Genome-Wide Identification and Expression Analysis of SnRK Gene Family under Abiotic Stress in Cucumber (Cucumis sativus L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Phylogenetic Analysis of the CsSnRK Gene Family in Cucumber
2.2. Gene Structure and Protein Motif Analysis
2.3. Chromosomal Location and Gene Duplication Analyses
2.4. Cis-Acting Elements Analysis CsSnRK Genes
2.5. Expression Analysis of CsSnRK2 Genes in Different Plant Tissues
2.6. Expression Analysis of CsSnRK Genes under Different Abiotic Stresses
2.6.1. Plant Materials and Treatments
2.6.2. RNA Extraction and Quantitative RT-PCR
3. Results
3.1. Identification of CsSnRK Gene Family in Cucumber
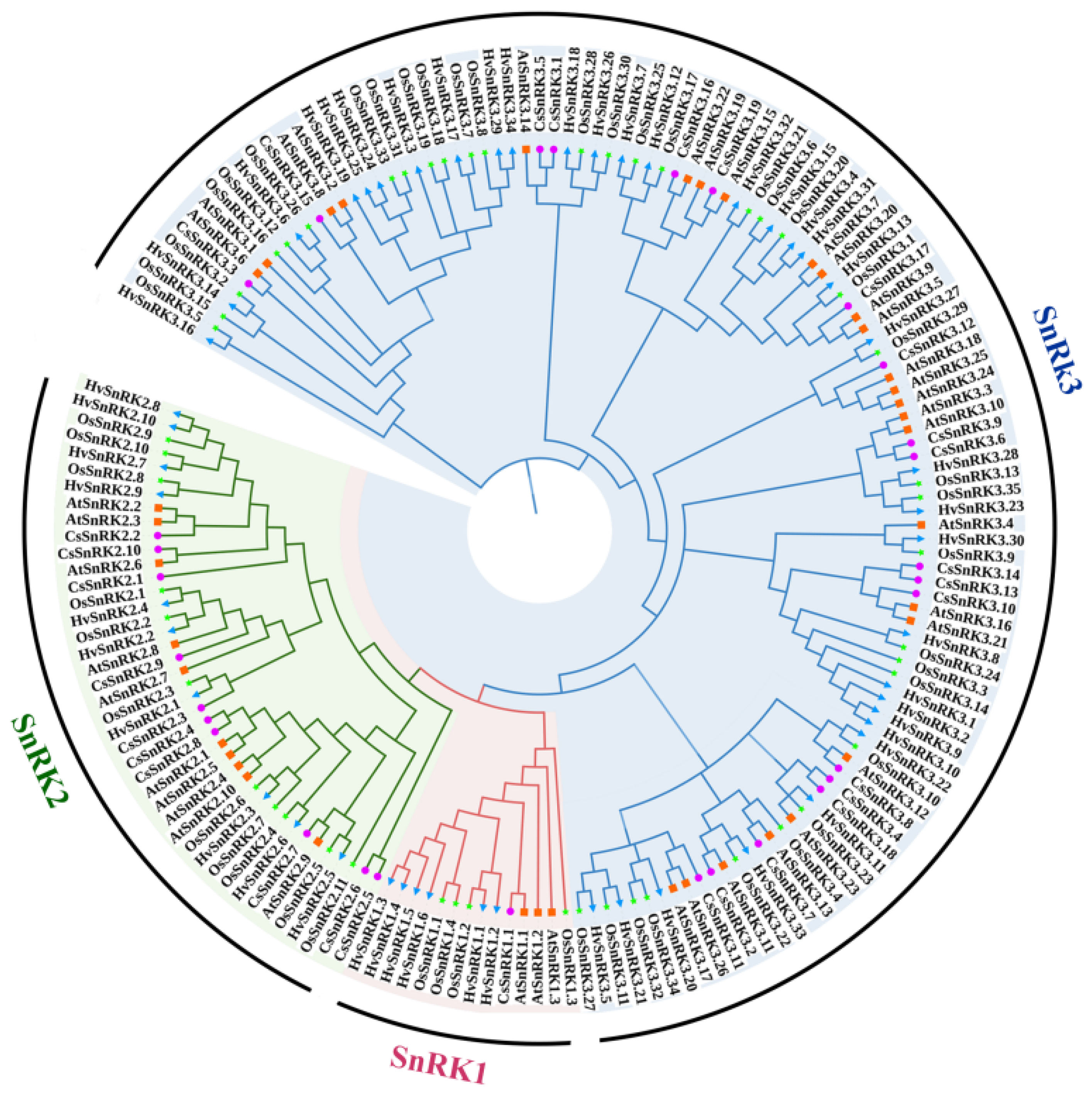
3.2. Phylogeny Analysis of CsSnRK Gene Family
3.3. Gene Structure and Conserved Motifs Analysis of CsSnRK Gene Family
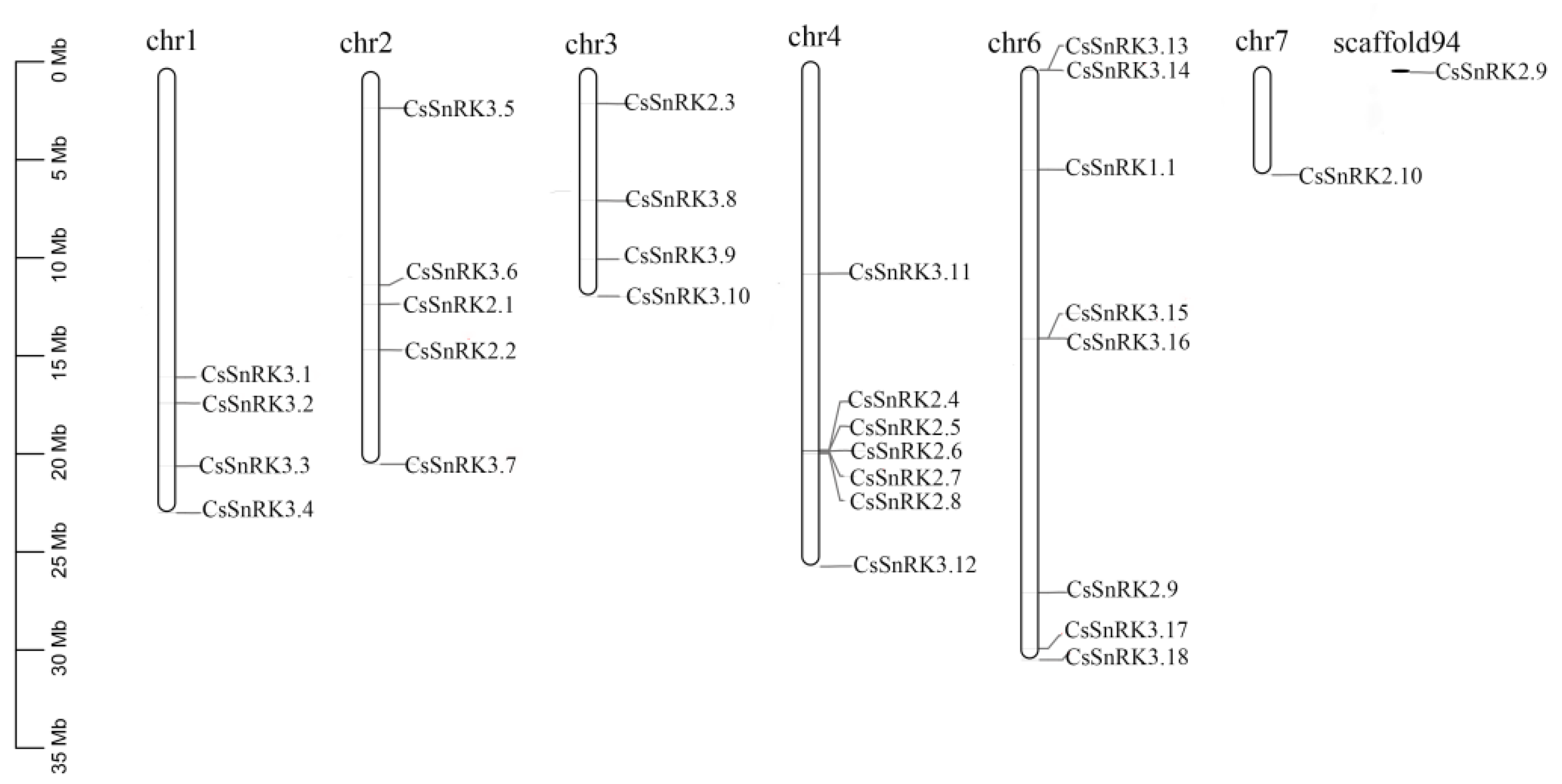
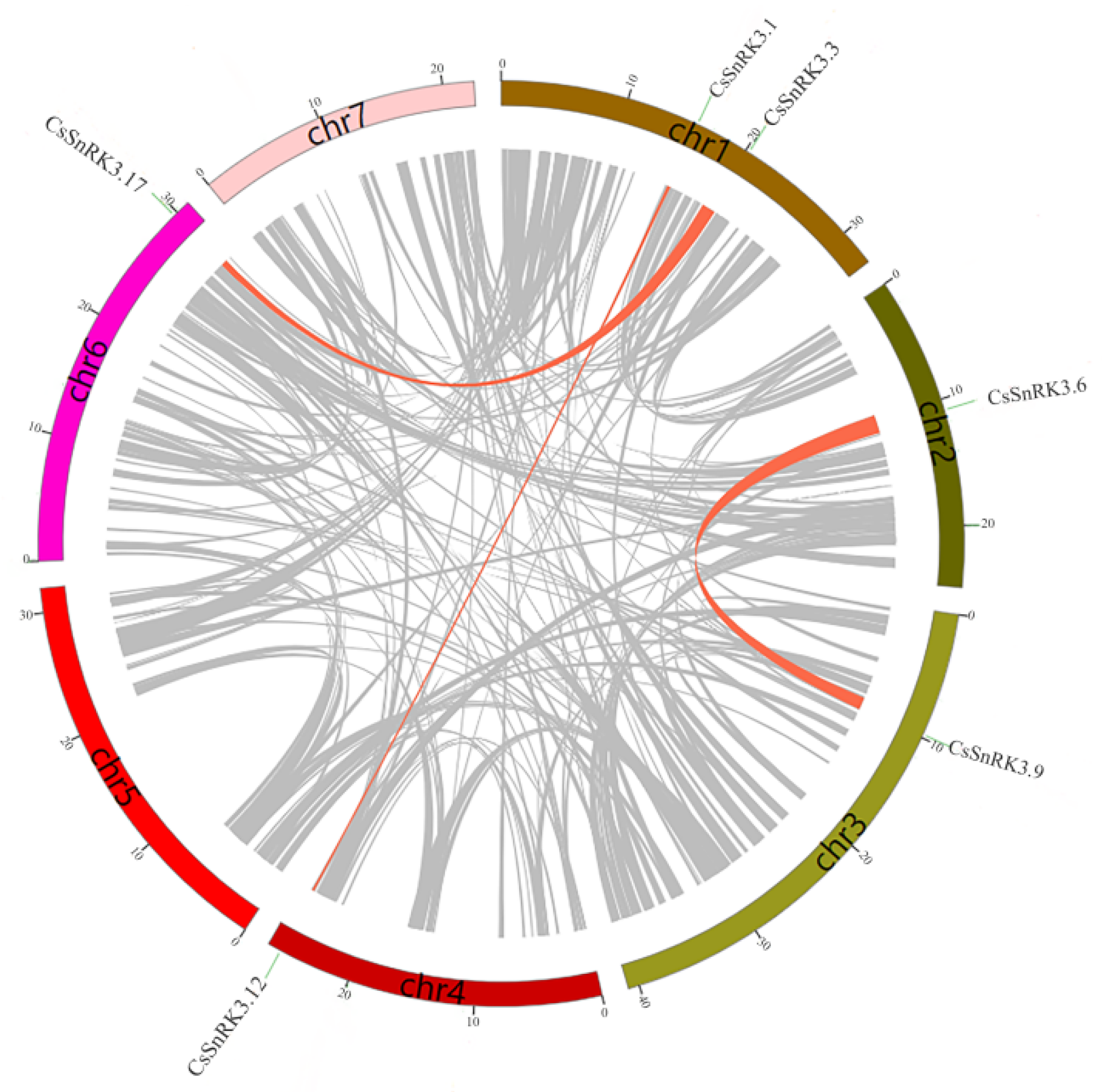
3.4. Chromosomal Location and Gene Duplication Analyses
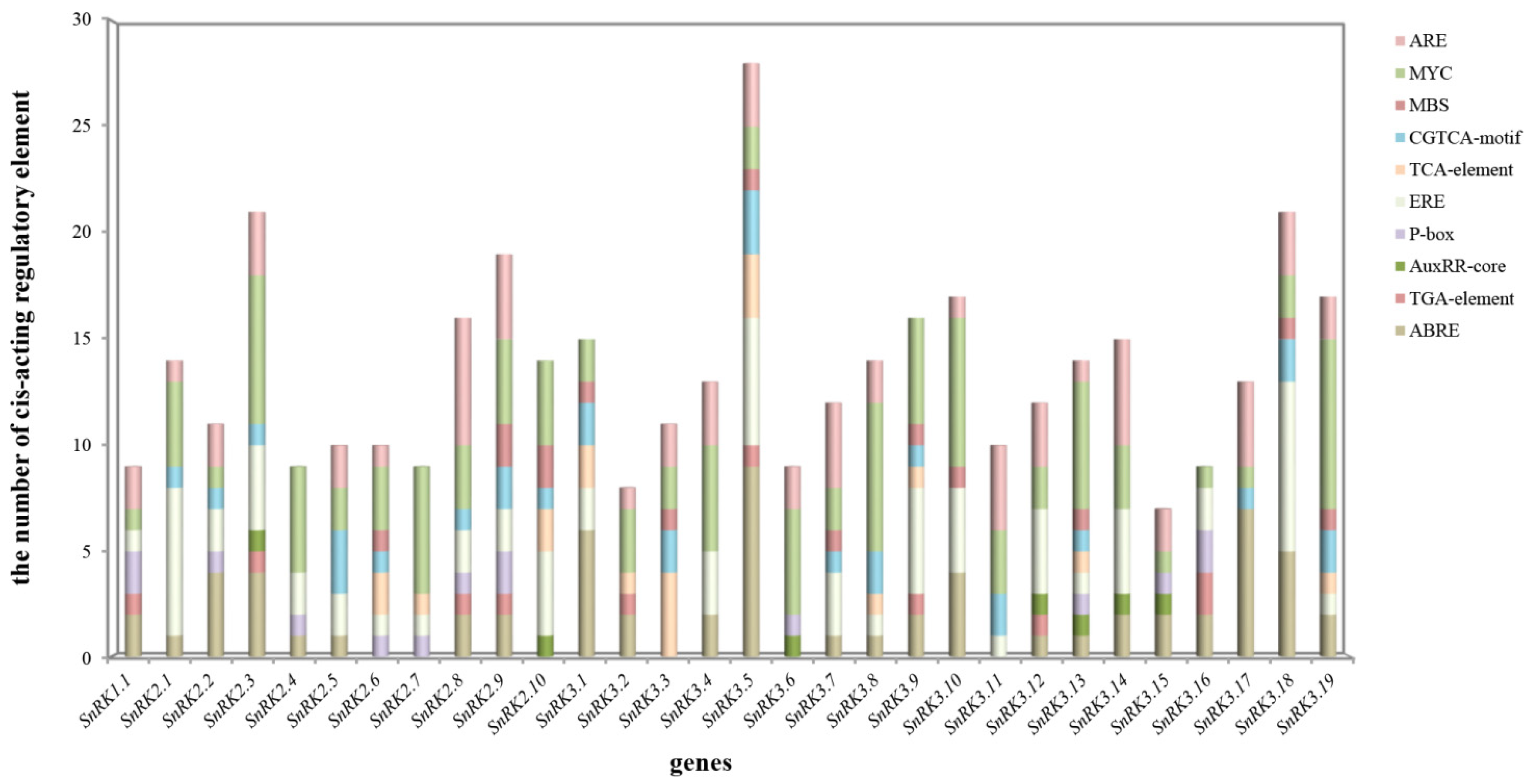
3.5. Cis-Acting Regulatory Elements Analysis in the Promoter Region of CsSnRK Genes
3.6. Expression Profiles Analysis of CsSnRK2 Genes
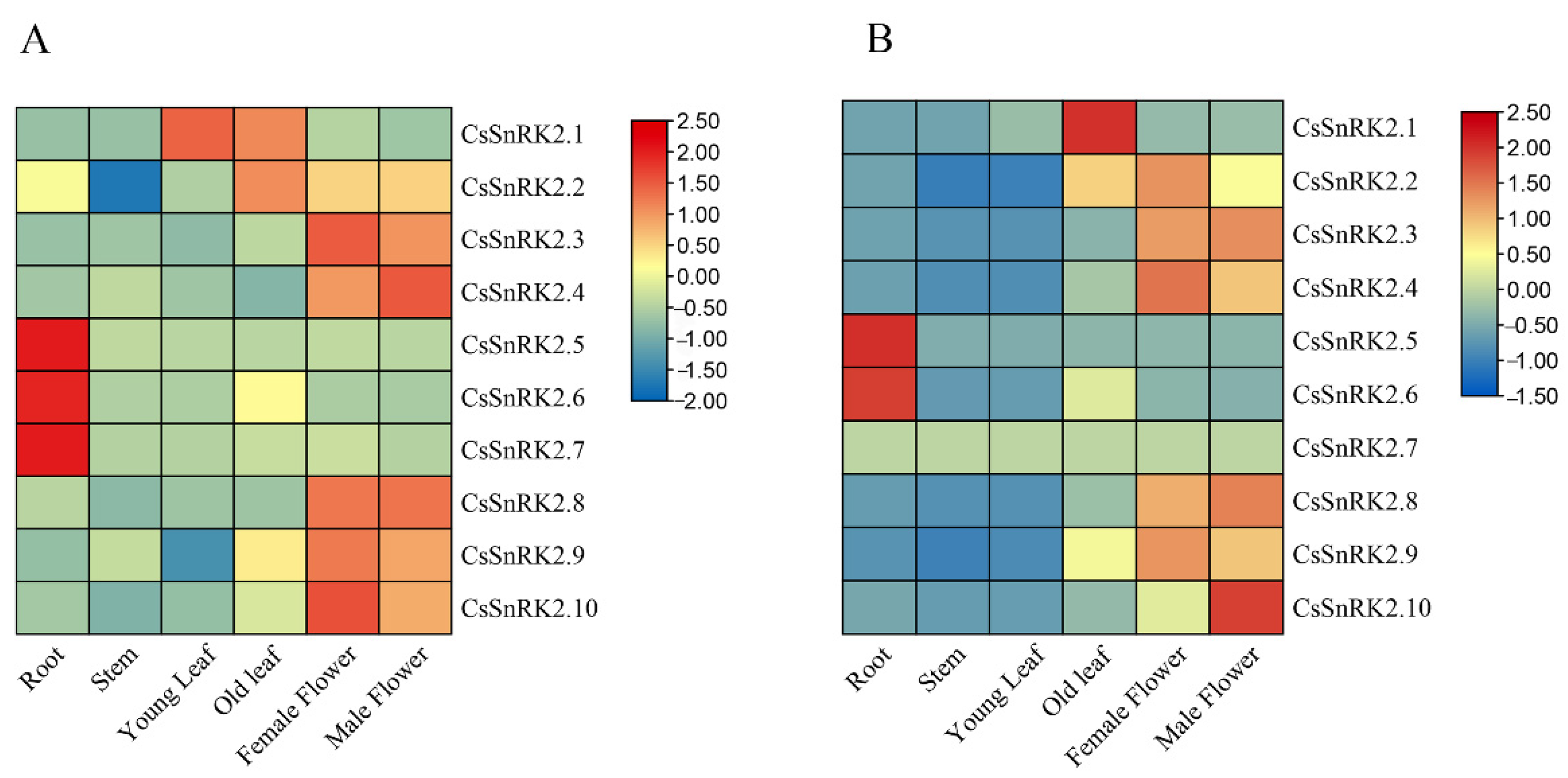
3.6.1. Expression Patterns of CsSnRK2 Gene Family in Different Tissues
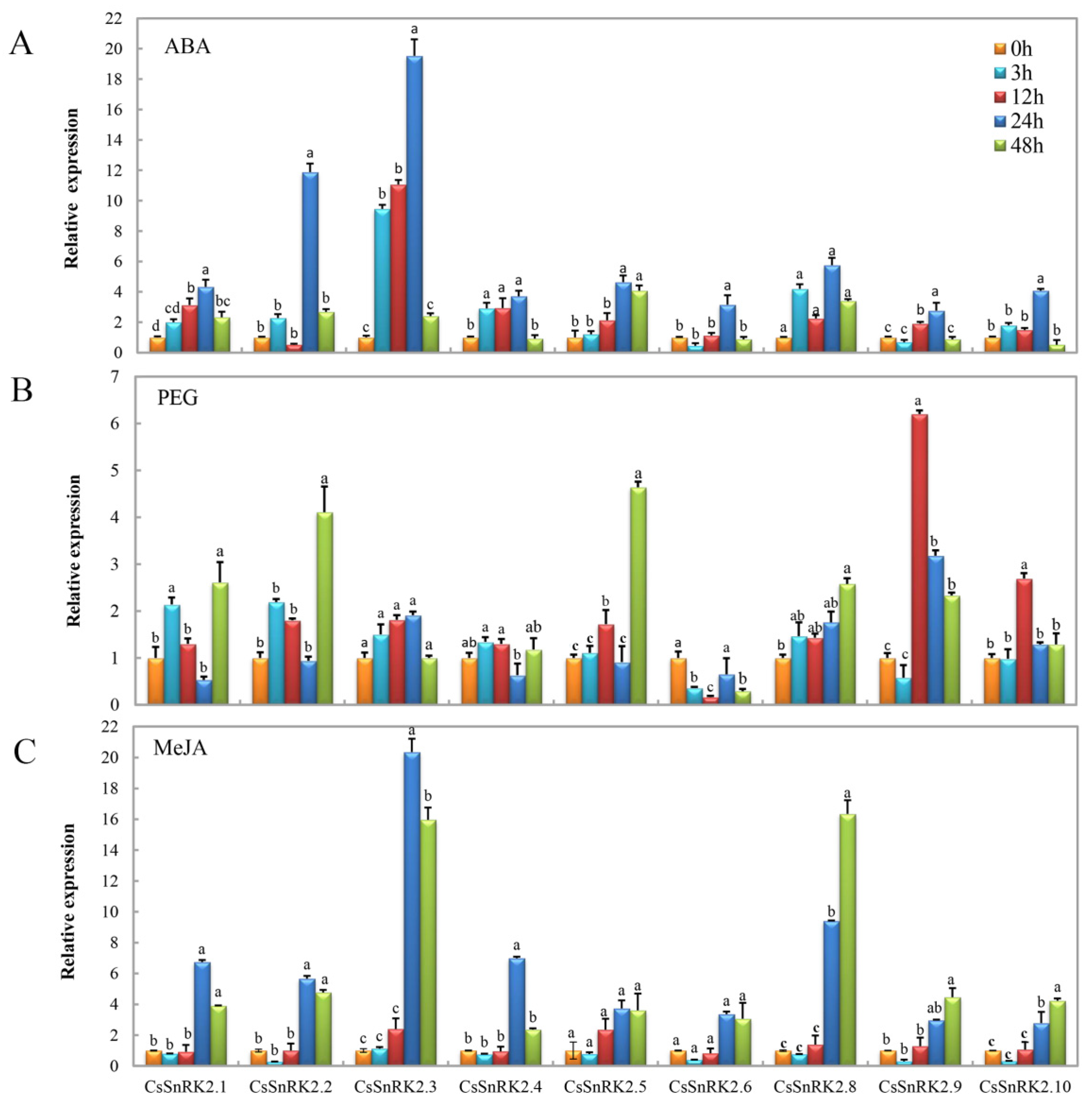
3.6.2. Expression Profiles of CsSnRK2 Gene Family under Different Abiotic Stresses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Demidchik, V.; Maathuis, F.; Voitsekhovskaja, O. Unravelling the plant signalling machinery: An update on the cellular and genetic basis of plant signal transduction. Funct. Plant Boil. 2018, 45, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bohnert, H.J.; Gong, Q.Q.; Li, P.H.; Ma, S.S. Unraveling abiotic stress tolerance mechanisms-getting genomics going. Curr. Opin. Plant Biol. 2006, 9, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hu, W.; Sun, J.; Liang, X.; Yang, X.; Wei, S.; He, G. Genome-wide analysis of SnRK gene family in Brachypodium distachyon and functional characterization of BdSnRK2. 