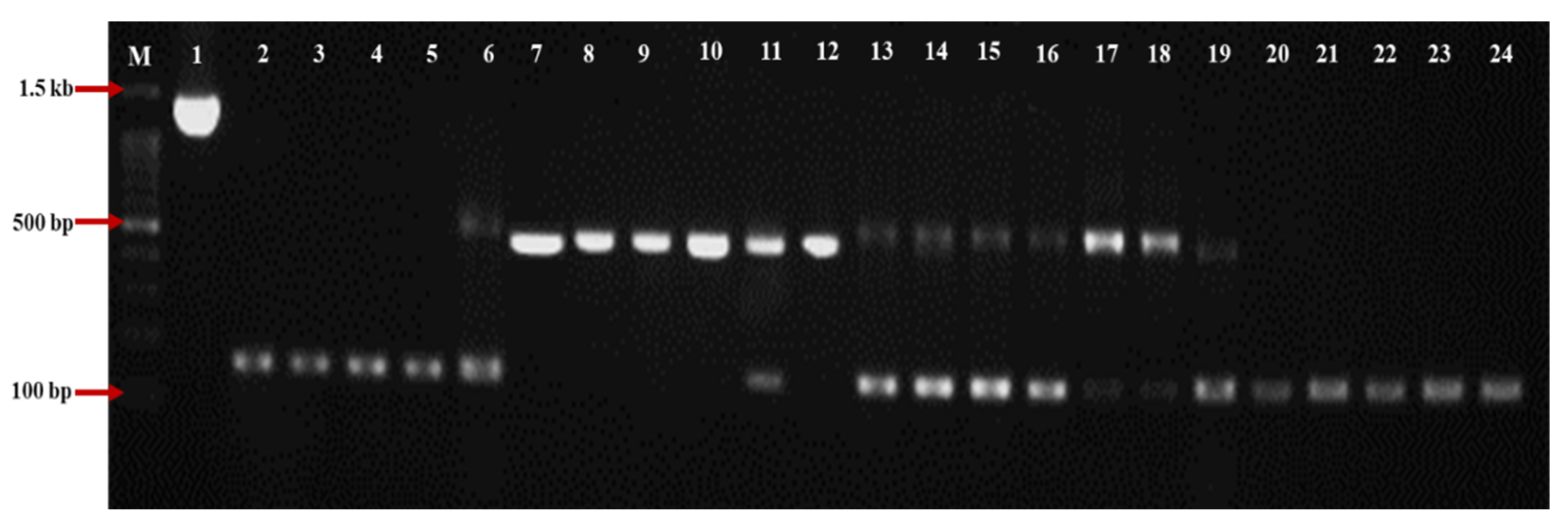
3.1. The Pattern of Loss/Gain Introns Distributed within the P. brassicae Populations in Japan
3.2. Intron Analysis of Small (18S) and Large (28S) Subunit rRNA Genes
3.2.1. Small Subunit (18S) rRNA Gene
3.2.2. Large Subunit (28S) rRNA Gene
4. Discussion
4.1. Early or Late Theory for the Intronic Distribution in P. brassicae?
4.2. The Intronic Evolution Equates to Speciation Event of P. brassicae Populations?
4.3. Prospective Research for Intragenic Sequences of P. brassicae
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tanaka, S.; Ito, S.; Kameya-Iwaki, M.; Katumoto, K.; Nishi, Y. Occurrence and distribution of clubroot disease on two cruciferous weeds, Cardamine flexuosa and C. scutata, in Japan. Trans. Mycol. Soc. Jpn. 1993, 34, 381–388. [Google Scholar]
- Tanaka, S.; Mizui, Y.; Terasaki, H.; Ito, S. Distribution of clubroot disease of a cruciferous weed, Cardamine flexuosa, in major isolated islands, Hokkaido and Okinawa, in Japan. Mycoscience 2006, 47, 72–77. [Google Scholar] [CrossRef]
- Osaki, K.; Fujiyama, S.; Nakayama, A.; Shimizu, Y.; Ito, S.; Tanaka, S. Relation between pathogenicity and genetic variation within Plasmodiophora brassicae. J. Gen. Plant Pathol. 2008, 74, 281–288. [Google Scholar] [CrossRef]
- Flipphi, M.; Fekete, E.; Ág, N.; Scazzocchio, C.; Karaffa, L. Spliceosome twin introns in fungal nuclear transcripts. Fungal Genet. Biol. 2013, 57, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Flipphi, M.; Ág, N.; Karaffa, L.; Kavalecz, N.; Cerqueira, G.; Scazzocchio, C.; Fekete, E. Emergence and loss of spliceosomal twin introns. Fungal Biol. Biotechnol. 2017, 4, 7. [Google Scholar] [CrossRef]
- Machouart, M.; Gueidan, C.; Khemisti, A.; Dulongcourty, R.; Sudhadham, M.; de Hoog, G.S. Use of ribosomal introns as new markers of genetic diversity in Exophiala dermatitidis. Fungal Biol. 2011, 115, 1038–1050. [Google Scholar] [CrossRef]
- Niwa, R.; Kawahara, A.; Murakami, H.; Tanaka, S.; Ezawa, T. Complete structure of nuclear rDNA of the obligate plant parasite Plasmodiophora brassicae: Intraspecific polymorphisms in the exon and group I intron of the large subunit rDNA. Protist 2011, 162, 423–434. [Google Scholar] [CrossRef]
- Schwelm, A.; Berney, C.; Dixelius, C.; Bass, D.; Neuhauser, S. The Large Subunit rDNA sequence of Plasmodiophora brassicae does not contain intra-species polymorphism. Protist 2016, 16, 544–554. [Google Scholar] [CrossRef]
- Laila, R.; Robin, A.H.; Yang, K.; Choi, G.J.; Park, J.I.; Nou, I.S. Detection of ribosomal DNA sequence polymorphisms in the protist Plasmodiophora brassicae for the identification of geographical isolates. Int. J. Mol. Sci. 2017, 18, 84. [Google Scholar] [CrossRef]
- Schwelm, A.; Neuhauser, S. Letter to the Editor: Detection of ribosomal DNA sequence polymorphisms in the protist Plasmodiophora brassicae for the identification of geographical isolates. Int. J. Mol. Sci. 2017, 18, 1454. [Google Scholar] [CrossRef]
- Ito, S.; Maehara, T.; Tanaka, S.; Kameya-Iwaki, M.; Yano, S.; Kishi, F. Cloning of a single-copy DNA sequence unique to Plasmodiophora brassicae. Physiol. Mol. Plant Pathol. 1997, 50, 289–300. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Rogozin, I.B.; Carmel, L.; Csuros, M.; Koonin, E.V. Origin and evolution of spliceosomal introns. Biol. Direct 2012, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- de Souza, S.J.; Long, M.; Klein, R.J.; Roy, S.; Lin, S. Toward a resolution of the introns early/late debate: Only phase zero introns are correlated with the structure of ancient proteins. Proc. Natl. Acad. Sci. USA 1998, 95, 5094–5099. [Google Scholar] [CrossRef]
