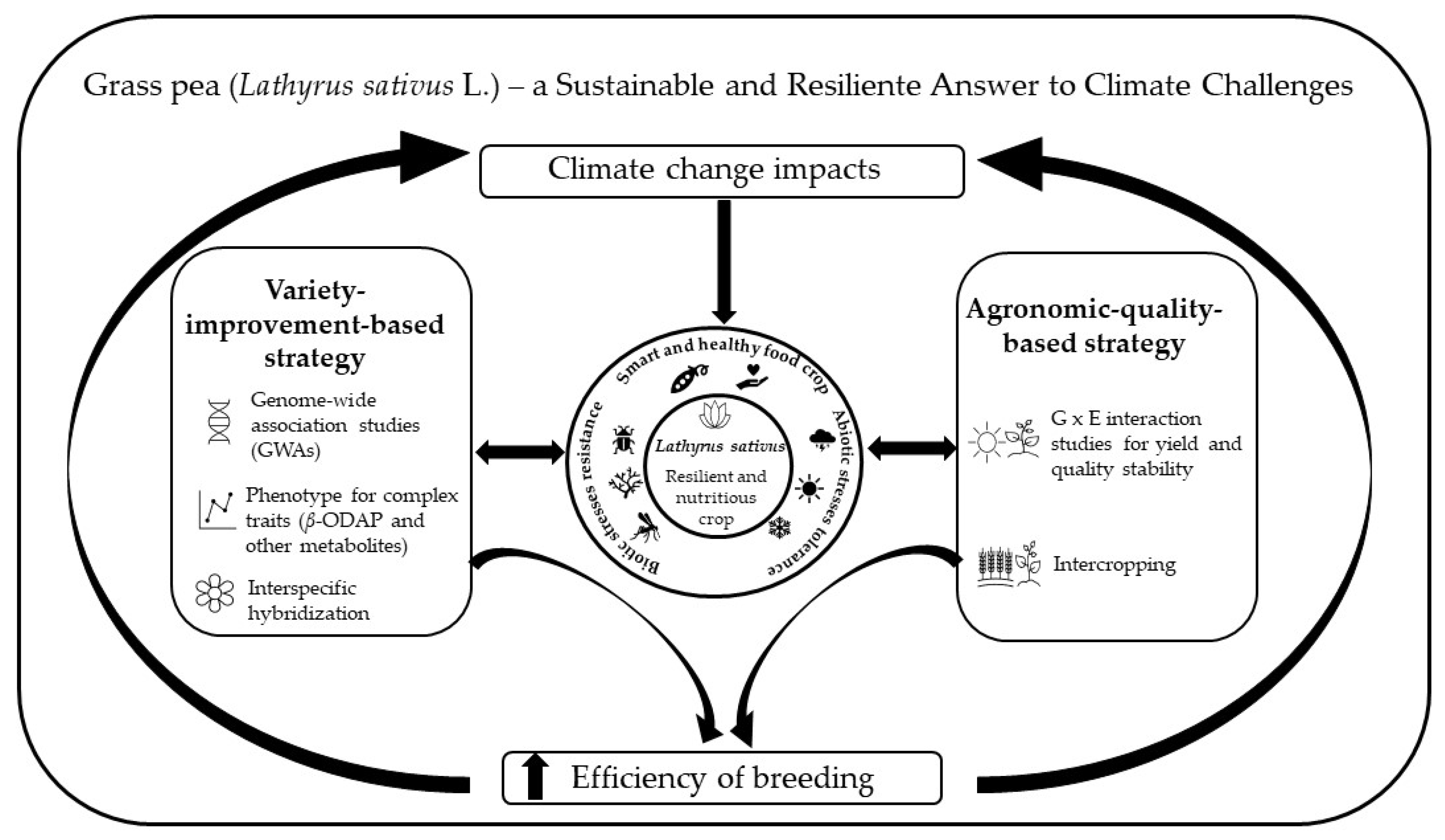
Grass Pea (Lathyrus sativus L.)—A Sustainable and Resilient Answer to Climate Challenges
Abstract
:1. Introduction
2. New Challenges and Opportunities Due to the Impacts of Climate Change in Grass Pea Production
3. Grass Pea as a Source of Important Traits to Tackle Climate Change
3.1. Grass Pea Abiotic Stresses Tolerance
3.2. Grass Pea Biotic Stresses Resistance
4. Yield vs. Quality Stability in Grass Pea
4.1. Grass Pea, a Smart and Healthy Food Crop
4.2. The Influence of the Environment on Quality-Related Traits with a Putative Rule in Grass Pea Resilience
5. Breeding and Agroecological Transition on Grass Pea Production Systems: Strategies toward Improved Resilience
5.1. Approaches for Diversity Creation and Increase Genetic Gains
5.1.1. Diversity Creation
5.1.2. Increase Genetic Gain
5.2. Transition to More Sustainable and Resilient Production Systems
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pielke, R. What is climate change? Incompatibility between the definitions used by science and policy organizations is an obstacle to effective action. Issues Sci. Technol. 2004, 20, 1–4. [Google Scholar]
- Ortiz-Bobea, A.; Ault, T.R.; Carrillo, C.M.; Chambers, R.G.; Lobell, D.B. Anthropogenic climate change has slowed global agricultural productivity growth. Nat. Clim. Chang. 2021, 11, 306–312. [Google Scholar] [CrossRef]
- Langridge, P.; Braun, H.; Hulke, B.; Ober, E.; Prasanna, B.M. Breeding crops for climate resilience. Theor. Appl. Genet. 2021, 134, 1607–1611. [Google Scholar] [CrossRef] [PubMed]
- Rubiales, D.; Araújo, S.S.; Vaz Patto, M.C.; Rispail, N.; Valdés-López, O. Editorial: Advances in legume research. Front. Plant Sci. 2018, 9, 501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duc, G.; Agrama, H.; Bao, S.; Berger, J.; Bourion, V.; De Ron, A.M.; Gowda, C.L.L.; Mikic, A.; Millot, D.; Singh, K.B.; et al. Breeding annual grain legumes for sustainable agriculture: New methods to approach complex traits and target new cultivar ideotypes. CRC Crit. Rev. Plant Sci. 2015, 34, 381–411. [Google Scholar] [CrossRef] [Green Version]
- Rubiales, D.; Annicchiarico, P.; Vaz Patto, M.C.; Julier, B. Legume breeding for the agroecological transition of global agri-food systems: A European perspective. Front. Plant Sci. 2021, 2, 782574. [Google Scholar] [CrossRef] [PubMed]
- Arnoldi, A.; Zanoni, C.; Lammi, C.; Boschin, G. The role of grain legumes in the prevention of hypercholesterolemia and hypertension. CRC Crit. Rev. Plant Sci. 2015, 34, 144–168. [Google Scholar] [CrossRef]
- Vaz Patto, M.C.; Mecha, E.; Pereira, A.B.; Leitão, S.T.; Alves, M.L.; Bronze, M.R. Deciphering grain legumes quality riddle: The genomics of bioactive compounds. In Breeding Grasses and Protein Crops in the Era of Genomics; Brazauskas, G., Statkevičiūtė, G., Jonavičienė, K., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 1, pp. 118–120. [Google Scholar] [CrossRef]
- Vaz Patto, M.C. Grain legume protein quality: A hot subject. Arbor 2016, 192, a314. [Google Scholar] [CrossRef] [Green Version]
- Almeida, N.F.; Leitão, S.T.; Caminero, C.; Torres, A.M.; Rubiales, D.; Vaz Patto, M.C. Transferability of molecular markers from major legumes to Lathyrus spp. for their application in mapping and diversity studies. Mol. Biol. Rep. 2014, 41, 269–283. [Google Scholar] [CrossRef] [Green Version]
- Vaz Patto, M.C.; Skiba, B.; Pang, E.C.K.; Ochatt, S.J.; Lambein, F.; Rubiales, D. Lathyrus improvement for resistance against biotic and abiotic stresses: From classical breeding to marker assisted selection. Euphytica 2006, 147, 133–147. [Google Scholar] [CrossRef]