9. Plant Sci. 2015, 237, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Baba, A.I.; Rigó, G.; Ayaydin, F.; Rehman, A.U.; Andrási, N.; Zsigmond, L.; Cséplő, Á. Functional analysis of the Arabidopsis thaliana CDPK-related kinase family: AtCRK1 regulates responses to continuous light. Int. J. Mol. Sci. 2018, 19, 1282. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Zhang, S. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 2015, 20, 56–64. [Google Scholar] [CrossRef]
- Colina, F.; Amaral, J.; Carbó, M.; Pinto, G.; Soares, A.; Cañal, M.J.; Valledor, L. Genome-wide identification and characterization of CKIN/SnRK gene family in Chlamydomonas reinhardtii. Sci. Rep. 2019, 9, 350. [Google Scholar] [CrossRef]
- Wang, Y.; Yan, H.; Qiu, Z.; Hu, B.; Zeng, B.; Zhong, C.; Fan, C. Comprehensive analysis of SnRK gene family and their responses to salt stress in Eucalyptus grandis. Int. J. Mol. Sci. 2019, 20, 2786. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Ye, Y.; Jiang, L.; Lin, Y.; Gu, X.; Chen, Q.; Sun, B.; Zhang, Y.; Luo, Y.; Wang, Y.; et al. Genome-wide characterization of Snf1-Related Protein Kinases (SnRKs) and expression analysis of SnRK1. 1 in strawberry. Genes 2020, 11, 427. [Google Scholar] [CrossRef]
- Hrabak, E.M.; Chan, C.W.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Harmon, A.C. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef] [Green Version]
- Coello, P.; Hey, S.J.; Halford, N.G. The sucrose non-fermenting-1-related (SnRK) family of protein kinases: Potential for manipulation to improve stress tolerance and increase yield. J. Exp. Bot. 2011, 62, 883–893. [Google Scholar] [CrossRef] [Green Version]
