Classification Method of Significant Rice Pests Based on Deep Learning
Abstract
1. Introduction
2. Related Work
3. Materials and Methods
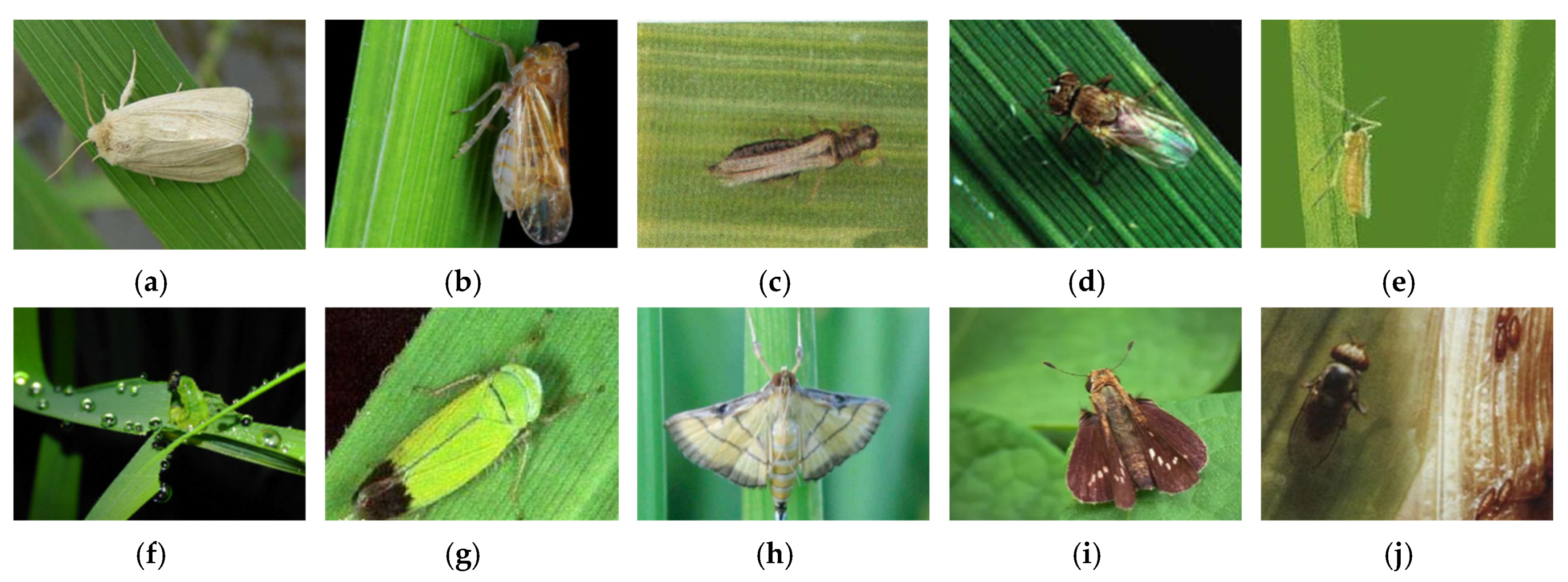
3.1. Image Acquisition
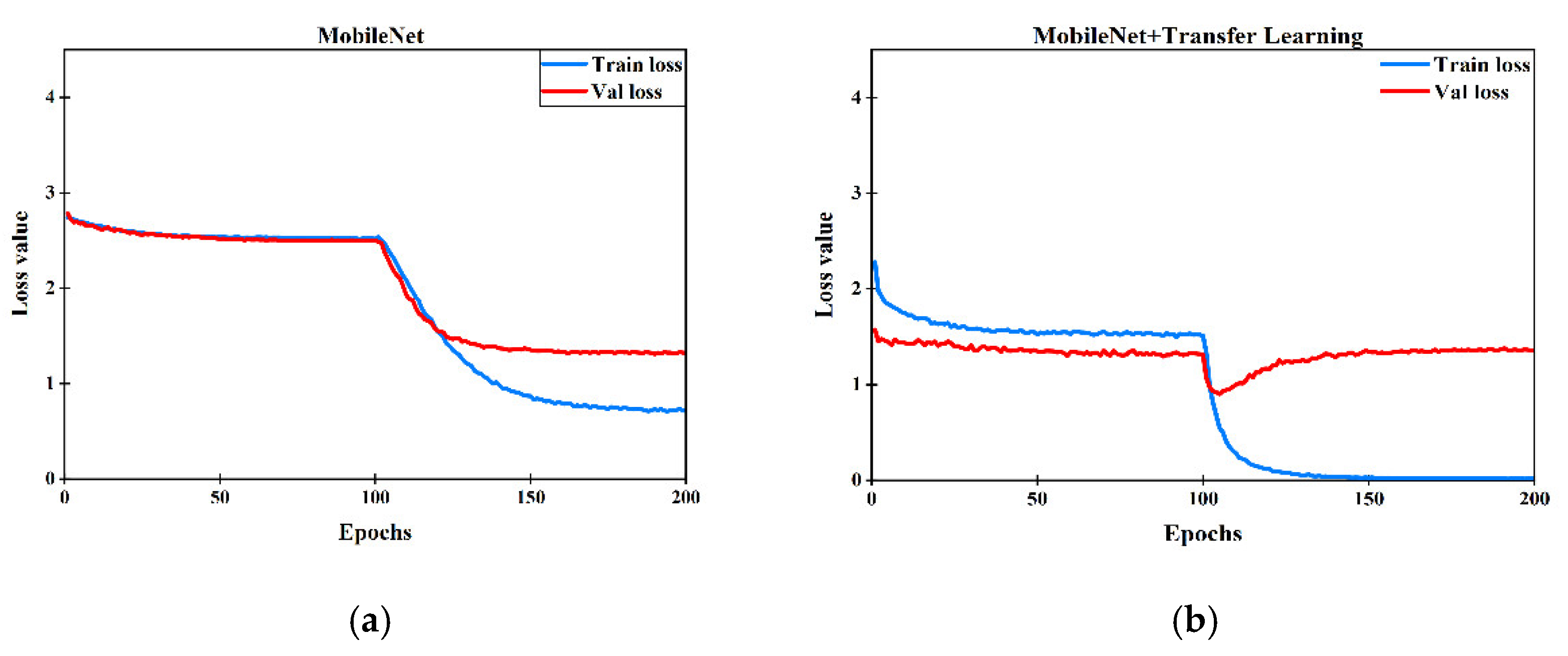
3.2. Transfer Learning
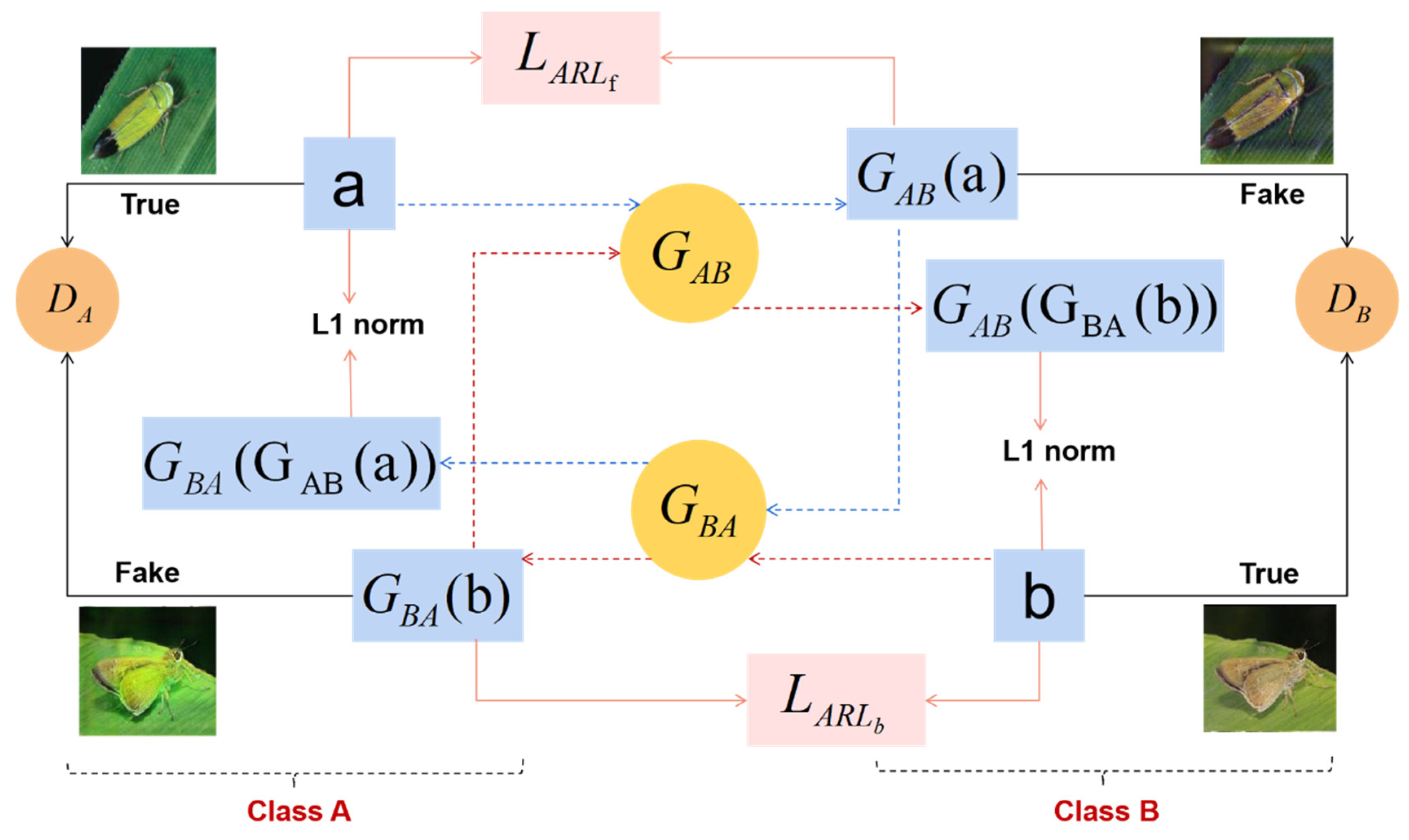
3.3. ARGAN Data Augmentation
3.4. Evaluation Indicators
3.5. Training Environment
4. Results