- Campbell, C.G.; Mehra, R.B.; Agrawal, S.K.; Chen, Y.Z.; Abd El Moneim, A.M.; Khawaja, H.I.T.; Yadov, C.R.; Tay, J.U.; Araya, W.A. Current status and future strategy in breeding grasspea (Lathyrus sativus). Euphytica 1993, 1, 167–175. [Google Scholar] [CrossRef]
- Hanbury, C.; White, C.; Mullan, B.; Siddique, K.H. A review of the potential of Lathyrus sativus L. and L. cicera L. grain for use as animal feed. Anim. Feed Sci. Technol. 2000, 87, 1–27. [Google Scholar] [CrossRef]
- Sarkar, A.; Emmrich, P.M.F.; Sarker, A.; Zong, X.; Martin, C.; Wang, T.L. Grass pea: Remodelling an ancient insurance crop for climate resilience. In Genomic Designing of Climate-Smart Pulse Crops; Kole, C., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 425–469. [Google Scholar] [CrossRef]
- Dempewolf, H.; Eastwood, R.J.; Guarino, L.; Khoury, C.K.; Müller, J.V.; Toll, J. Adapting agriculture to climate change: A global initiative to collect, conserve, and use crop wild relatives. Agroecol. Sustain. Food Syst. 2014, 38, 369–377. [Google Scholar] [CrossRef] [Green Version]
- Lambein, F.; Travella, S.; Kuo, Y.H.; Van Montagu, M.; Heijde, M. Grass pea (Lathyrus sativus L.): Orphan crop, nutraceutical or just plain food? Planta 2019, 250, 821–838. [Google Scholar] [CrossRef] [Green Version]
- Rathi, D.; Chakraborty, S.; Chakraborty, N. Grass pea, a critical recruit among neglected and underutilized legumes, for tapping genomic resources. Curr. Plant Biol. 2021, 26, 100200. [Google Scholar] [CrossRef]
- Das, A.; Parihar, A.K.; Barpete, S.; Kumar, S.; Gupta, S. Current Perspectives on reducing the β-ODAP content and improving potential agronomic traits in grass pea (Lathyrus sativus L.). Front. Plant Sci. 2021, 12, 703275. [Google Scholar] [CrossRef]
- Arora, N.K. Impact of climate change on agriculture production and its sustainable solutions. Environ. Sustain. 2019, 2, 95–96. [Google Scholar] [CrossRef] [Green Version]
- Burdonid, J.J.; Zhanid, J. Climate change and disease in plant communities. PLoS Biol. 2020, 18, e3000949. [Google Scholar] [CrossRef]
- Anderson, R.; Bayer, P.E.; Edwards, D. Climate change and the need for agricultural adaptation. Curr. Opin. Plant Biol. 2020, 56, 197–202. [Google Scholar] [CrossRef]
- Ray, D.K.; Gerber, J.S.; Macdonald, G.K.; West, P.C. Climate variation explains a third of global crop yield variability. Nat. Commun. 2015, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Johansen, C.; Ali, M.; Gowda, C.L.L.; Ramakrishna, A.; Nigam, S.N.; Chauhan, Y.S. Regional opportunities for warm season grain legumes in the Indo-Gangetic Plain. In Legumes in Rice and Wheat Cropping Systems of the Indo-Gangetic Plain-Constraints and Opportunities; Johansen, C., Duxbury, J.M., Virmani, S.M., Gowda, C.L.L., Pande, S., Joshi, P.K., Eds.; International Crops Research Institute for the Semi-Arid Tropics: Patancheru, India; Cornell University: Ithaca, NY, USA, 2000; pp. 185–199. [Google Scholar]
- Chattopadhyay, C.; Birah, A.; Jalali, B.L. Climate change: Impact on biotic stresses afflicting crop plants. In Natural Resource Management: Ecological Perspectives; Peshin, R., Dhawan, A.K., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 133–146. [Google Scholar] [CrossRef]
- Kislev, M.E. Origins of the cultivation of Lathyrus sativus and L. cicera (Fabaceae). Econ. Bot. 1989, 43, 262–270. [Google Scholar] [CrossRef]
- Dixit, G.P.; Parihar, A.K.; Bohra, A.; Singh, N.P. Achievements and prospects of grass pea (Lathyrus sativus L.) improvement for sustainable food production. Crop J. 2016, 4, 407–416. [Google Scholar] [CrossRef] [Green Version]
- Campbell, C.G. Grass Pea, Lathyrus sativus L.; International Plant Genetics Resources Institute: Rome, Italy, 1997; Volume 18. [Google Scholar]