- Halford, N.G.; Hey, S.J. Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem. J. 2009, 419, 247–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hey, S.J.; Byrne, E.; Halford, N.G. The interface between metabolic and stress signalling. Ann. Bot. 2009, 105, 197–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alderson, A.; Sabelli, P.A.; Dickinson, J.R.; Cole, D.; Richardson, M.; Kreis, M.; Halford, N.G. Complementation of snf1, a mutation affecting global regulation of carbon metabolism in yeast, by a plant protein kinase cDNA. Proc. Natl. Acad. Sci. USA 1991, 88, 8602–8605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jossier, M.; Bouly, J.P.; Meimoun, P.; Arjmand, A.; Lessard, P.; Hawley, S.; Thomas, M. SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J. 2009, 59, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Radchuk, R.; Radchuk, V.; Weschke, W.; Borisjuk, L.; Weber, H. Repressing the expression of the SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiol. 2006, 140, 263–278. [Google Scholar] [CrossRef] [Green Version]
- Baena-González, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef]
- Saha, J.; Chatterjee, C.; Sengupta, A.; Gupta, K.; Gupta, B. Genome-wide analysis and evolutionary study of sucrose non-fermenting 1-related protein kinase 2 (SnRK2) gene family members in Arabidopsis and Oryza. Comput. Boil. Chem. 2014, 49, 59–70. [Google Scholar] [CrossRef]
- Kulik, A.; Wawer, I.; Krzywińska, E.; Bucholc, M.; Dobrowolska, G. SnRK2 protein kinases-key regulators of plant response to abiotic stresses. Omics 2011, 15, 859–872. [Google Scholar] [CrossRef]
- Boudsocq, M. Barbier-Brygoo, H., Lauriere, C. Identification of nine sucrose non-fermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 41758–41766. [Google Scholar] [CrossRef] [Green Version]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 1717–1722. [Google Scholar] [CrossRef] [Green Version]