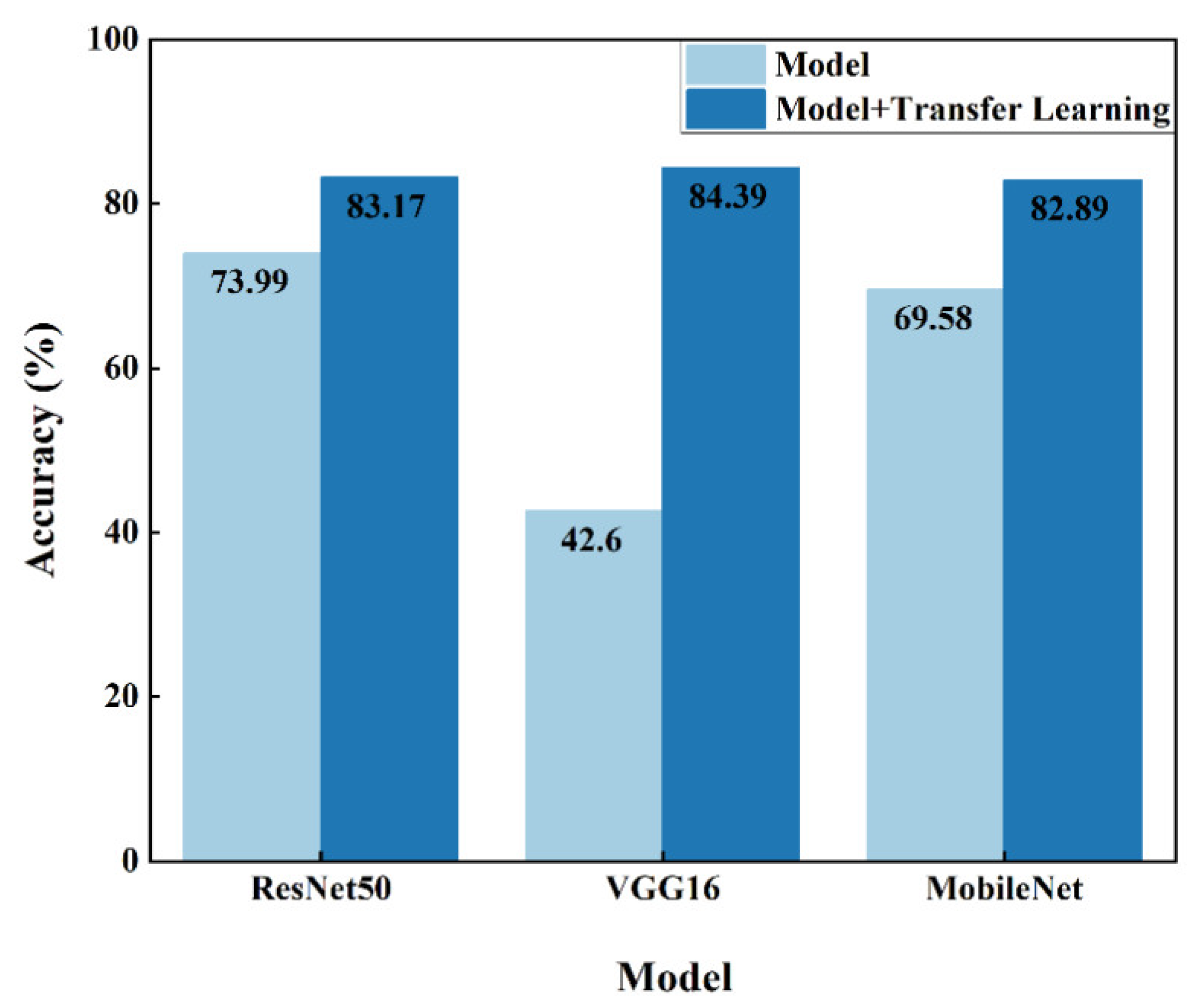
4.1. Parameter Fine-Tuning of ResNet, VGGNet and MobileNet on ImageNet Dataset
4.2. Evaluation of the IP_RicePests Dataset
4.3. ARGAN Data Augmentation Evaluation
5. Discussion
- Firstly, we hand-selected a total of 8417 images from 14 categories of rice pest samples in IP102 for two weeks, creating a higher quality rice pest data sample 1.0;
- Secondly, we used crawler technology to expand a large number of images for each category of pest samples, and obtained a higher quality rice pest data sample 2.0 after hand-screening;
- Thirdly, after reviewing the literature and receiving expert guidance, we manually screened the rice pest data sample 2.0 again, and finally formed a large-scale dataset on rice major pest classification, IP_RicePests.