- Mikić, A.; Mihailović, V.; Ćupina, B.; Ethurić, B.; Krstić, D.; Vasić, M.; Vasiljević, S.; Karagić, D.; EthorEthević, V. Towards the re-introduction of grass pea (Lathyrus sativus) in the West Balkan Countries: The case of Serbia and Srpska (Bosnia and Herzegovina). Food Chem. Toxicol. 2011, 49, 650–654. [Google Scholar] [CrossRef] [PubMed]
- Polignano, G.B.; Bisignano, V.; Tomaselli, V.; Uggenti, P.; Alba, V.; Gatta, C. Della. Genotype × Environment interaction in grass pea (Lathyrus sativus L.) lines. Int. J. Agron. 2009, 2009, 898396. [Google Scholar] [CrossRef] [Green Version]
- Rubiales, D.; Emeran, A.A.; Flores, F. Adaptation of grass pea (Lathyrus sativus) to Mediterranean environments. Agronomy 2020, 10, 1295. [Google Scholar] [CrossRef]
- Kumar, S.; Gupta, P.; Barpete, S.; Sarker, A.; Amri, A.; Mathur, P.N.; Baum, M. Grass pea. In Genetic and Genomic Resources of Grain Legumes Improvement; Singh, M., Upadhyaya, H.D., Bisht, I.S., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2013; pp. 269–292. [Google Scholar] [CrossRef]
- Rizvi, A.H.; Sarker, A.; Dogra, A. Enhancing grasspea (Lathyrus sativus L.) production in problematic soils of South Asia for nutritional security. Indian J. Genet. Plant Breed. 2016, 76, 583–592. [Google Scholar] [CrossRef]
- Kumar, S.; Bejiga, G.; Ahmed, S.; Nakkoul, H.; Sarker, A. Genetic improvement of grass pea for low neurotoxin (β-ODAP) content. Food Chem. Toxicol. 2011, 49, 589–600. [Google Scholar] [CrossRef]
- Hopkin, M. Climate change: World round-up. Nat. News 2005, 16. [Google Scholar] [CrossRef]
- Mousavi-Derazmahalleh, M.; Bayer, P.E.; Hane, J.K.; Valliyodan, B.; Nguyen, H.T.; Nelson, M.N.; Erskine, W.; Varshney, R.K.; Papa, R.; Edwards, D. Adapting legume crops to climate change using genomic approaches. Plant Cell Environ. 2019, 42, 6–19. [Google Scholar] [CrossRef] [Green Version]
- Vinke, K.; Martin, M.A.; Adams, S.; Baarsch, F.; Bondeau, A.; Coumou, D.; Donner, R.V.; Menon, A.; Perrette, M.; Rehfeld, K.; et al. Climatic risks and impacts in South Asia: Extremes of water scarcity and excess. Reg. Environ. Chang. 2017, 17, 1569–1583. [Google Scholar] [CrossRef]
- Wheeler, T.; Von Braun, J. Climate change impacts on global food security. Science 2013, 341, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, L.; Rubiales, D.; Vaz Patto, M.C. Grass pea prospectives at the Mediterranean Basin. Legume Perspect. 2015, 10, 8–9. [Google Scholar]
- Rubiales, D.; Barilli, E.; Flores, F. Broomrape as a major constraint for grass pea (Lathyrus sativus) production in Mediterranean rain-fed environments. Agronomy 2020, 10, 1931. [Google Scholar] [CrossRef]
- Lionello, P.; Malanotte-Rizzoli, P.; Boscolo, R.; Alpert, P.; Artale, V.; Li, L.; Luterbacher, J.; May, W.; Trigo, R.; Tsimplis, M.; et al. The Mediterranean climate: An overview of the main characteristics and issues. In Mediterranean; Lionello, P., Malanotte-Rizzoli, P., Boscolo, R., Eds.; Developments in Earth and Environmental Sciences; Elsevier: Amsterdam, The Netherlands, 2006; Volume 4, pp. 1–26. [Google Scholar] [CrossRef]
- Rotter, R.; Van de Geijin, S.C. Climate change effects on plant growth, crop yield and livestock. Clim. Chang. 1999, 43, 651–681. [Google Scholar] [CrossRef]
- Palutikof, J.P.; Wigley, T.M.L. Developing climate change scenarios for the Mediterranean region. In Climate Change and the Mediterranean; Jeftic, L., Pernetta, J.C., Keckes, S., Eds.; Edward Arnold: London, UK, 1996; Volume 2, pp. 27–56. [Google Scholar]
- Yadav, S.S.; Redden, R.; McNeil, D.L.; Patil, S.A. (Eds.) Climate change and management of cool season grain legume crops. In Climate Change Management of Cool Seaon. Grain Legume Crops; Springer: Dordrecht, The Netherlands, 2010; pp. 1–460. [Google Scholar] [CrossRef]