- Albrecht, V.; Ritz, O.; Linder, S.; Harter, K.; Kudla, J. The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 2001, 20, 1051–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, Y.; Huang, Y.; Xiong, L. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 2007, 144, 1416–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Ren, F.; Zhou, L.; Wang, Q.Q.; Zhong, H.; Li, X.B. The Brassica napus calcineurin B-Like 1/CBL-interacting protein kinase 6 (CBL1/CIPK6) component is involved in the plant response to abiotic stress and ABA signalling. J. Exp. Bot. 2012, 63, 6211–6222. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, X.L.; Zhao, Y.Z. Selection for cold-resistance cucumber varieties in solar greenhouse in Beijing and Tianjin. Tianjin Agric. Sci. 2019, 25, 26–28+33. [Google Scholar]
- Fujii, H.; Zhu, J.K. Osmotic stress signaling via protein kinases. Cell. Mol. Life Sci. 2012, 69, 3165–3173. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Qin, Y.; Zou, Y.; Ma, F. Genome-wide identification and expression profiling of the SnRK2 gene family in Malus prunifolia. Gene 2014, 552, 87–97. [Google Scholar] [CrossRef]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Lopez, R. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Paterson, A.H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Wei, G.; Tian, P.; Zhang, F.; Qin, H.; Miao, H.; Chen, Q.; Hu, Z.; Cao, L.; Wang, M.; Gu, X.; et al. Integrative analyses of nontargeted volatile profiling and transcriptome data provide molecular insight into VOC diversity in cucumber plants (Cucumis sativus). Plant Physiol. 2016, 172, 603–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, R.; Luo, Y.; Yun, F.; Wu, X.; Wang, P.; Liao, W. Genome-wide identification, expression profile, and alternative splicing analysis of CAMTA family genes in Cucumber (Cucumis sativus L.). Agronomy 2021, 11, 1827. [Google Scholar] [CrossRef]