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lou, Y.-G.; Zhang, G.-R.; Zhang, W.-Q.; Hu, Y.; Zhang, J. Biological control of rice insect pests in China. Biol. Control 2013, 67, 8–20. [Google Scholar] [CrossRef]
- Yao, Q.; Lv, J.; Liu, Q.-J.; Diao, G.-Q.; Yang, B.-J.; Chen, H.-M.; Tang, J. An Insect Imaging System to Automate Rice Light-Trap Pest Identification. J. Integr. Agric. 2012, 11, 978–985. [Google Scholar] [CrossRef]
- Mehrotra, R.; Namuduri, K.; Ranganathan, N. Gabor filter-based edge detection. Pattern Recognit. 1992, 25, 1479–1494. [Google Scholar] [CrossRef]
- Dalal, N.; Triggs, B. Histograms of oriented gradients for human detection. In Proceedings of the Computer Vision and Pattern Recognition, San Diego, CA, USA, 20–26 June 2005; pp. 886–893. [Google Scholar] [CrossRef]
- Oliva, A.; Torralba, A. Modeling the Shape of the Scene: A Holistic Representation of the Spatial Envelope. Int. J. Comput. Vis. 2001, 42, 145–175. [Google Scholar] [CrossRef]
- Lowe, D.G. Distinctive Image Features from Scale-Invariant Keypoints. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Bay, H.; Tuytelaars, T.; van Gool, L. SURF: Speeded up Robust Features. Lect. Notes Comput. Sci. 2006, 3951, 404–417. [Google Scholar] [CrossRef]
- Xiao, D.; Feng, J.; Lin, T.; Pang, C.; Ye, Y. Classification and recognition scheme for vegetable pests based on the BOF-SVM model. Int. J. Agric. Biol. Eng. 2018, 11, 190–196. [Google Scholar] [CrossRef]
- Chen, P.-H.; Lin, C.-J.; Schölkopf, B. A tutorial on ν-support vector machines. Appl. Stoch. Model. Bus. Ind. 2005, 21, 111–136. [Google Scholar] [CrossRef]
- Sethy, P.K.; Barpanda, N.K.; Rath, A.K.; Behera, S.K. Deep feature based rice leaf disease identification using support vector machine. Comput. Electron. Agric. 2020, 175, 105527. [Google Scholar] [CrossRef]
- Webb, G. Naïve Bayes. In Encyclopedia of Machine Learning and Data Mining; Springer: Berlin/Heidelberg, Germany, 2016; pp. 1–2. [Google Scholar] [CrossRef]
- Larijani, M.R.; Asli-Ardeh, E.A.; Kozegar, E.; Loni, R. Evaluation of image processing technique in identifying rice blast disease in field conditions based on KNN algorithm improvement by K-means. Food Sci. Nutr. 2019, 7, 3922–3930. [Google Scholar] [CrossRef]