- Hillocks, R.J.; Maruthi, M.N. Grass pea (Lathyrus sativus): Is there a case for further crop improvement? Euphytica 2012, 186, 647–654. [Google Scholar] [CrossRef]
- Milczak, M.; Pedzinski, M.; Mnichowska, H.; Szwed-Urbas, K.; Rybinski, W. Creative breeding of grass pea (Lathyrus sativus L.) in Poland. Lathyrus Lathyrism Newsl. 2001, 2, 85–88. [Google Scholar]
- Girma, D.; Korbu, L. Genetic improvement of grass pea (Lathyrus sativus) in Ethiopia: An unfulfilled promise. Plant Breed. 2012, 131, 231–236. [Google Scholar] [CrossRef]
- Vaz Patto, M.C.; Rubiales, D. Lathyrus diversity: Available resources with relevance to crop improvement—L. sativus and L. cicera as case studies. Ann. Bot. 2014, 113, 895–908. [Google Scholar] [CrossRef] [Green Version]
- Leakey, A.D.B.; Ferguson, J.N.; Pignon, C.P.; Wu, A.; Jin, Z.; Hammer, G.L.; Lobell, D.B. Water use efficiency as a constraint and target for improving the resilience and productivity of C3 and C4 crops. Annu. Rev. Plant Biol. 2019, 70, 781–808. [Google Scholar] [CrossRef]
- Eggels, S.; Blankenagel, S.; Schön, C.C.; Avramova, V. The carbon isotopic signature of C4 crops and its applicability in breeding for climate resilience. Theor. Appl. Genet. 2021, 134, 1663–1675. [Google Scholar] [CrossRef]
- Bertolino, L.T.; Caine, R.S.; Gray, J.E. Impact of stomatal density and morphology on water-use efficiency in a changing world. Front. Plant Sci. 2019, 10, 225. [Google Scholar] [CrossRef] [Green Version]
- Leport, L.; Turner, N.C.; French, R.J.; Tennant, D.; Thomson, B.D.; Siddique, K.H.M. Water relations, gas exchange and growth of cool-season grain legumes in a Mediterranean-type environment. Eur. J. Agron. 1998, 9, 295–303. [Google Scholar] [CrossRef]
- Thomson, B.D.; Siddique, K.H.M.; Barr, M.D.; Wilson, J.M. Grain legume species in low rainfall Mediterranean-type environments. I. Phenology and seed yield. Field Crops Res. 1997, 54, 173–187. [Google Scholar] [CrossRef]
- Siddique, K.H.M.; Regan, K.L.; Tennant, D.; Thomson, B.D. Water use and water use efficiency of cool season grain legumes in low rainfall Mediterranean-type environments. Eur. J. Agron. 2001, 15, 267–280. [Google Scholar] [CrossRef]
- Gusmão, M.; Siddique, K.H.M.; Flower, K.; Nesbitt, H.; Veneklaas, E.J. Water deficit during the reproductive period of grass pea (Lathyrus sativus L.) reduced grain yield but maintained seed size. J. Agron. Crop Sci. 2012, 198, 430–441. [Google Scholar] [CrossRef]
- Jiang, J.; Su, M.; Chen, Y.; Gao, N.; Jiao, C.; Sun, Z.; Li, F.; Wang, C. Correlation of drought resistance in grass pea (Lathyrus sativus) with reactive oxygen species scavenging and osmotic adjustment. Biologia 2013, 68, 231–240. [Google Scholar] [CrossRef]
- Choudhary, A.K.; Kumar, J.; Gupta, S.; Sultana, R. Breeding for adaptive traits in pulses. In Proceedings of the National Conference on Bringing Self-Sufficiency in Pulses for Eastern India, Kanpur, India, 5–6 August 2016; pp. 36–43. [Google Scholar]
- Lynch, J.P. Steep, Cheap and Deep: An ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 2013, 112, 347–357. [Google Scholar] [CrossRef] [Green Version]
- Mia, W.; Yamauchi, A.; Kono, Y. Root system structure of six food legume species: Inter- and intraspecific variations. Jpn. J. Crop Sci. 1996, 65, 131–140. [Google Scholar] [CrossRef] [Green Version]
- Rathi, D.; Verma, J.K.; Pareek, A.; Chakraborty, S.; Chakraborty, N. Dissection of grasspea (Lathyrus Sativus L.) root exoproteome reveals critical insights and novel rroteins. Plant Sci. 2022, 31, 111161. [Google Scholar] [CrossRef]
