Abstract
Nonedible materials such as agricultural wastes can serve as sources of antimicrobial peptides (AMPs) effective against bacterial plant pathogens. In this study, thirteen agricultural samples were collected and their protein hydrolysates obtained using pepsin. Peptides smaller than 3 kDa were purified by reverse-phase chromatography, cation exchange chromatography, and pI-based fractionation and tested for activity against plant pathogenic bacteria at each step. Active peptides were then analyzed for putative mechanisms using nanoLC–MS/MS and the Mascot program. Ultimately, eight candidate peptides originating from bagasse were selected and chemically synthesized for a comparative study of growth inhibition in plant pathogenic bacteria and plant growth-promoting rhizobacteria (PGPRs). Three synthesized peptides exhibited a potent activity against plant pathogenic bacteria while also supporting the growth of PGPRs. Proteomics analysis revealed the peptides PQLAVF (Pro-Gln-Leu-Ala-Val-Phe) and MDRFL (Met-Asp-Arg-Phe-Leu) to act against Xanthomonas oryzae pv. oryzae via membrane-active mechanisms, while peptide VQLMNSL (Val-Gln-Leu-Met-Asn-Ser-Leu) acted against Pectobacterium carotovorum and Agrobacterium rhizogenes through intracellular-active mechanisms. Further study remains necessary to customize peptides by amino acid substitution not only for a higher effective activity against these and other critical pathogens, but also for a higher stability of peptides in critical condition when applied in industrial processes in the future.
1. Introduction
Bacteria-caused diseases have been a crucial factor influencing agricultural plant production and food manufacturing for thousands of years. They still pose a considerable threat to the food supplies of many countries today [1]. Although advancements in science and technology have led to considerable decreases in the frequency and severity of disease outbreaks, 20–30% of actual production is still hampered each year because of plant diseases [2,3]. Bacterial plant diseases are less common than fungal or viral diseases, but the economic losses they induce are nonetheless devastating [4].
Currently, bacterial plant disease control relies mainly on chemical agents; however, the effects of such agents on long-term environmental pollution and as carcinogens in the food chain limit their future use [5]. Moreover, several strategies for plant disease control combined with many farm practices applied in modern cultivation have caused unintended troubles, including environmental degradation [6], loss of biodiversity [7,8], and the creation of advantageous habitats for the contamination, reproduction, transmission, and rapid evolution of plant pathogens [9,10,11]. In addition, chemical antibiotics such as streptomycin and oxytetracycline are applied for the treatment of plant bacterial diseases, but this practice enables the development of antibiotic resistance. Once antibiotic resistance is found in a plant pathogen population, it speedily becomes widespread [12], which increases the negative impacts of plant diseases on food security and human society [13]. Ultimately, to achieve sustainable plant pathogen control, it is necessary to develop effective alternatives for combating resistant pathogens that are also environmentally friendly.
Antimicrobial peptides (AMPs) are natural guardians against pathogen invaders and function in innate immune systems [14]. Over 1500 AMPs have been found in many living things, both eukaryotes and prokaryotes [15]. Commonly, AMPs exhibit broad activity against many types of organisms, namely fungi, bacteria, parasites, and viruses; hence, they are categorized as antifungal, antibacterial, antiparasitic, and antiviral, respectively [16]. The antibacterial activity of peptides is a result of amphiphilic composition and a high degree of positive charge within their structure. This quality supports peptide attachment and insertion into the bacterial cell membrane to create a pore, thereby bringing about membrane disruption and cell lysis [17,18]. A number of AMP families have been reported in plants, such as defensins, thionins, snakins, lipid transfer proteins, cyclotides, and hevein-like proteins. Some plant AMPs have had their structures and activities reported in the PhytAMP database. Overall, peptides in PhytAMP most commonly exhibit antifungal activity (51%), followed by antibacterial (33%) and antiviral (10%) activities [19].
Peptide size is also an important factor determining the efficacy of agricultural antibacterial agents [20]. Small synthetic antimicrobial peptides (ssAMPs), usually less than ten amino acids in size, could provide an acceptable alternative because their synthesis cost is significantly lower than the cost of producing long peptides. Choi et al. [12] set out to create new antibacterial hexapeptide ssAMPs with efficacy against Xanthomonas citri subsp. citri, the citrus canker pathogen. Of fourteen hexapeptides tested, they found three that were able to kill various X. citri strains when applied at a concentration of 10 µg/mL. In addition, soaking citrus leaves with the hexapeptides alongside pathogens significantly suppressed disease progress. Importantly, ssAMP sequences can be selected quickly through screens using proteomic techniques, while still retaining most of the functional aspects of native AMPs.
In this work, we chose agricultural wastes as ssAMP sources. Agricultural production has increased by more than three times over the past five decades due to the growth of the world population. Agricultural wastes are mainly generated from farming activities pertaining to crop production, such as planting, pruning, and harvesting. A large amount of waste also comes from agro-industrial production, with an annual increase in this area of about 7.5% [21]. The negative influence of agricultural wastes on human health, animal health, and bio-pollution is significant. In many developing countries, agricultural wastes are randomly discarded or burnt in public areas; this constitutes the beginning of air and soil pollution, and residue from wastes may seep into a water source, thereby causing water pollution [22].
The objective of this study was to investigate the antibacterial activity of ssAMPs from agricultural wastes (both wastes from farming areas and agro-industrial wastes) obtained through protein hydrolysis with pepsin. Only peptides of less than 3 kDa were retained and purified with reverse-phase chromatography, cation exchange chromatography, and off-gel fractionation. Final fractions containing peptides active against bacterial plant pathogens were analyzed using LC–MS/MS and the Mascot program, and small peptides with high Mascot scores were selected for synthesis and experimental evaluation of activity. Three peptides that demonstrated efficacy against bacterial plant pathogens but not against plant growth-promoting rhizobacteria (PGPRs), which have important roles in biocontrol, were finally selected for characterization of their antibacterial mechanisms via proteomic profiling of expressed bacterial proteins after exposure to peptides.
2. Materials and Methods
2.1. Time and Place of Research
This research was conducted during 2020–2022 at the Functional Proteomics Technology Laboratory, Functional Ingredients and Food Innovation Research Group Laboratory, National Center for Genetics Engineering and Biotechnology, and National Science and Technology Development Agency in Pathumthani, Thailand.