- Li, D.; Wang, R.; Xie, C.; Liu, L.; Zhang, J.; Li, R.; Wang, F.; Zhou, M.; Liu, W. A Recognition Method for Rice Plant Diseases and Pests Video Detection Based on Deep Convolutional Neural Network. Sensors 2020, 20, 578. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhou, Z.; Tian, L.; Liu, Y.; Luo, X. Brown rice planthopper (Nilaparvata lugens Stal) detection based on deep learning. Precis. Agric. 2020, 21, 1385–1402. [Google Scholar] [CrossRef]
- Rahman, C.R.; Arko, P.S.; Ali, M.E.; Khan, M.A.I.; Apon, S.H.; Nowrin, F.; Wasif, A. Identification and recognition of rice diseases and pests using convolutional neural networks. Biosyst. Eng. 2020, 194, 112–120. [Google Scholar] [CrossRef]
- Liu, Z.; Gao, J.; Yang, G.; Zhang, H.; He, Y. Localization and Classification of Paddy Field Pests using a Saliency Map and Deep Convolutional Neural Network. Sci. Rep. 2016, 6, 20410. [Google Scholar] [CrossRef] [PubMed]
- Alfarisy, A.A.; Chen, Q.; Guo, M. Deep learning based classification for paddy pests & diseases recognition. In Proceedings of the 2018 International Conference on Mathematics and Artificial Intelligence, Chengdu, China, 20–22 April 2018; pp. 22–25. [Google Scholar] [CrossRef]
- Jia, Y.; Shelhamer, E.; Donahue, J.; Karayev, S.; Long, J.; Girshick, R.; Guadarrama, S.; Darrell, T. Caffe: Convolutional architecture for fast feature embedding. In Proceedings of the 2014 ACM Multimedia Conference, Orlando, FL, USA, 3–7 November 2014; pp. 675–678. [Google Scholar] [CrossRef]
- Burhan, S.A.; Minhas, S.; Tariq, A.; Hassan, M.N. Comparative study of deep learning algorithms for disease and pest detection in rice crops. In Proceedings of the 12th International Conference on Electronics, Computers and Artificial Intelligence (ECAI), Bucharest, Romania, 16 October 2020; pp. 1–5. [Google Scholar] [CrossRef]
- Wu, X.; Zhan, C.; Lai, Y.-K.; Cheng, M.-M.; Yang, J. IP102: A large-scale benchmark dataset for insect pest recognition. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 8779–8788. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Sagiroglu, S.; Sinanc, D. Big data: A review. In Proceedings of the 2013 International Conference on Collaboration Technologies and Systems (CTS), San Diego, CA, USA, 20–24 May 2013; pp. 42–47. [Google Scholar]