- Malek, M.A.; Afzal, A.; Rahman, M.M.; Salahuddin, A.B.M. Lathyrus sativus: A crop for harsh environments. In Linking Research and Marketing Opportunities for Pulses in the 21st Century. Current Plant Science and Biotechnology in Agriculture; Knight, R., Ed.; Springer: Dordrecht, The Netherlands, 2000; Volume 34, pp. 369–373. [Google Scholar] [CrossRef]
- Munns, R.; Gilliham, M. Salinity tolerance of crops—What is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef] [Green Version]
- Corwin, D.L. Climate change impacts on soil salinity in agricultural areas. Eur. J. Soil Sci. 2021, 72, 842–862. [Google Scholar] [CrossRef]
- Piwowarczyk, B.; Tokarz, K.; Kamińska, I. Responses of grass pea seedlings to salinity stress in vitro culture conditions. Plant Cell. Tissue Organ Cult. 2016, 124, 227–240. [Google Scholar] [CrossRef]
- Tokarz, K.M.; Wesołowski, W.; Tokarz, B.; Makowski, W.; Wysocka, A.; Jędrzejczyk, R.J.; Chrabaszcz, K.; Malek, K.; Kostecka-Gugała, A. Stem photosynthesis—A key element of grass pea (Lathyrus sativus L.) acclimatisation to salinity. Int. J. Mol. Sci. 2021, 22, 685. [Google Scholar] [CrossRef] [PubMed]
- Tokarz, B.; Wójtowicz, T.; Makowski, W.; Jedrzejczyk, R.J.; Tokarz, K.M. What is the difference between the response of grass pea (Lathyrus sativus L.) to salinity and drought stress?—A physiological study. Agronomy 2020, 10, 833. [Google Scholar] [CrossRef]
- Sita, K.; Sehgal, A.; Hanumantharao, B.; Nair, R.M.; Vara Prasad, P.V.; Kumar, S.; Gaur, P.M.; Farooq, M.; Siddique, K.H.M.; Varshney, R.K.; et al. Food legumes and rising temperatures: Effects, adaptive functional mechanisms specific to reproductive growth stage and strategies to improve heat tolerance. Front. Plant Sci. 2017, 8, 1658. [Google Scholar] [CrossRef] [Green Version]
- Shunmugam, A.S.K.; Kannan, U.; Jiang, Y.; Daba, K.A.; Gorim, L.Y. Physiology based approaches for breeding of next-generation food legumes. Plants 2018, 7, 72. [Google Scholar] [CrossRef] [Green Version]
- Kumar, G.; Tripathi, R. Influence of heat stress on genome of grass pea (Lathyrus sativus). J. Environ. Biol. 2009, 30, 405–408. [Google Scholar]
- Allard, R.W. History of plants population genetics. Annu. Rev. Genet. 1999, 33, 1–27. [Google Scholar] [CrossRef]
- Sampaio, A.M.; Vitale, S.; Turrà, D.; Di Pietro, A.; Rubiales, D.; van Eeuwijk, F.; Vaz Patto, M.C. A diversity of resistance sources to Fusarium oxysporum f. sp. pisi found within grass pea germplasm. Plant Soil 2021, 463, 19–38. [Google Scholar] [CrossRef]
- Rao, S.C.; Northup, B.K. Grass pea (Lathyrus sativus L.) as a pre-plant nitrogen source for continuous conventionally tilled winter wheat. Crop Sci. 2011, 51, 1325–1333. [Google Scholar] [CrossRef]
- Robertson, L.D.; Abd El-Moneim, A.M. Lathyrus germplasm collection, conservation and utilization for crop improvement. In Proceedings of the a Regional Workshop on Lathyrus genetic Resources in Asia. Indira Gandhi Agricultural University, Raipur, India, 27–29 December 1995; IPGRI Office for South Asia: New Delhi, India, 1996; p. 97. [Google Scholar]
- Vaz Patto, M.C.; Fernández-Aparicio, M.; Moral, A.; Rubiales, D. Characterization of resistance to powdery mildew (Erysiphe pisi) in a germplasm collection of Lathyrus sativus. Plant Breed. 2006, 125, 308–310. [Google Scholar] [CrossRef]
- Martins, D.C.; Santos, C.; Sampaio, A.M.; Rubiales, D.; Vaz Patto, M. Lathyrus sativus resistance against existing and emerging powdery mildews (Erysiphe pisi and Erysiphe trifolii): A case of commonalities or total discrepancy? Pest Manag. Sci. 2022. submitted. [Google Scholar]
- Vaz Patto, M.C.; Rubiales, D. Identification and characterization of partial resistance to rust in a germplasm collection of Lathyrus sativus L. Plant Breed. 2009, 128, 495–500. [Google Scholar] [CrossRef]