2.2. Sample Collection
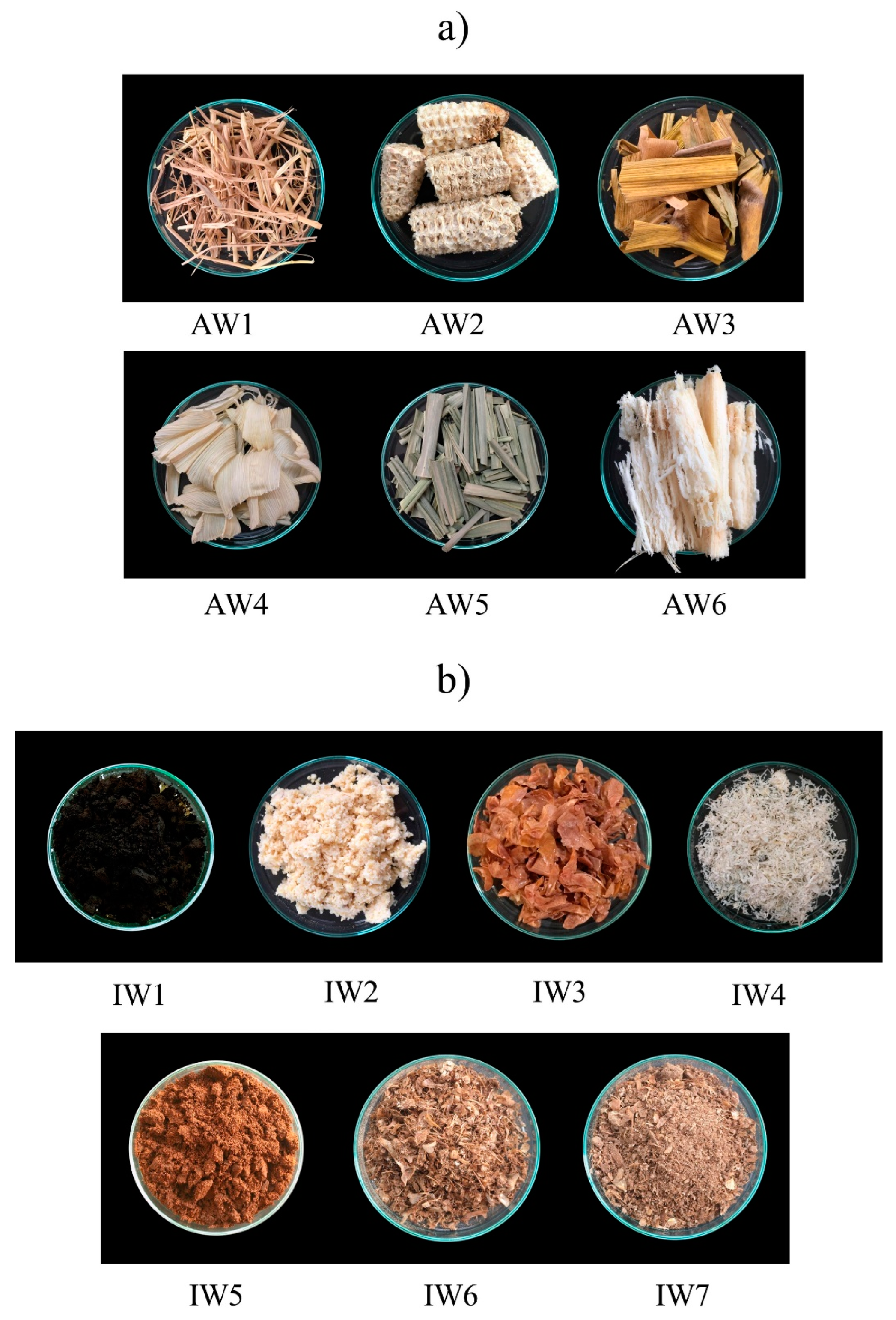
Waste samples were collected and classified into two groups, agricultural wastes and agro-industrial wastes (Table 1 and Figure 1).

Table 1.
Waste sample classification.

Figure 1.
Images of the waste samples, by group: (a) agricultural wastes; (b) agro-industrial wastes.
2.3. Preparation of Protein Hydrolysates and Small Peptides (<3 kDa)
Crude proteins were prepared by extracting each waste sample using 0.05 M sodium acetate, pH 4.0 with mechanical shaking at 25 ± 2 °C for 1 h, followed by autoclaving at 121 °C for 15 min to select only heat-tolerance proteins that show high stability when applied in industrial condition, and the autoclave condition could also eliminate all microbes that might be contaminated in samples. The total protein concentration in the supernatant was measured by a Lowry assay [23] using bovine serum albumin (BSA) as the protein standard. The absorbance at 750 nm (OD750) was measured and the protein concentration calculated from a calibration curve. The crude proteins were then hydrolyzed with pepsin (Sigma–Aldrich, St. Luis, MO, USA) in a ratio of 1:25 (pepsin:sample), and incubated with shaking at 200 rpm for 12 h at 37 °C. Next, the reaction was terminated by boiling for 10 min. The supernatant of the crude hydrolysate was retained after centrifugation at 10,000× g for 10 min at RT. The resulting hydrolysates were collected and diluted by five times with 0.5 M sodium acetate (NaOAc), then filtrated (cut-off) through a semipermeable membrane (Vivaspin 20, 3 kDa MWCO, GE Healthcare, Chicago, UK) to yield peptides smaller than 3 kDa, which were frozen at −20 °C until use.
2.4. Bacterial Plant Pathogens and Antimicrobial Activity Assays
Four plant pathogens were selected for investigation: Xanthomonas oryzae pv. oryzae (isolated from rice), Xanthomonas citri DOA-BC902, Pectobacterium carotovorum DOA-BC681, and Agrobacterium rhizogenes TISTR511. When assaying antibacterial activity, each pathogen was first prepared in tryptic soy agar (TSA) (Difco BBL, USA) and cultured for 24 h at 28 °C. Then, a single colony was picked and cultured in tryptic soy broth (TSB) (Difco BBL, Sparks, MD, USA) for 12–16 h to achieve an inoculum of 0.05 at OD600 (4 × 107 CFU/mL). The previously filtrated less-than-3 kDa peptides were then assayed for antibacterial activity against the pathogens in triplicate using the broth dilution method, for which bacteria in TSB, phosphate-buffered saline (PBS), and antibiotics (ampicillin and kanamycin) were used as controls. Antibiotics and hydrolysates/peptides were used at a final concentration of 100 µg/mL. OD600 values after incubation for 0, 2, 4, and 6 h were recorded using a microplate reader (Synergy H1 Hybrid Multi-Mode Reader, Biotek, Winusky, VT, USA). The inhibitory percentage was calculated from [(OD600 control − OD600 test)/OD600 control] × 100.