- Mikołajczyk, A.; Grochowski, M. Data augmentation for improving deep learning in image classification problem. In Proceedings of the International Interdisciplinary PhD Workshop (IIPhDW), Swinoujscie, Poland, 9–12 May 2018; pp. 117–122. [Google Scholar] [CrossRef]
- Taylor, L.; Nitschke, G. Improving deep learning with generic data augmentation. In Proceedings of the 2018 IEEE Symposium Series on Computational Intelligence (SSCI), Bangalore, India, 18–21 November 2018; pp. 1542–1547. [Google Scholar] [CrossRef]
- Perez, L.; Wang, J. The Effectiveness of Data Augmentation in Image Classification Using Deep Learning. arxiv 2017, arXiv:1712.04621. [Google Scholar]
- Nazki, H.; Lee, J.; Yoon, S.; Park, D.S. Synthetic Data Augmentation for Plant Disease Image Generation Using GAN. Proc. Korea Contents Assoc. Conf. 2018, 459–460. [Google Scholar]
- Ding, B.; Long, C.; Zhang, L.; Xiao, C. ARGAN: Attentive recurrent generative adversarial network for shadow detection and removal. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Korea, 27 October–2 November 2019. [Google Scholar] [CrossRef]
- Nazki, H.; Yoon, S.; Fuentes, A.; Park, D.S. Unsupervised image translation using adversarial networks for improved plant disease recognition. Comput. Electron. Agric. 2020, 168, 105117. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Wang, J.; Lin, C.; Ji, L.; Liang, A. A new automatic identification system of insect images at the order level. Knowledge-Based Syst. 2012, 33, 102–110. [Google Scholar] [CrossRef]
- Xie, C.; Zhang, J.; Li, R.; Li, J.; Hong, P.; Xia, J.; Chen, P. Automatic classification for field crop insects via multiple-task sparse representation and multiple-kernel learning. Comput. Electron. Agric. 2015, 119, 123–132. [Google Scholar] [CrossRef]
- Samanta, R.K.; Ghosh, I. Tea Insect Pests Classification Based on Artificial Neural Networks. Int. J. Comput. Eng. Sci. 2012, 2, 1–13. [Google Scholar]
- Deng, L.; Wang, Y.; Han, Z.; Yu, R. Research on insect pest image detection and recognition based on bio-inspired methods. Biosyst. Eng. 2018, 169, 139–148. [Google Scholar] [CrossRef]
- Venugoban, K.; Ramanan, A. Image Classification of Paddy Field Insect Pests Using Gradient-Based Features. Int. J. Mach. Learn. Comput. 2014, 4, 1–5. [Google Scholar] [CrossRef][Green Version]