- Almeida, N.F.; Leitão, S.T.; Krezdorn, N.; Rotter, B.; Winter, P.; Rubiales, D.; Vaz Patto, M.C. Allelic diversity in the transcriptomes of contrasting rust-infected genotypes of a lasting resource for smart breeding. BMC Plant Biol. 2014, 14, 376. [Google Scholar] [CrossRef] [Green Version]
- Martins, D.C.; Rubiales, D.; Vaz Patto, M.C. Association mapping of Lathyrus sativus L. disease response to Uromyces pisi reveals novel loci underlying partial resistance. Front. Plant Sci. 2022, 13, 842545. [Google Scholar] [CrossRef]
- Barilli, E.; Cobos, M.J.; Rubiales, D. Clarification on host range of Didymella pinodes the causal agent of pea Ascochyta blight. Front. Plant Sci. 2016, 7, 592. [Google Scholar] [CrossRef]
- Infantino, A.; Zaccardelli, M.; Costa, C.H.; Ozkilinc, H.; Habibi, A.; Peever, T. A new disease of grass pea (Lathyrus sativus) caused by Ascochyta lentis var. lathyri. J. Plant. Phatol. 2016, 98, 541–548. [Google Scholar]
- Gurung, A.M.; Pang, E.C.K.; Taylor, P.W.J. Examination of Pisum and Lathyrus species as sources of Ascochyta blight resistance for field pea (Pisum sativum). Australas. Plant Pathol. 2002, 31, 41–45. [Google Scholar] [CrossRef]
- Skiba, B.; Ford, R.; Pang, E.C.K. Construction of a linkage map based on a Lathyrus sativus backcross population and preliminary investigation of QTLs associated with resistance to Ascochyta blight. Theor. Appl. Genet. 2004, 109, 1726–1735. [Google Scholar] [CrossRef]
- Almeida, N.F.; Krezdorn, N.; Rotter, B.; Winter, P.; Rubiales, D.; Vaz Patto, M.C. Lathyrus sativus transcriptome resistance response to Ascochyta lathyri investigated by Deep Super SAGE analysis. Front. Plant Sci. 2015, 6, 178. [Google Scholar] [CrossRef] [Green Version]
- Sampaio, A.M.; Alves, M.L.; Pereira, P.; Valiollahi, E.; Santos, C.; Šatović, Z.; Rubiales, D.; Araújo, S.d.S.; van Eeuwijk, F.; Vaz Patto, M.C. Grass pea natural variation reveals oligogenic resistance to Fusarium oxysporum f. sp. pisi. Plant Genome 2021, 14, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Aparicio, M.; Flores, F.; Rubiales, D. Escape and true resistance to crenate broomrape (Orobanche crenata Forsk.) in grass pea (Lathyrus sativus L.) germplasm. Field Crops Res. 2012, 125, 92–97. [Google Scholar] [CrossRef]
- Pandey, R.L.; Sharma, R.N.; Chitale, M.W. Status of Lathyrus genetic resources in India. In Lathyrus Genetic Resources Network, Proceedings of the a IPGRI-ICARDA-ICAR Regional Group Meeting, New Delhi, India, 8–10 Dezember 1997; Mathur, P.N., Ramanatha Rao, V., Arora, R.K., Eds.; National Bureau of Plant Genetic Resources: New Delhi, India, 1999; pp. 7–14. [Google Scholar]
- Lambein, F. Homeopathy, longevity and Lathyrus sativus toxicity. Lathyrus Latthyrism Newsl. 2000, 1, 4–5. [Google Scholar]
- Lambein, F.; Kuo, Y.H.; Kusama-Eguchi, K.; Ikegami, F. 3-N-oxalyl-L-2,3-diaminopropanoic acid, a multifunctional plant metabolite of toxic reputation. Arkivoc 2007, 2007, 45–52. [Google Scholar] [CrossRef] [Green Version]
- Rao, S.L.N.; Adiga, P.R.; Sarma, P.S. The isolation and characterization of β-N-oxalyl-l-α,β-diaminopropionic acid: A neurotoxin from the seeds of Lathyrus sativus. Biochemistry 1964, 3, 432–436. [Google Scholar] [CrossRef]
- Getahun, H.; Lambein, F.; Vanhoorne, M.; Van Der Stuyft, P. Food-aid cereals to reduce neurolathyrism related to grass-pea preparations during famine. Lancet 2003, 362, 1808–1810. [Google Scholar] [CrossRef]
- Sellami, M.H.; Pulvento, C.; Amarowicz, R.; Lavini, A. Field phenotyping and quality traits of grass pea genotypes in South Italy. J. Sci. Food Agric. 2020. [Google Scholar] [CrossRef]
- Chatterjee, C.; Debnath, M.; Karmakar, N.; Sadhukhan, R. Stability of grass pea (Lathyrus sativus L.) genotypes in different agroclimatic zone in Eastern part of India with special reference to West Bengal. Genet. Resour. Crop Evol. 2019, 66, 1515–1531. [Google Scholar] [CrossRef]