- He, M.W.; Wang, Y.; Wu, J.Q.; Shu, S.; Sun, J.; Guo, S.R. Isolation and characterization of S-Adenosylmethionine synthase gene from cucumber and responsive to abiotic stress. Plant Physiol. Biochem. 2019, 141, 431–445. [Google Scholar] [CrossRef] [PubMed]
- Noman, M.; Jameel, A.; Qiang, W.D.; Ahmad, N.; Liu, W.C.; Wang, F.W.; Li, H.Y. Overexpression of GmCAMTA12 enhanced drought tolerance in Arabidopsisand soybean. Int. J. Mol. Sci. 2019, 20, 4849. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Hu, L.; Wu, H.; Jiang, L.; Liu, S. Genome-wide identification and transcriptional expression analysis of cucumber superoxide dismutase (SOD) family in response to various abiotic stresses. Int. J. Genom. 2017, 2017, 7243973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, L.M.; Hua, W.P.; Cao, X.Y.; Yan, J.A.; Chen, C.; Wang, Z.Z. Genome-wide identification and expression analysis of the superoxide dismutase (SOD) gene family in Salvia miltiorrhiza. Gene 2020, 742, 144603. [Google Scholar] [CrossRef]
- Emanuelle, S.; Doblin, M.S.; Stapleton, D.I.; Bacic, A.; Gooley, P.R. Molecular insights into the Enigmatic Metabolic Regulator, SnRK1. Trends Plant Sci. 2016, 21, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Lyzenga, W.J.; Liu, H.; Schofield, A.; Muise-Hennessey, A.; Stone, S.L. Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin-proteasome system. J. Exp. Bot. 2013, 64, 2779–2791. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Zhou, L.; Jiang, P.; Lu, R.; Halford, N.G.; Liu, C. Genome-wide identification of sucrose nonfermenting-1-related protein kinase (SnRK) genes in barley and RNA-seq analyses of their expression in response to abscisic acid treatment. BMC Genom. 