- Al Hiary, H.; Ahmad, S.B.; Reyalat, M.; Braik, M.; Alrahamneh, Z. Fast and Accurate Detection and Classification of Plant Diseases. Int. J. Comput. Appl. 2011, 17, 31–38. [Google Scholar] [CrossRef]
- Xie, C.; Wang, R.; Zhang, J.; Chen, P.; Dong, W.; Li, R.; Chen, T.; Chen, H. Multi-level learning features for automatic classification of field crop pests. Comput. Electron. Agric. 2018, 152, 233–241. [Google Scholar] [CrossRef]
- Fernández, A.; García, S.; del Jesus, M.J.; Herrera, F. A study of the behaviour of linguistic fuzzy rule based classification systems in the framework of imbalanced data-sets. Fuzzy Sets Syst. 2007, 159, 2378–2398. [Google Scholar] [CrossRef]
- Pan, S.J.; Tsang, I.W.; Kwok, J.T.; Yang, Q. Domain Adaptation via Transfer Component Analysis. IEEE Trans. Neural Netw. 2011, 22, 199–210. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Johnson, J.; Alahi, A.; Fei-Fei, L. Perceptual losses for real-time style transfer and super-resolution. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 8–16 October 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 694–711. [Google Scholar]
- Cha, M.; Gwon, Y.; Kung, H.T. Adversarial nets with perceptual losses for text-to-image synthesis. In Proceedings of the 2017 IEEE 27th International Workshop on Machine Learning for Signal Processing (MLSP), Tokyo, Japan, 25–28 September 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Jay, F.; Renou, J.-P.; Voinnet, O.; Navarro, L. Unpaired image-to-image translation using cycle-consistent adversarial networks Jun-Yan. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 183–202. [Google Scholar]
- Cao, X.; Wei, Z.; Gao, Y.; Huo, Y. Recognition of Common Insect in Field Based on Deep Learning. J. Phys. Conf. Ser. 2020, 1634, 012034. [Google Scholar] [CrossRef]








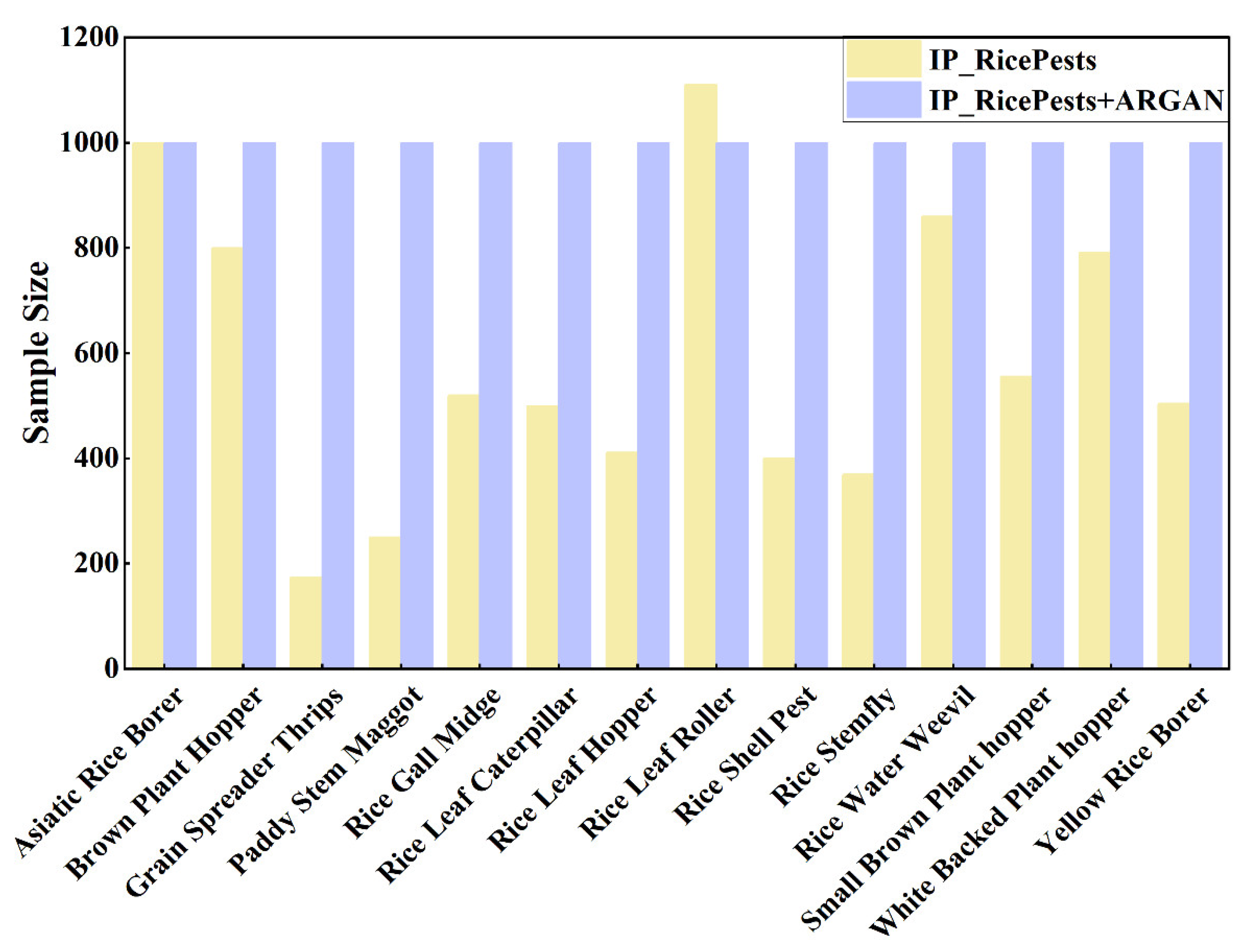


| Categories | IP102 | IP_RicePests |
|---|---|---|
| Asiatic rice borer | 1073 | 1000 |
| Brown plant hopper | 834 | 800 |
| Grain spreader thrips | 173 | 174 |
| Paddy stem maggot | 241 | 250 |
| Rice gall midge | 506 | 520 |
| Rice leaf caterpillar | 487 | 500 |
| Rice leaf hopper | 404 | 411 |
| Rice leaf roller | 1115 | 1110 |
| Rice shell pest | 409 | 401 |
| Rice stemfly | 369 | 370 |
| Rice water weevil | 856 | 860 |