- Vaz Patto, M.C.; Amarowicz, R.; Aryee, A.N.A.; Boye, J.I.; Chung, H.J.; Martín-Cabrejas, M.A.; Domoney, C. Achievements and challenges in improving the nutritional quality of food legumes. CRC Crit. Rev. Plant Sci. 2015, 34, 105–143. [Google Scholar] [CrossRef]
- Fikre, A.; Negwo, T.; Kuo, Y.H.; Lambein, F.; Ahmed, S. Climatic, edaphic and altitudinal factors affecting yield and toxicity of Lathyrus sativus grown at five locations in Ethiopia. Food Chem. Toxicol. 2011, 49, 623–630. [Google Scholar] [CrossRef]
- Jiao, C.J.; Jiang, J.L.; Ke, L.M.; Cheng, W.; Li, F.M.; Li, Z.X.; Wang, C.Y. Factors affecting β-ODAP content in Lathyrus sativus and their possible physiological mechanisms. Food Chem. Toxicol. 2011, 49, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Mecha, E.; Erny, G.L.; Guerreiro, A.C.L.; Feliciano, R.P.; Barbosa, I.; Bento da Silva, A.; Leitão, S.T.; Veloso, M.M.; Rubiales, D.; Rodriguez-Mateos, A.; et al. Metabolomics profile responses to changing environments in a common bean (Phaseolus vulgaris L.) germplasm collection. Food Chem. 2022, 370, 131003. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Moneim, A.M.; Cocks, P.S. Adaptation and yield stability of selected lines of Lathyrus spp. under rainfed conditions in West Asia. Euphytica 1993, 66, 89–97. [Google Scholar] [CrossRef]
- Malosetti, M.; Ribaut, J.M.; van Eeuwijk, F.A. The statistical analysis of multi-environment data: Modelling Genotype-by-Environment interaction and its genetic basis. Front. Physiol. 2013, 4, 44. [Google Scholar] [CrossRef] [Green Version]
- Hanbury, C.D.; Siddique, K.H.M.; Galwey, N.W.; Cocks, P.S. Genotype-Environment interaction for seed yield and ODAP concentration of Lathyrus sativus L. and L. cicera L. in Mediterranean-type environments. Euphytica 1999, 110, 45–60. [Google Scholar] [CrossRef]
- Grela, E.R.; Rybiński, W.; Klebaniuk, R.; Matras, J. Morphological characteristics of some accessions of grass pea (Lathyrus sativus L.) grown in Europe and nutritional traits of their seeds. Genet. Resour. Crop Evol. 2010, 57, 693–701. [Google Scholar] [CrossRef]
- Fikre, A.; Korbu, L.; Kuo, Y.H.; Lambein, F. The contents of the neuro-excitatory amino acid β-ODAP (β-N-oxalyl-l-α,β-diaminopropionic acid), and other free and protein amino acids in the seeds of different genotypes of grass pea (Lathyrus sativus L.). Food Chem. 2008, 110, 422–427. [Google Scholar] [CrossRef]
- Rao, S.L.N. A look at the brighter facets of β-N-oxalyl-l-α,β-diaminopropionic acid, homoarginine and the grass pea. Food Chem. Toxicol. 2011, 49, 620–622. [Google Scholar] [CrossRef]
- Jammulamadaka, N.; Burgula, S.; Medisetty, R.; Ilavazhagan, G.; Rao, S.L.N.; Singh, S.S. β-N-oxalyl-l-α,β-diaminopropionic acid regulates mitogen-activated protein kinase signalling by down-regulation of phosphatidylethanolamine-binding protein 1. J. Neurochem. 2011, 118, 176–186. [Google Scholar] [CrossRef]
- Pastor-Cavada, E.; Juan, R.; Pastor, J.E.; Alaiz, M.; Vioque, J. Chemical composition and nutritional characteristics of the seed oil of wild Lathyrus, Lens and Pisum species from Southern Spain. J. Am. Oil Chem. Soc. 2009, 86, 329–335. [Google Scholar] [CrossRef]
- Aspinwall, M.J.; Loik, M.E.; Resco De Dios, V.; Tjoelker, M.G.; Payton, P.R.; Tissue, D.T. Utilizing intraspecific variation in phenotypic plasticity to bolster agricultural and forest productivity under climate change. Plant Cell Environ. 2015, 38, 1752–1764. [Google Scholar] [CrossRef] [PubMed]