2.5. Experimental Design and Statistical Analysis
A completely randomized experimental design was used. Thirteen kinds of agricultural wastes were tested, and the experimental units were bacterial plant pathogens (X. oryzae pv. oryzae, X. citri DOA-BC902, Pectobacterium carotovorum DOA-BC681, and A. rhizogenes TISTR511) and plant growth-promoting rhizobacteria (PGPRs) (Bacillus subtilis ATCC6633, Pseudomonas aeruginosa ATCC27853, and Pseudomonas fluorescens TISTR2630), grown in tryptic soy broth (TSB) (Difco BBL, USA) at 28 °C in 96-well plates. Experiments were run in triplicate, and all results are presented as mean ± standard deviation.
2.6. Peptide Purification by Reverse-Phase Chromatography
The active peptides were initially purified by reverse-phase chromatography using a Delta-Pak C18 column (100 Å, 3.9 mm × 150 mm; Interlink Scientific Services Ltd., Kent, UK) previously equilibrated with 0.1% trifluoroacetic acid (TFA) in acetonitrile (ACN). The column was washed with 0.1% TFA in sterile water, after which a sample containing 0.1% TFA was loaded to bind the column. The hydrophilic fraction (coded UBR: unbound fraction of reverse-phase chromatography) was eluted from the column with 0.1% TFA in sterile water. Next, the hydrophobic fraction (coded BR: bound fraction of reverse-phase chromatography) was eluted stepwise with 0.1% TFA in ACN. All steps were carried out at an adjusted flow rate of 1 mL/min. Both UBR and BR fractions were further evaluated for antimicrobial activity.
2.7. Peptide Purification by Cation Exchange Chromatography
Prior to this purification step, the conductivity of all active fractions from the reverse-phase purification was examined. If conductivity was observed, contaminants were first eliminated using a P-6 desalting column (50 mL; Bio-Rad Laboratories, Inc., Heracles, CA, USA). Afterward, salt-removed samples were adjusted to pH 4. Then, salt-removed and nonconductive samples were ion-separated by cation exchange chromatography using AKTATM start (GE Healthcare, Chicago, IL, USA) through a HiTrap SP Sepharose FF (1 mL; Cytiva, Marlborough, MA, USA) cation chromatography exchange column with a flow rate of 1 mL/min and fraction volume of 1 mL. The column was then washed out with 50 mM NaOAc (pH 4) at 3 column volumes (CV) at a flow rate of 0.5 mL/min and fraction volume of 1 mL; this fraction was labeled UBC (the unbound fraction of cation exchange chromatography). Subsequently, the final fraction BC (bound fraction of cation exchange chromatography) was eluted with 1 M NaCl, a gradient of 0–100, linear 3 CV, and fixed fraction volume of 0.5 mL. Each fraction was buffer-exchanged from 50 mM NaOAc, pH 4 with a gradient of 1 M NaCl to sterile water prior to antibacterial activity determination.
2.8. Peptide Purification by pI-Based Fractionation
Active fractions from cation exchange chromatography were separated according to their isoelectric points (pI). For pI-based peptide separation, the 3100 OFFGEL Fractionator was utilized with an 18-well (18 cm) setup and pH interval from 3 to 10 according to the supplier’s protocol; details are listed in Table 2. The peptides in each resultant fraction with demonstrated bioactivity were further analyzed by nanoLC–MS/MS and the Mascot software (Matrix Science, London, UK) [24].

Table 2.
Parameters used when running the 18 cm OFFGEL unit.
2.9. Peptide Synthesis and Determination of Antibacterial Activity
Peptides were synthesized following the solid-phase peptide synthesis method reported by Hansen and Oddo [25]. After synthesis, the peptide samples were prepared for antimicrobial activity testing.
2.10. Study of Peptide–Microbe Interaction Mechanisms
After treatment of the selected plant pathogenic microbes with active peptides for 6 h, all control and experimental samples were tryptic-digested and their protein profiles determined by LC–MS/MS using the Ultimate 3000 Nano/Capillary LC System (Thermo Scientific, Waltham, MA, USA) coupled to an ESI-Ion Trap MS HCT ultra PTM Discovery System (Bruker Daltonics GmbH, Billica, MA, USA) with electrospray. The obtained MS/MS spectra were analyzed with the DeCyder MS 2.0 differential analysis software (GE Healthcare, Chicago, IL, USA), and the resulting output was searched against the NCBI database using the Mascot software (Matrix Science, London, UK) [24]. Similarities and differences of protein expression profiles obtained under different treatments were visualized by a Venn diagram [26] and protein functionality annotations were retrieved from UniProt (https://www.uniprot.org/id-mapping accessed on 1 April 2022.).
3. Results
3.1. Antibacterial Activity of Peptides Less Than 3 kDa in Size
Out of the thirteen waste samples investigated here, four hydrolysates (AW6, IW4, IW3, and AW1) showed obviously superior potential, with inhibitory percentages of 50% or higher (Table 3). Notably, AW6 ranked first for every targeted bacterial pathogen, while IW4 ranked second for all targets except X. oryzae pv. oryzae. In addition, IW3 ranked second against X. oryzae pv. oryzae and third against X. citri, while AW1 ranked fourth for X. citri.

Table 3.
Ranking of <3 kDa peptide samples according to inhibitory activity against plant pathogenic bacteria at 6 h after treatment.
3.2. Antibacterial Activity after Peptide Purification
After antibacterial screening, four peptide samples (AW1, AW6, IW3, and IW4) were selected for purification in three steps: reverse-phase chromatography, cation exchange chromatography, and pI-based fractionation. Bacterial growth inhibition was re-evaluated in triplicate after each purification step (Table 4).

Table 4.
Observed inhibitory percentages of purified peptide samples against growth of plant pathogenic bacteria.
The reverse-phase chromatography method yielded two fractions; hence, antibacterial activity was subsequently assayed in a total of eight samples. As indicated in Table 4, the UBR fraction exhibited a higher inhibitory activity than the corresponding BR fraction for every sample; that is, the hydrophilic fractions gave better results. While all BR samples showed inhibitory activity, values were consistently less than 15. The single most effective sample was AW6 UBR, which demonstrated the best inhibitory percentage (around 60%) for every bacterium tested. Meanwhile, the samples AW1 UBR, IW3 UBR, and IW4 UBR all showed a low percent inhibition against Pectobacterium carotovorum and A. rhizogenes, but better efficacy against X. oryzae pv. oryzae and X. citri. Ultimately, the samples chosen for the next purification step were AW1 UBR, AW6 UBR, IW3 UBR, and IW4 UBR.