2021, 22, 300. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Lin, J.; Li, H.; Li, X.; Yang, Q.; Cheng, Z.M.; Chang, Y. Characterization of CIPK family in Asian pear (Pyrus bretschneideri Rehd) and co-expression analysis related to salt and osmotic stress responses. Front. Plant Sci. 2016, 7, 1361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Ma, H.; DePamphilis, C.W. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef]
- Hadiarto, T.; Tran, L.S.P. Progress studies of drought-responsive genes in rice. Plant Cell Rep. 2011, 30, 297–310. [Google Scholar] [CrossRef]
- Xue, T.; Wang, D.; Zhang, S.; Ehlting, J.; Ni, F.; Jakab, S.; Zheng, C.; Zhong, Y. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genom. 2008, 9, 550. [Google Scholar] [CrossRef] [Green Version]
- Fujii, H.; Zhu, J.K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Xue, L.; Batelli, G.; Lee, S.; Hou, Y.J.; van Oosten, M.J.; Zhang, H.; Tao, W.A.; Zhu, J.K. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 2013, 110, 11205–11210. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Ge, X.; Yang, Z.; Zhang, C.; Zhao, G.; Chen, E.; Liu, J.; Zhang, X.; Li, F. Genome-wide identification and characterization of SnRK2 gene family in cotton (Gossypium hirsutum L.). BMC Genet. 2017, 18, 54. [Google Scholar] [CrossRef]
- Wu, Z.; Cheng, J.; Hu, F.; Qin, C.; Xu, X.; Hu, K. The SnRK2 family in pepper (Capsicum annuum L.): Genome-wide identification and expression analyses during fruit development and under abiotic stress. Genes Genom. 2020, 42, 1117–1130. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K.A.Z.U.K.O. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, L.; Dong, B.; Song, Z.; Meng, D.; Fu, Y. Genome-wide identification and characterization of CIPK family and analysis responses to various stresses in apple (Malus domestica). Int. J. Mol. Sci. 2018, 19, 2131. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Gene Name | Gene ID | Amino Acid (aa) | Exons | MW (kDa) | pI | Subcellular Localization |