- Koonin, E.V. The origin of introns and their role in eukaryogenesis: A compromise solution to the introns-early versus introns-late debate? Biol. Direct 2006, 1, 22. [Google Scholar] [CrossRef]
- Belshaw, R.; Bensasson, D. The rise and falls of introns. Heredity 2006, 96, 208–213. [Google Scholar] [CrossRef]
- Long, M.; Rosenberg, C. Testing the “proto-splice sites” model of intron origin: Evidence from analysis of intron phase correlations. Mol. Biol. Evol. 2000, 17, 1789–1796. [Google Scholar] [CrossRef]
- Roy, S.W.; Gilbert, W. The pattern of intron loss. Proc. Natl. Acad. Sci. USA 2005, 102, 713–718. [Google Scholar] [CrossRef]
- Roy, S.W.; Hartl, D.L. Very little intron loss/gain in Plasmodium: Intron loss/gain mutation rates and intron number. Genome Res. 2006, 16, 750–756. [Google Scholar] [CrossRef]
- Roy, S.W.; Penny, D. Widespread intron loss suggests retrotransposon activity in ancient apicomplexans. Mol. Biol. Evol. 2007, 24, 1926–1933. [Google Scholar] [CrossRef]
- Stjelja, S.; Fogelqvist, J.; Tellgren-Roth, C. The architecture of the Plasmodiophora brassicae nuclear and mitochondrial genomes. Sci. Rep. 2019, 9, 15753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flint-Garcia, S.A. Genetics and consequences of crop domestication. J. Agric. Food Chem. 2013, 61, 8267–8276. [Google Scholar] [CrossRef] [PubMed]
- Al-Shehbaz, I.A.; Beilstein, M.A.; Kellogg, E.A. Systematics and phylogeny of the Brassicaceae (Cruciferae): An overview. Plant Syst. Evol. 2006, 259, 89–120. [Google Scholar] [CrossRef]
- Šlenker, M.; Zozomová-Lihová, J.; Mandáková, T.; Kudoh, H.; Zhao, Y.; Soejima, A.; Yahara, T.; Skokanová, K.; Španiel, S.; Marhold, K. Morphology and genome size of the widespread weed Cardamine occulta: How it differs from cleistogamic C. kokaiensis and other closely related taxa in Europe and Asia. Bot. J. Linn. Soc. 2018, 187, 456–482. [Google Scholar]
- Rutland, C.A.; Hall, N.D.; McElroy, J.S. The impact of polyploidization on the evolution of weed species: Historical understanding and current limitations. Front. Agron. 2021, 3, 626454. [Google Scholar] [CrossRef]
- Ebert, D.; Fields, P.D. Host-parasite co-evolution and its genomic signature. Nat. Rev. Genet. 2020, 21, 754–768. [Google Scholar] [CrossRef]
- Märkle, H.; John, S.; Cornille, A.; Fields, P.D.; Tellier, A. Novel genomic approaches to study antagonistic coevolution between hosts and parasites. Mol. Ecol. 2021, 30, 3660–3676. [Google Scholar] [CrossRef]
- Mandáková, T.; Zozomová-Lihová, J.; Kudoh, H.; Zhao, Y.P.; Lysak, M.A.; Marhold, K. The story of promiscuous crucifers: Origin and genome evolution of an invasive species, Cardamine occulta (Brassicaceae), and its relatives. Ann. Bot. 2019, 124, 209–220. [Google Scholar] [CrossRef]
- Crow, K.D.; Wagner, G.P. What is the role of genome duplication in the evolution of complexity and diversity? Mol. Biol. Evol. 2006, 23, 887–892. [Google Scholar] [CrossRef]
- Rolfe, S.A.; Strelkov, S.E.; Links, M.G. The compact genome of the plant pathogen Plasmodiophora brassicae is adapted to intracellular interactions with host Brassica spp. BMC Genom. 2016, 17, 272. [Google Scholar] [CrossRef]