| Small brown plant hopper | 553 | 556 |
| White backed plant hopper | 893 | 792 |
| Yellow rice borer | 504 | 504 |
| Total Quantity | 8417 | 8248 |
| Parameters | ARGAN | ResNet50/VGG16/MobileNet (Freeze Process) | ResNet50/VGG16/MobileNet (Unfreeze Process) |
|---|---|---|---|
| Batch Size | 8 | 4 | 8 |
| Epoch | 100000 | 100 | 100 |
| Optimizer | Adam | Adam | Adam |
| Learning rate | 0.0001 | 0.001 | 0.0001 |
| Model | Transfer Learning | Pre (%) | Rec (%) | F1 (%) | Acc (%) |
|---|---|---|---|---|---|
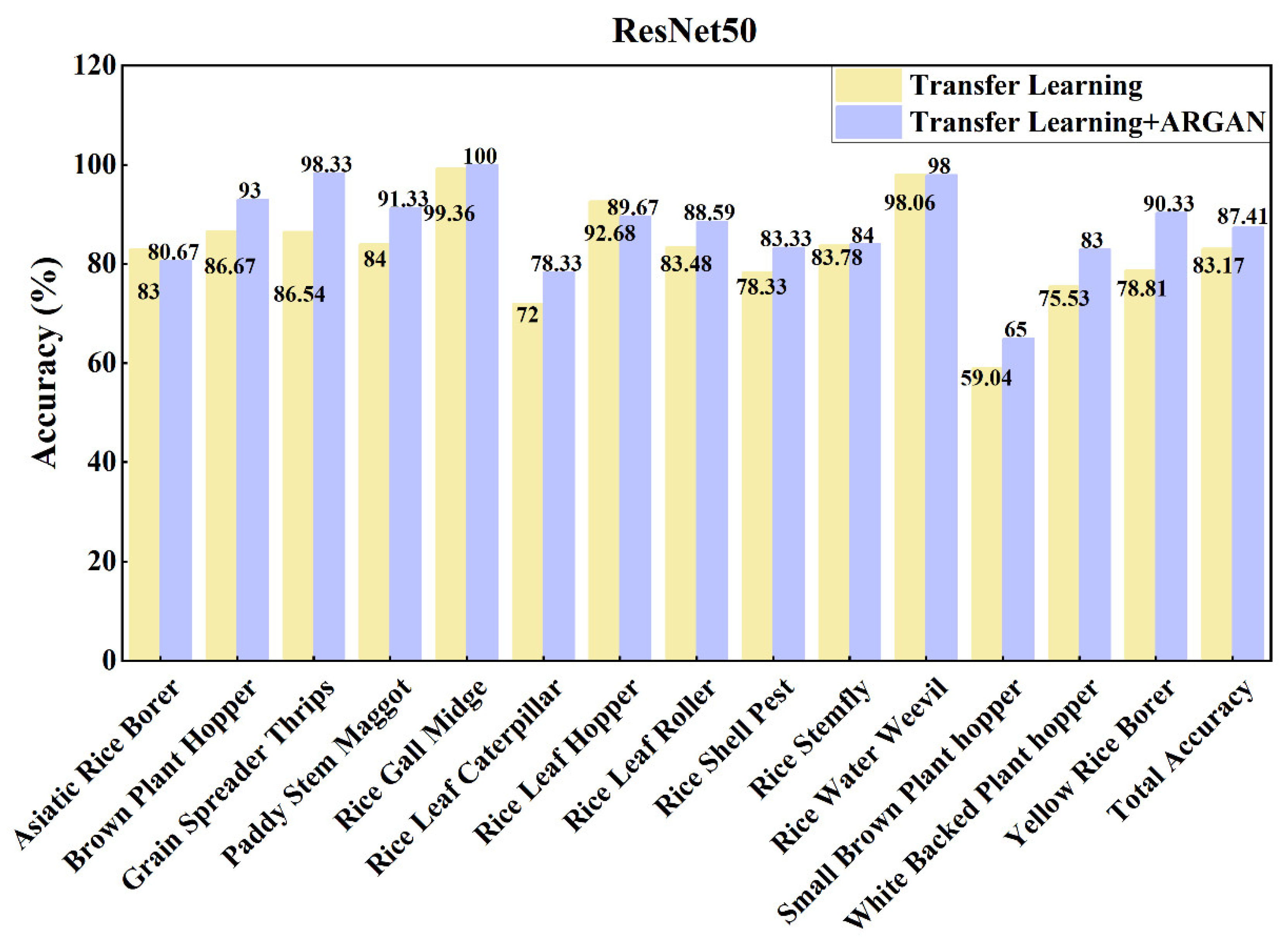
| ResNet50 | No | 73.74 | 72.10 | 72.70 | 73.99 |
| Yes | 83.41 | 82.95 | 83.02 | 83.17 | |
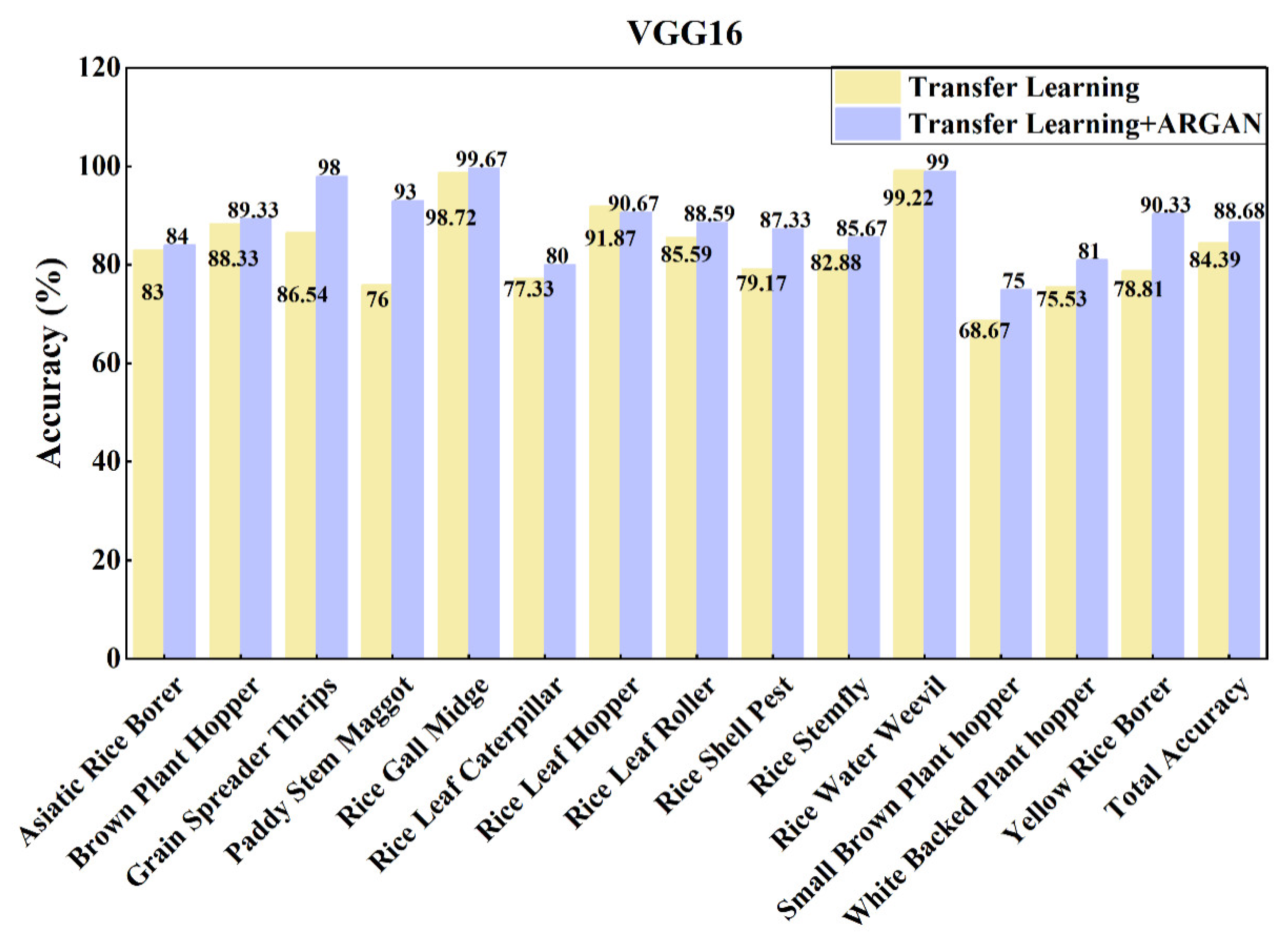
| VGG16 | No | 44.65 | 38.23 | 39.47 | 42.60 |
| Yes | 84.70 | 83.69 | 84.09 | 84.39 | |
| MobileNet | No | 67.85 | 66.22 | 66.61 | 69.58 |
| Yes | 83.19 | 82.43 | 82.62 | 82.89 |
| Datasets | Pre (%) | Rec (%) | F1 (%) | Acc (%) |
|---|---|---|---|---|
| IP102 | 65.83 | 64.85 | 65.10 | 65.35 |
| IP_RicePests | 83.41 | 82.95 | 83.02 | 83.17 |
| Model | ARGAN | Pre (%) | Rec (%) | F1 (%) | Acc (%) |
|---|---|---|---|---|---|
| ResNet50 | No | 83.41 | 82.95 | 83.02 | 83.17 |
| Yes | 87.81 | 87.40 | 87.35 | 87.41 | |
| VGG16 | No | 84.70 | 83.69 | 84.09 | 84.39 |
| Yes | 88.87 | 88.68 | 88.69 | 88.68 | |
| MobileNet | No | 83.19 | 82.43 | 82.62 | 82.89 |
| Yes | 86.73 | 86.41 | 86.22 | 86.44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Jiang, X.; Jia, X.; Duan, X.; Wang, Y.; Mu, J. Classification Method of Significant Rice Pests Based on Deep Learning. Agronomy 2022, 12, 2096. https://doi.org/10.3390/agronomy12092096
Li Z, Jiang X, Jia X, Duan X, Wang Y, Mu J. Classification Method of Significant Rice Pests Based on Deep Learning. Agronomy. 2022; 12(9):2096. https://doi.org/10.3390/agronomy12092096
Chicago/Turabian StyleLi, Zhiyong, Xueqin Jiang, Xinyu Jia, Xuliang Duan, Yuchao Wang, and Jiong Mu. 2022. "Classification Method of Significant Rice Pests Based on Deep Learning" Agronomy 12, no. 9: 2096. https://doi.org/10.3390/agronomy12092096
APA StyleLi, Z., Jiang, X., Jia, X., Duan, X., Wang, Y., & Mu, J. (2022). Classification Method of Significant Rice Pests Based on Deep Learning. Agronomy, 12(9), 2096. https://doi.org/10.3390/agronomy12092096