|---|---|---|---|---|---|---|
| CsSnRK1.1 | CsaV3_6G006250.1 | 515 | 10 | 58.81 | 8.59 | cytoplasm |
| CsSnRK2.1 | CsaV3_2G014200.1 | 361 | 9 | 41.24 | 4.44 | nucleus |
| CsSnRK2.2 | CsaV3_2G016930.1 | 363 | 10 | 41.07 | 4.45 | nucleus |
| CsSnRK2.3 | CsaV3_3G002220.1 | 340 | 9 | 38.38 | 5.95 | cytoplasm |
| CsSnRK2.4 | CsaV3_4G030050.1 | 355 | 9 | 40.90 | 6.38 | nucleus |
| CsSnRK2.5 | CsaV3_4G030080.1 | 673 | 18 | 77.01 | 7.02 | cytoplasm |
| CsSnRK2.6 | CsaV3_4G030100.1 | 372 | 12 | 42.44 | 8.78 | nucleus |
| CsSnRK2.7 | CsaV3_4G030120.1 | 486 | 13 | 55.73 | 5.53 | nucleus |
| CsSnRK2.8 | CsaV3_4G030260.1 | 355 | 10 | 40.94 | 5.83 | nucleus |
| CsSnRK2.9 | CsaV3_6G045260.1 | 344 | 9 | 38.95 | 5.07 | cytoplasm |
| CsSnRK2.10 | CsaV3_7G008840.1 | 365 | 9 | 41.22 | 4.52 | cytoplasm |
| CsSnRK3.1 | CsaV3_1G028820.1 | 430 | 1 | 48.51 | 9.44 | cytoplasm |
| CsSnRK3.2 | CsaV3_1G030280.1 | 356 | 15 | 40.56 | 5.74 | chloroplast |
| CsSnRK3.3 | CsaV3_1G033260.1 | 463 | 2 | 52.40 | 9.63 | chloroplast |
| CsSnRK3.4 | CsaV3_1G036820.1 | 464 | 16 | 52.05 | 9.55 | cytoplasm |
| CsSnRK3.5 | CsaV3_2G003670.1 | 433 | 2 | 48.69 | 9.52 | cytoplasm |
| CsSnRK3.6 | CsaV3_2G013250.1 | 430 | 1 | 47.88 | 9.45 | chloroplast |
| CsSnRK3.7 | CsaV3_2G030480.1 | 446 | 14 | 50.85 | 7.79 | cytoplasm |
| CsSnRK3.8 | CsaV3_3G007670.1 | 441 | 14 | 50.08 | 8.42 | cytoplasm |
| CsSnRK3.9 | CsaV3_3G012720.1 | 433 | 1 | 48.60 | 9.64 | chloroplast |
| CsSnRK3.10 | CsaV3_3G015590.1 | 442 | 12 | 49.58 | 6.05 | cytoplasm |
| CsSnRK3.11 | CsaV3_4G017300.1 | 639 | 20 | 72.87 | 7.7 | cytoplasm |