The second purification employed cation exchange chromatography. All samples featured both unbound (coded-UBC) and bound (coded-BC) peaks except AW1 UBR, for which only an unbound peak was obtained (AW1 UBR-UBC). Evaluating the samples for antibacterial activity revealed AW6 BC to have the best inhibitory percentage for every bacterial pathogen (Table 4). Accordingly, AW6 BC was chosen for the third purification step, pI-based or off-gel fractionation. After the 7-day running time, the quantity of protein obtained in each well was only enough to assay antibacterial activity against two pathogens with one replicate each, the results of which comprise the last section of Table 4. The fractions from wells 5, 6, 7, and 18 all inhibited the growth of X. oryzae pv. oryzae, while that from well 13 inhibited P. carotovorum. Consequently, AW6 UBR-BC wells 5, 6, 7, 13, and 18 were selected for peptide sequencing and further analysis using LC–MS/MS and the Mascot software.
3.3. Antibacterial Activity of Small Synthetic Peptides
In this study, thousands of peptides were analyzed and sequenced by Mascot; eight peptides having high peptide scores and fewer than ten amino acid residues were selected for determination of antibacterial activity (Table 5). The active peptides were selected from those more effective against plant pathogens and less effective against PGPRs. The results showed very obviously that peptides no. 1 (PQLAVF) and no. 5 (MDRFL) were most effective against X. oryzae pv. oryzae, while peptide no. 2 (VQLMNSL) exhibited the greatest efficacy against Pectobacterium carotovorum and A. rhizogenes. In addition, peptide no. 2 did not demonstrate any effect against PGPRs (Bacillus subtilis and Pseudomonas fluorescens). These three effective peptides were used in the subsequent determination of peptide–microbe mechanisms.

Table 5.
Inhibitory percentages of small synthetic peptides derived from purified protein hydrolysates of bagasse (Saccharum sp.; AW6).
3.4. Determination of Peptide–Microbe Interaction Mechanisms
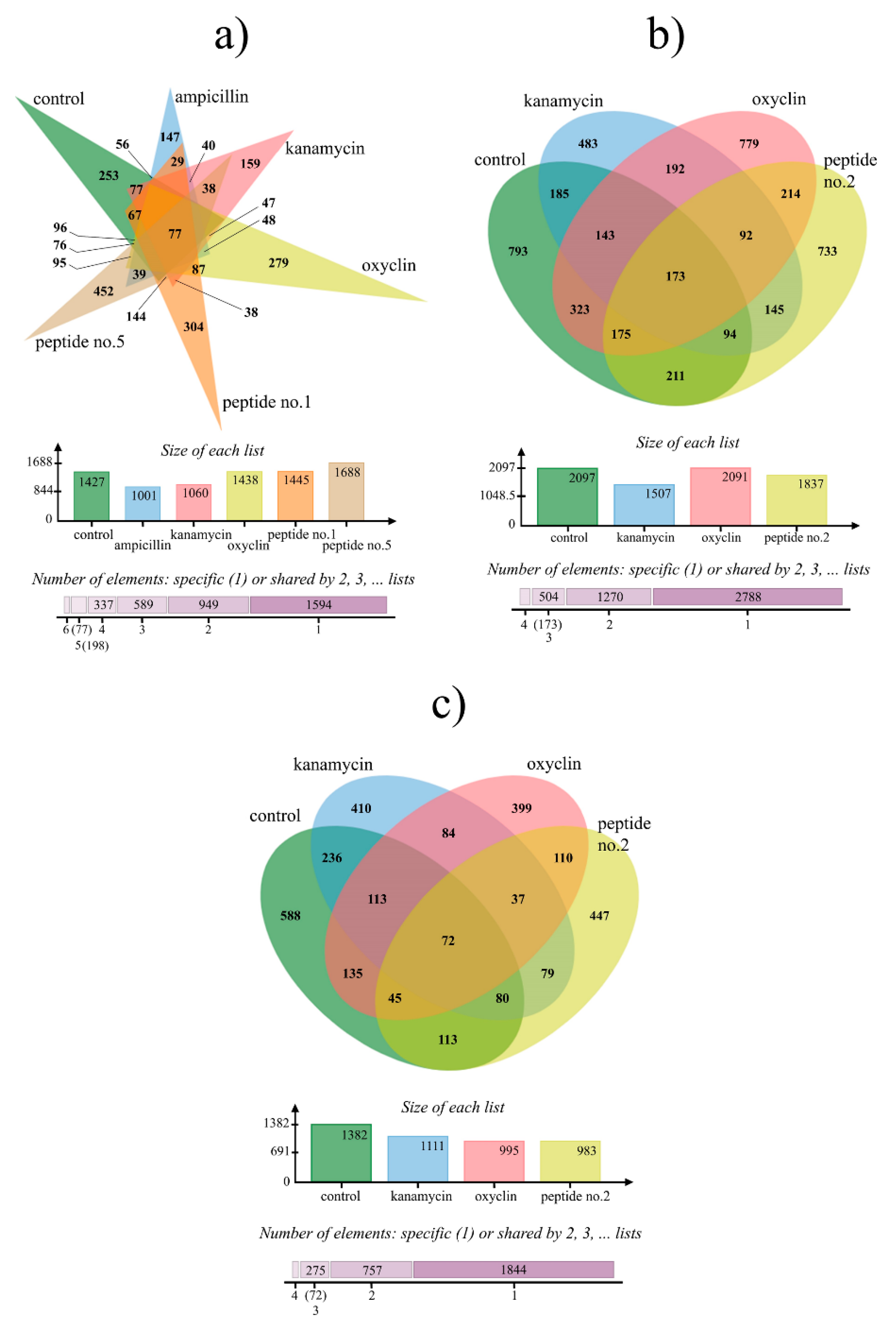
The peptide–microbe pairs for which interaction mechanisms were characterized are listed in Table 6. In these assays, untreated microbes were used as the negative control and antibiotic-treated microbes as the positive control. Many differentially expressed proteins were identified between control and treated samples, and patterns of protein expression under different treatments were straightforwardly visualized by a Venn diagram [26] (Figure 2).

Table 6.
Selected peptides and bacterial plant pathogens used for the determination of antibacterial mechanisms by shotgun proteomics.

Figure 2.
Venn diagram summary of protein expression in bacterial plant pathogens treated with selected peptides and antibiotics: (a) X. oryzae pv. oryzae, (b) Pectobacterium carotovorum, and (c) A. rhizogenes. The number of elements means the number of proteins detected in each treatment.