- Bulman, S.; Ridgway, H.J.; Eady, C.; Conner, A.J. Intron-rich gene structure in the intracellular plant parasite Plasmodiophora brassicae. Protist 2007, 158, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Rogers, S.O. Integrated evolution of ribosomal RNAs, introns, and intron nurseries. Genetica 2019, 147, 103–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, C.S.; Weinstein, B.N.; Roy, S.W.; Brown, C.M. Analysis of fungal genomes reveals commonalities of intron gain or loss and functions in intron-poor species. Mol. Biol. Evol. 2021, 38, 4166–4186. [Google Scholar] [CrossRef]
- Edgell, D.R.; Chalamcharla, V.R.; Belfort, M. Learning to live together: Mutualism between self-splicing introns and their hosts. BMC Biol. 2011, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Sibley, C.R.; Blazquez, L.; Ule, J. Lessons from non-canonical splicing. Nat. Rev. Genet. 2016, 17, 407–421. [Google Scholar] [CrossRef] [PubMed]
- Schwelm, A.; Ludwig-Müller, J. Molecular pathotyping of Plasmodiophora brassicae-genomes, marker genes, and obstacles. Pathogens 2021, 10, 259. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Qin, L.; Zhou, Z.; Hendriks, W.; Liu, S.; Wei, Y. Refining the life cycle of Plasmodiophora brassicae. Phytopathology 2020, 110, 1704–1712. [Google Scholar] [CrossRef] [PubMed]



| Intronic Sequences | Cardamine Group | Crop Group | |
|---|---|---|---|
| 18S | I | Variable | Variable |
| II | Absence | Presence | |
| III | Absence | Presence | |
| 28S | I | Variable | Presence |
| II | Absence | Presence | |
| Mutated Positions Identified in 28S Intron II | ||||||
|---|---|---|---|---|---|---|
| AT isolate (KX011115) | 30 | 45 | 200 | 282 | 342–345 * | 345–347 |
| C | G | G | G | ATC | ATA | |
| Capsella bursa-pastoris | G | G | A | deleted | deleted | ATA |
| Hagi and Onga | G | T | A | deleted | ATC | deleted |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lam, A.T.P.; Sasaki, K.; Yanagi, Y.; Tanaka, S.; Ito, S.-i. Intraspecific Variability and Distribution Difference within the Ribosomal Introns of the Discrete Plasmodiophora brassicae Group in Japan: A Case Study for Complex Dynamics of Intron Evolution. Agronomy 2022, 12, 2154. https://doi.org/10.3390/agronomy12092154
Lam ATP, Sasaki K, Yanagi Y, Tanaka S, Ito S-i. Intraspecific Variability and Distribution Difference within the Ribosomal Introns of the Discrete Plasmodiophora brassicae Group in Japan: A Case Study for Complex Dynamics of Intron Evolution. Agronomy. 2022; 12(9):2154. https://doi.org/10.3390/agronomy12092154
Chicago/Turabian StyleLam, Anh Tung Phan, Kazunori Sasaki, Yukiko Yanagi, Shuhei Tanaka, and Shin-ichi Ito. 2022. "Intraspecific Variability and Distribution Difference within the Ribosomal Introns of the Discrete Plasmodiophora brassicae Group in Japan: A Case Study for Complex Dynamics of Intron Evolution" Agronomy 12, no. 9: 2154. https://doi.org/10.3390/agronomy12092154
APA StyleLam, A. T. P., Sasaki, K., Yanagi, Y., Tanaka, S., & Ito, S.-i. (2022). Intraspecific Variability and Distribution Difference within the Ribosomal Introns of the Discrete Plasmodiophora brassicae Group in Japan: A Case Study for Complex Dynamics of Intron Evolution. Agronomy, 12(9), 2154. https://doi.org/10.3390/agronomy12092154