| CsSnRK3.12 | CsaV3_4G036630.1 | 443 | 1 | 50.29 | 8.61 | cytoplasm |
| CsSnRK3.13 | CsaV3_6G000230.1 | 436 | 12 | 49.38 | 6.45 | cytoplasm |
| CsSnRK3.14 | CsaV3_6G000240.1 | 431 | 12 | 49.03 | 8.34 | chloroplast |
| CsSnRK3.15 | CsaV3_6G019060.1 | 408 | 4 | 46.14 | 8.41 | nucleus |
| CsSnRK3.16 | CsaV3_6G019070.1 | 424 | 1 | 48.14 | 8.86 | cytoplasm |
| CsSnRK3.17 | CsaV3_6G050960.1 | 467 | 1 | 53.01 | 8.06 | cytoplasm |
| CsSnRK3.18 | CsaV3_6G051970.1 | 454 | 15 | 51.17 | 9.22 | chloroplast |
| CsSnRK3.19 | CsaV3_UNG100750.1 | 432 | 1 | 48.94 | 6.63 | cytoplasm |
| Duplicated Genes | Ka | Ks | Ka/Ks |
|---|---|---|---|
| CsSnRK3.1/CsSnRK3.12 | 0.313425 | 4.27516 | 0.073313 |
| CsSnRK3.3/CsSnRK3.17 | 0.322712 | 4.26853 | 0.075603 |
| CsSnRK3.6/CsSnRK3.9 | 0.23843 | 1.75127 | 0.136146 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Y.; Niu, Y.; Gao, R.; Wang, C.; Liao, W. Genome-Wide Identification and Expression Analysis of SnRK Gene Family under Abiotic Stress in Cucumber (Cucumis sativus L.). Agronomy 2022, 12, 1550. https://doi.org/10.3390/agronomy12071550
Luo Y, Niu Y, Gao R, Wang C, Liao W. Genome-Wide Identification and Expression Analysis of SnRK Gene Family under Abiotic Stress in Cucumber (Cucumis sativus L.). Agronomy. 2022; 12(7):1550. https://doi.org/10.3390/agronomy12071550
Chicago/Turabian StyleLuo, Yanyan, Yuan Niu, Rong Gao, Chunlei Wang, and Weibiao Liao. 2022. "Genome-Wide Identification and Expression Analysis of SnRK Gene Family under Abiotic Stress in Cucumber (Cucumis sativus L.)" Agronomy 12, no. 7: 1550. https://doi.org/10.3390/agronomy12071550
APA StyleLuo, Y., Niu, Y., Gao, R., Wang, C., & Liao, W. (2022). Genome-Wide Identification and Expression Analysis of SnRK Gene Family under Abiotic Stress in Cucumber (Cucumis sativus L.). Agronomy, 12(7), 1550. https://doi.org/10.3390/agronomy12071550