In total, 1445 proteins with altered expression were identified in X. oryzae pv. oryzae treated with peptide no. 1, and 1688 in the same pathogen treated with peptide no. 5 (Figure 2a). Among these, 304 and 452 proteins were uniquely expressed in association with peptides no. 1 and 5, respectively, and only a small number overlapped with expression profiles obtained under kanamycin, ampicillin, and oxycline treatments. Pectobacterium carotovorum was treated with peptide no. 2, in which condition 1837 proteins were found to be expressed (Figure 2b). Among those proteins, 733 were uniquely present under treatment with peptide no. 2, 214 were also expressed under oxycline treatment, and 145 also under kanamycin treatment. A. rhizogenes was likewise treated with peptide no. 2, which yielded an expression profile consisting of 983 proteins. A total of 447 proteins were unique to treatment with peptide no. 2, while 110 were also expressed under oxycline and 79 under kanamycin.
The proteins expressed in peptide-treated pathogen cells were further analyzed by consulting the UniProt database to identify Gene Ontology or functional annotations.
4. Discussion
4.1. Preparation of Protein Hydrolysates and Screening of Antibacterial Activity
In the initial protein extraction, sample proteins were extracted with 0.05 M sodium acetate, pH 4 for 1 h in keeping with previous reports that mild-acid extraction is appropriate for isolating plant proteins due to minimizing oxidation, polymerization of phenolic compounds, and irreversible protein binding [27]. Lay et al. [28] similarly extracted proteins from ornamental tobacco and petunia using 50 mM sulfuric acid; Pickardt et al. [27] found that increasing the concentration of sodium chloride enhanced the relative protein yield; Taniguchi et al. [29] adjusted the pH to 2.0 with 1 M HCl before hydrolyzing with pepsin. Following extraction, peptides in this study were heated in an autoclave (121 °C for 15 min) to remove heat-intolerant and heat-labile proteins, conformant to the method of Lay et al. [30].
Plant AMPs are normally smaller than 10 kDa [31]; accordingly, we used a semipermeable membrane to filter the diluted hydrolysates before purification. Notably, the shorter peptides are more cost-effective to synthesize; Gordon et al. [20] highlighted that peptides of less than ten amino acids could be a good option because the cost of synthesizing them is much lower than for long peptides. Therefore, we used a Vivaspin 20 with 3 kDa MWCO to retain only peptides of less than 3 kDa (approximately 27–28 amino acids). In 2017, Choi et al. [12] tried to identify an ssAMP effective against X. citri subsp. Citri using the PS-SPCL (positioning scanning of a synthetic peptide combinatorial library) technique; among fourteen investigated ssAMPs, they found three hexapeptides that showed bactericidal activity against X. citri subsp. Citri strains. Taniguchi et al. [29] also selected on peptide size using dialysis tubing.
In terms of inhibitory percentage against plant pathogenic bacteria as shown in Table 3, the protein hydrolysate from bagasse (AW6) ranked first for every tested pathogen. These results agree with a report by Velazquez-Martinez et al. [32] that found sugarcane bagasse with 2.2% crude protein to have a high antimicrobial activity against Escherichia coli, Bacillus cereus, and Staphylococcus aureus. That coconut residue hydrolysate ranked second in this work is consistent with a report that coconut AMPs (Cn-AMPs) have an extremely efficient antimicrobial activity against both Gram-positive and Gram-negative pathogenic bacteria including E. coli, Bacillus subtilis, S. aureus, and Pseudomonas aeruginosa [33]. Moreover, the antimicrobial activity of rice straw hydrolysate observed here aligns with the work of Park et al. [34], which reported rice straw extract to inhibit growth of the bloom-forming cyanobacterium Microcystis aeruginosa. Finally, these results are consistent with our last study, in which we found coconut residue, peanut seed coat, and rice straw protein hydrolysates to have strong antimicrobial activity against the plant pathogens Ralstonia solanacearum and Burkholderia cepacia, achieving over 74% inhibition [35].
4.2. Peptide Purification
Antibacterial screening identified four effective hydrolysates (AW1, AW6, IW3, and IW4), after which the key next step was peptide purification. Normally, purification methods rely on intrinsic physio-biochemical properties such as size, overall net charge, solvent tolerance, and thermostability. Scott et al. [36] highlighted that some protein mixtures from waste samples should be separated according to the presence of polar and nonpolar side groups, while others may be separated on the basis of basic and acidic amino acids. A complex mixture of amino acids can also be separated using chromatographic purification techniques, with ion exchange chromatography being particularly important to develop in the separation of peptides from complex mixtures. This study accordingly employed multiple purification steps, namely reverse-phase chromatography to separate hydrophobic and hydrophilic fractions followed by cation exchange chromatography and pI-based purification (off-gel fractionation). Each purification step functioned to narrow the scope of bioactive peptides. After the initial reverse-phase chromatography, one fraction for each of the four samples exhibited antibacterial activity (the UBR fractions). However, after cation exchange chromatography, only one fraction from one sample, AW6 UBR-BC, demonstrated antibacterial activity. Following the final pI-based purification, five fractions exhibited antibacterial activity (wells 5, 6, 7, 13, and 18) and were selected for further analysis using LC–MS/MS and the Mascot software. Of the multitude of peptides identified as having high peptide scores and sizes smaller than ten amino acids, only eight were selected for synthesis and characterization of the mechanisms of their antimicrobial activity against plant pathogen cells.
4.3. Peptide–Microbe Interaction Mechanisms
Protein expression Venn diagrams (Figure 2) revealed that the majority of proteins expressed by bacterial plant pathogens treated with the selected peptides were unique (that is, they were not also expressed in cultures treated with antibiotics). Thus, it could be concluded that peptides no. 1, 2, and 5 have mechanisms of action distinct from those of the tested antibiotics. Data from UniProt provided further insight into these mechanisms.
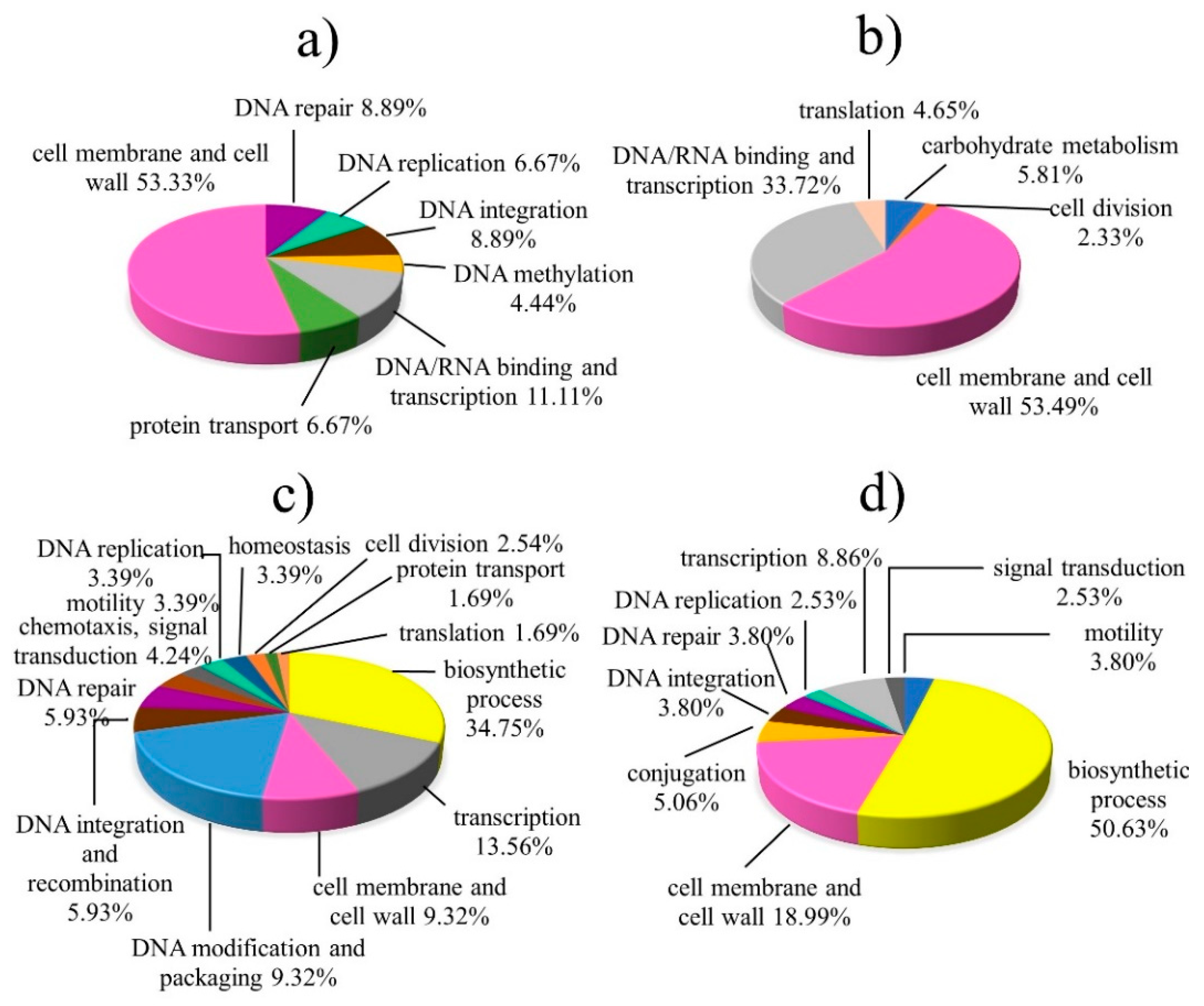
First, we examined the Gene Ontology annotations (biological process, molecular function, and cellular compartment) of all proteins expressed under peptide treatment (but not found in antibiotic treatment or control) and organized them into groups (Figure 3). For X. oryzae pv. oryzae treated with peptide no. 1 (Figure 3a), most expressed proteins (53.33%) related to the cell membrane and cell wall, and a smaller proportion related to DNA-related biological processes. These results suggest that the antimicrobial mechanism of peptide no. 1 in X. oryzae pv. oryzae is primarily related to the cell membrane and cell wall. It is possible that the peptide may have caused cell leakage and that some part of it also affected DNA-related processes.

Figure 3.
Functional profiles of identified proteins expressed in peptide-treated plant pathogens (but not found in antibiotic treated or control samples): (a) X. oryzae pv. oryzae with peptide no. 1, (b) X. oryzae pv. oryzae with peptide no. 5, (c) Pectobacterium carotovorum with peptide no. 2, and (d) A. rhizogenes with peptide no. 2.
Similarly, in X. oryzae pv. oryzae treated with peptide no. 5 (Figure 3b), the majority of expressed proteins (53.49%) related to the cell membrane and cell wall; this group included proteins involved in cell wall organization and integral components of membranes. Other expressed proteins had functions relating to DNA/RNA binding, transcription and translation, carbohydrate metabolism, and cell division. Overall, the functional profiles of proteins expressed in X. oryzae pv. oryzae treated with peptides no. 1 and 5 were similar, and those functions suggest these peptides primarily interact with the cell membrane directly, and secondarily may have some feature that interacts with intracellular targets such as DNA, RNA, and proteins. Thus, peptides no. 1 and 5 can be considered membrane-active AMPs.
In Pectobacterium carotovorum treated with peptide no. 2, expressed proteins were most commonly related to biosynthetic processes (34.75%); annotations in this group included the general biosynthetic process, peptidoglycan catabolic process, phosphopyruvate-dependent sugar phosphotransferase system, and polysaccharide biosynthetic process. Other prominent function groups included regulation of transcription, the cell membrane, and the cell wall, and terms relating to DNA. These results imply that peptide no. 2 mostly interacts with proteins involved in intracellular processes including biosynthesis, transcription and translation, DNA repair, and signal transduction. However, some part of the peptide may interact with the cell membrane and cell wall of Pectobacterium carotovorum.
Finally, we examined the functional classifications of proteins expressed by A. rhizogenes after treatment with peptide no. 2 (Figure 3d). Again, more than half were related to biosynthetic processes (50.63%); specific terms included the carbohydrate metabolic process and transport, fatty acid and lipid biosynthesis, and amino acid metabolism and peptidoglycan biosynthesis. In addition, a small number of proteins had functions relating to the cell membrane and cell wall. Overall, the functional classifications of proteins expressed in P. carotovorum and A. rhizogenes after treatment with peptide no. 2 are similar in that most related to metabolic processes. Thus, peptide no. 2 could be called an intracellular-active AMP.
Among the eight peptides investigated, three were found to effectively inhibit the growth of X. oryzae pv. oryzae, P. carotovorum, and A. rhizogenes. Peptides no. 1 and 5 were determined to be membrane-active AMPs, and peptide no. 2 an intracellularly active AMP. In general, membrane-active AMPs are usually unstructured when in aqueous solution, and form an α-helical structure in the presence of a lipid membrane. Under appropriate conditions, membrane-active AMPs create transmembrane pores or channels that allow leakage of intracellular molecules, eventually leading to cell death [37]. Various modes of action have been proposed on the basis of the arrangement of AMPs on a membrane: namely, the barrel-stave pore, toroidal pore, or carpet mechanism. Furthermore, the precise peptide–microbe mechanism is dependent on lipid membrane composition, peptide structure, peptide concentration, temperature, and pH [38].
In contrast, intracellularly active AMPs can inhibit or kill microbial cells without causing membrane disruption. This type of AMP interacts with targets inside the cells, namely DNA, RNA, or proteins [39].
Several AMPs are reported to have only one mode of action regardless of their concentration; for example, apidaecin exhibits a nonmembrane-lytic mode of action in every concentration and condition [40]. However, many AMPs are reported to have dual mechanisms contingent on peptide concentration. Typically, concentrations above the minimum inhibitory concentration (MIC) lead to membrane lysis with the AMP acting as a detergent, whereas concentrations lower than the MIC cause membrane penetration and targeting of macromolecules within cells [41]. This intracellular action causes inhibition of metabolic processes and also leads to the death of the bacteria. As illustrated in Figure 3c,d, the intracellular mechanism of peptide no. 2 is related to nucleic acids (DNA/RNA), lipids, and proteins. This is consistent with other findings regarding the mechanisms of intracellular-active AMPs in general. These results conform to several previous reports.
Interestingly, while peptides no. 1 and 5 have primarily membrane-active mechanisms of action, they were also associated with expression of some proteins related to metabolic processes. At the same time, peptide no. 2 is primarily an intracellular-active AMP, but is also implied to affect the cell wall and cell membrane. These results are similar to some previous findings. Shi et al. [42] reported that melittin, an AMP from Apis mellifera, disrupts the cell membrane, making holes that result in cytoplasm leakage, but it also may inhibit the biosynthesis of both DNA and proteins. In addition, the bactericidal peptide indolicidin has exhibited more than one mechanism of antimicrobial action. In aqueous solution, it takes on an amphipathic and globular conformation, distinctly unlike the structures adopted in lipids, and these different structures feature different functions. Gel retardation and fluorescence quenching experiments revealed indolicidin to have DNA-binding properties and to interact with lipid bilayers at different concentrations [43].
5. Conclusions
Protein hydrolysates of AW1 (rice straw), AW6 (bagasse), IW3 (peanut seed coat), and IW4 (coconut residue) exhibited antibacterial activity against bacterial plant pathogens including X. oryzae pv. oryzae, X. citri, Pectobacterium carotovorum, and A. rhizogenes. Peptides PQLAVF, VQLMNSL, and MDRFL were identified as novel peptides derived from AW6 (bagasse) that inhibited the growth of X. oryzae pv. oryzae, Pectobacterium carotovorum, and A. rhizogenes. Of those, VQLMNSL did not show any effect against the PGPRs Bacillus subtilis and Pseudomonas fluorescens. Regarding the mechanism of effect, PQLAVF and MDRFL might cause cell leakage and interfere with DNA-related processes in X. oryzae pv. oryzae, while VQLMNSL interferes with the biological processes of Pectobacterium carotovorum and A. rhizogenes.
Author Contributions
Conceptualization, T.D., N.P. (Nonglak Parinthawong), and S.R.; methodology, T.D., S.C., S.T., and N.P. (Narumon Phaonakrop); software, N.P. (Narumon Phaonakrop); validation, T.D., N.P. (Nonglak Parinthawong), and S.R.; formal analysis, T.D.; investigation, T.D.; writing—original draft preparation, T.D.; writing—review and editing, T.D., N.P. (Nonglak Parinthawong), and S.R.; visualization, S.R.; supervision, N.P. (Nonglak Parinthawong); project administration, T.D. and N.P. (Nonglak Parinthawong); funding acquisition, N.P. (Nonglak Parinthawong) and S.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Faculty of Agricultural Technology, King Mongkut’s Institute of Technology Ladkrabang, grant number 2564-02-04-018; Functional ingredient and Food innovation program, National Science and Technology Development Agency, grant number P-19-50420.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All-new research data were presented in this contribution.
Acknowledgments
We thank the Functional Proteomics Technology Laboratory, National Center for Genetic Engineering and Biotechnology for providing laboratory facilities. We also thank Rajabhat Rajanagarindra University for support of T.D.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Palmgren, M.G.; Edenbrandt, A.K.; Vedel, S.E.; Andersen, M.M.; Landes, X.; Østerberg, J.T.; Falhof, J.; Olsen, L.I.; Christensen, S.B.; Sandøe, P.; et al. Are we ready for back-to-nature crop breeding? Trends Plant Sci. 2015, 20, 155–164. [Google Scholar] [CrossRef]
- Oerke, E.; Dehne, H.W. Safeguarding production—Losses in major crops and the role of crop protection. Crop Prot. 2004, 23, 275–285. [Google Scholar] [CrossRef]
- Oerke, E. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Vidaver, A.K. Uses of antimicrobials in plant agriculture. Clin. Infect. Dis. 2002, 3, S107–S110. [Google Scholar] [CrossRef] [PubMed]
- Daoubi, M.; Hernandez-Galan, R.; Benharref, A.; Collado, I.G. Screening study of lead compoundds for natural product-based fungicides: Antifungal activity and biotransformation of 6α, 7α-dihydroxy-β-himachalene by Botrytis cinerea. J. Agric. Food Chem. 2005, 53, 6673–6677. [Google Scholar] [CrossRef] [PubMed]
- Enserink, M.; Pamela, J.; Hines, P.J.; Sacha, N.; Vignieri, S.N.; Wigginton, N.S.; Yeston, J.S. The pesticide paradox. Science 2013, 341, 728–729. [Google Scholar] [CrossRef]
- Lucas, J.A. Advances in plant disease and pest management. J. Agric. Sci. 2011, 149, 91–114. [Google Scholar] [CrossRef]
- Gonthier, D.J.; Ennis, K.K.; Farinas, S.; Hsieh, H.Y.; Iverson, A.L.; Batáry, P.; Rudolphi, J.; Tscharntke, T.; Cardinale, B.J.; Perfecto, I. Biodiversity conservation in agriculture requires a multi-scale approach. Proc. R. Soc. B 2014, 281, 1358. [Google Scholar] [CrossRef] [PubMed]
- Zhan, J.; Mundt, C.C.; Hoffer, M.H.; McDonald, B.A. Local adaptation and effect of host genotype on the evolution of pathogen: An experimental test in a plant pathosystem. J. Evol. Biol. 2002, 15, 634–647. [Google Scholar] [CrossRef]
- Sommerhalder, R.J.; McDonald, B.A.; Mascher, F.; Zhan, J.S. Sexual recombinants make a significant contribution to epidemics caused by the wheat pathogen Phaeosphaeria nodorum. Phytopathology 2010, 100, 855–862. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhan, J.; McDonald, B.A. Experimental measures of pathogen competition and relative fitness. Annu. Rev. Phytopathol. 2013, 51, 131–153. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Park, E.; Lee, S.W.; Hyun, J.W.; Baek, K.H. Selection of small synthetic antimicrobial peptides inhibiting Xanthomonas citri subsp. citri causing citrus canker. Plant Pathol. J. 2017, 33, 87–94. [Google Scholar] [CrossRef] [PubMed][Green Version]
- He, D.-C.; Zhan, J.-S.; Xie, L.-H. Problems, challenges and future of plant disease management: From an ecological point of view. J. Integr. Agric. 2016, 15, 705–715. [Google Scholar] [CrossRef]
- Park, I.Y.; Cho, J.H.; Kim, K.S.; Kim, Y.B.; Kim, M.S.; Kim, S.C. Helix stability confers salt resistance upon helical antimicrobial peptides. J. Biol. Chem. 2004, 279, 13896–13901. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, G. ADP: The antimicrobial peptide database. Nucleic Acids Res. 2004, 32, D590–D592. [Google Scholar] [CrossRef]
- Zhang, L.J.; Gallo, R.L. Antimicrobial peptides. Curr. Biol. 2006, 26, R14–R19. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.P.S.; Rosek, A.; Hancock, R.E.W. Structure activity relationship for the β-hairpin cationic antimicrobial peptide polyphemusin I. Biochim. Biophys. Acta Proteins Proteom. 2004, 1698, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Hall, K.N.; Aguilar, M.I. Antimicrobial peptide structure and mechanism of action: A focus on the role of membrane structure. Curr. Top. Med. Chem. 2016, 16, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Hammami, R.B.; Hamida, J.; Vergoten, G.; Fliss, I. PhytAMP: A database dedicated to antimicrobial plant peptides. Nucleic Acids Res. 2008, 37, D963–D968. [Google Scholar] [CrossRef] [PubMed]
- Gordon, Y.J.; Romanowski, E.G.; McDermott, A.M. A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs. Curr. Eye Res. 2005, 30, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Adejumo, I.O.; Adebiyi, O.A. Agricultural Solid Wastes: Causes, Effects, and Effective Management. In Strategies of Sustainable Solid Waste Management; IntechOpen: London, UK, 2015. [Google Scholar] [CrossRef]
- Varma, V.S.; Yadav, J.; Das, S.; Kalamdhad, A.S. Potential of waste carbide sludge addition on earthworm growth and organic matter degradation during vermicomposting of agricultural wastes. Ecol. Eng. 2015, 83, 90–95. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.C.; Creasy, D.M.; Cottrel, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Hansen, P.R.; Oddo, A. Fmoc Solid-Phase Peptide Synthesis. Methods Mol. Biol. 2015, 1348, 33–50. [Google Scholar] [CrossRef] [PubMed]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [PubMed]
- Pickardt, C.; Neidhart, S.; Griesbach, C.; Dube, M.; Knauf, U.; Kammerer, D.R.; Carle, R. Optimisation of mild-acidic protein extraction from defatted sunflower (Helianthus annuus L.) meal. Food Hydrocoll. 2009, 23, 1966–1973. [Google Scholar] [CrossRef]
- Lay, F.T.; Brugliera, F.; Anderson, M.A. Isolation and Properties of Floral Defensins from Ornamental Tobacco and Petunia. Plant Physiol. 2003, 131, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Kameda, M.; Namae, T.; Ochiai, A.; Saitoh, E.; Tanaka, T. Identification and characterization of multifunctional cationic peptides derived from peptic hydrolysates of rice bran protein. J. Funct. Foods 2017, 34, 287–296. [Google Scholar] [CrossRef]
- Lay, F.T.; Anderson, M.A. Defensins—Components of the Innate Immune System in Plants. Curr. Protein Pept. Sci. 2005, 6, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Broekaert, W.F.; Cammue, B.P.A.; De Bolle, M.F.C.; Thevissen, K.; De Samblanx, G.W.; Osborn, R.W. Antimicrobial peptides from plants. Crit. Rev. Plant Sci. 1997, 16, 297–323. [Google Scholar] [CrossRef]
- Velazquez-Martinez, V.; Valles-Rosales, D.; Rodriguez-Uribe, L.; Holguin, O.; Quintero-Quiroz, J.; Reyes-Jaquez, D.; Rodriguez-Borbon, M.I.; Villagrán-Villegas, L.Y.; Delgado, E. Antimicrobial, shelf-life stability, and effect of maltodextrin and gum arabic on the encapsulation efficiency of sugarcane bagasse bioactive compounds. Foods 2021, 10, 116. [Google Scholar] [CrossRef]
- Mandal, S.M.; Migliolo, L.; Franco, O.L.; Ghosh, A.K. Identification of an antifungal peptide from Trapa natans fruits with inhibitory effects on Candida tropicalis biofilm formation. Peptides 2011, 32, 1741–1747. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Han, M.S.; Ahn, C.Y.; Kim, H.S.; Yoon, B.D.; Oh, H.M. Growth inhibition of bloom-forming cyanobacterium Microcystis aeruginosa by rice straw extract. Lett. Appl. Microbiol. 2006, 43, 307–312. [Google Scholar] [CrossRef]
- Ditsawanon, T.; Parinthawong, N.; Phaonakrob, N.; Roytrakul, S. Protein hydrolysates from agricultural wastes for plant bacterial disease control. IJAT 2022, 18, 479–488. [Google Scholar]
- Scott, E.; Peter, F.; Sander, J. Biomass in the manufacture of industrial products-the use of proteins and amino acids. Appl. Microbiol. Biotechnol. 2007, 75, 751–762. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [PubMed]
- Sani, M.A.; Separovic, F. How Membrane-Active Peptides Get into Lipid Membranes. Acc. Chem. Res. 2017, 49, 1130–1138. [Google Scholar] [CrossRef] [PubMed]
- Otvos, L. Antibacterial peptides and proteins with multiple cellular targets. J. Pept. Sci. 2005, 11, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Casteels, P.; Ampe, C.; Jacobs, F.; Vaeck, M.; Tempst, P. Apidaecins: Antibacterial peptides from honeybees. EMBO J. 1989, 8, 2387–2391. [Google Scholar] [CrossRef]
- Cudic, M.; Otvos, J.L. Intracellular target.ts of antibacterial peptides. Curr. Drug Targets 2002, 3, 101–106. [Google Scholar] [CrossRef]
- Shi, W.; Li, C.; Li, M.; Zong, X.; Han, D.; Chen, Y. Antimicrobial peptide melittin against Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen in rice. Appl. Microbiol. Biotechnol. 2016, 100, 5059–5067. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.H.; Chen, C.; Jou, M.L.; Lee, A.Y.; Lin, Y.C.; Yu, Y.P.; Huang, W.T.; Wu, S.H. Structural and DNA-binding studies on the bovine antimicrobial peptide, indolicidin: Evidence for multiple conformations involved in binding to membranes and DNA. Nucleic Acids Res. 2005, 33, 4053–4064. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).