- Pilbeam, D.J. Breeding crops for improved mineral nutrition under climate change conditions. J. Exp. Bot. 2015, 66, 3511–3521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piergiovanni, A.R.; Lupo, F.; Zaccardelli, M. Environmental effect on yield, composition and technological seed traits of some Italian ecotypes of grass pea (Lathyrus sativus L.). J. Sci. Food Agric. 2011, 91, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Moneim, A.M.; van Dorrestein, B.; Baum, M.; Mulugeta, W. Role of ICARDA in improving the nutritional quality and yield potential of grasspea (Lathyrus sativus L.) for subsistence farmers in developing countries. Int. Food Policy Res. Inst. 1999, 2, 9. [Google Scholar]
- Piwowarczyk, B.; Kamińska, I.; Rybiński, W. Influence of PEG generated osmotic stress on shoot regeneration and some biochemical parameters in Lathyrus culture. Czech J. Genet. Plant Breed. 2014, 50, 77–83. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.C.; Xing, G.M.; Li, F.M.; Wang, S.M.; Fan, X.W.; Li, Z.X.; Wang, Y.F. Abscisic acid promotes accumulation of toxin ODAP in relation to free spermine level in grass pea seedlings (Lathyrus sativus L.). Plant Physiol. Biochem. 2006, 44, 161–169. [Google Scholar] [CrossRef]
- Kong, H.Y.; Zhu, H.; Zhou, R.; Akram, N.A.; Wang, Y.B.; Jiao, C.J.; Xiong, Y.C. Role of abscisic acid in modulating drought acclimation, agronomic characteristics and β-N-Oxalyl-L-α,β-diaminopropionic acid (β-ODAP) Accumulation in grass pea (Lathyrus Sativus L.). J. Sci. Food Agric. 2021, 102, 2553–2562. [Google Scholar] [CrossRef]
- Piwowarczyk, B.; Tokarz, K.; Makowski, W.; Łukasiewicz, A. Different acclimatization mechanisms of two grass pea cultivars to osmotic stress in vitro culture. Acta Physiol. Plant. 2017, 39, 96. [Google Scholar] [CrossRef] [Green Version]
- Xing, G.; Cui, K.; Li, J.; Wang, Y.; Li, Z. Water stress and accumulation of β-N-oxalyl-L-α,β-diaminopropionic acid in grass pea (Lathyrus sativus). J. Agric. Food Chem. 2001, 49, 216–220. [Google Scholar] [CrossRef]
- Verma, A.; Nidhi, N.; Kaur, G.; Mantri, S.; Sharma, T.R.; Pandey, A.K.; Kandoth, P.K. Contrasting β-ODAP content correlates with stress gene expression in Lathyrus cultivars. Physiol. Plant. 2022, 174, e13616. [Google Scholar] [CrossRef]
- Cooper, M.; Voss-Fels, K.P.; Messina, C.D.; Tang, T.; Hammer, G.L. Tackling G × E × M interactions to close on-farm yield-gaps: Creating novel pathways for crop improvement by predicting contributions of genetics and management to crop productivity. Theor. Appl. Genet. 2021, 134, 1625–1644. [Google Scholar] [CrossRef]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of climate change on crops adaptation and strategies to tackle its outcome: A review. Plants 2019, 8, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liliane, T.N.; Charles, M.S. Factors affecting yield of crops. In Agronomy—Climate Change and Food Security; Amanullah, A., Ed.; IntechOpen: London, UK, 2020; pp. 1–16. [Google Scholar] [CrossRef]
- Kole, C.; Muthamilarasan, M.; Henry, R.; Edwards, D.; Sharma, R.; Abberton, M.; Batley, J.; Bentley, A.; Blakeney, M.; Bryant, J.; et al. Application of genomics-assisted breeding for generation of climate resilient crops: Progress and prospects. Front. Plant Sci. 2015, 6, 563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzafera, P.; Favarin, J.L.; de Andrade, S.A.L. Editorial: Intercropping systems in sustainable agriculture. Front. Sustain. Food Syst. 2021, 5, 1–3. [Google Scholar] [CrossRef]
- Wang, F.; Yang, T.; Burlyaeva, M.; Li, L.; Jiang, J.; Fang, L.; Redden, R.; Zong, X. Genetic diversity of grasspea and its relative species revealed by SSR markers. PLoS ONE 2015, 10, e0118542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dempewolf, H.; Baute, G.; Anderson, J.; Kilian, B.; Smith, C.; Guarino, L. Past and future use of wild relatives in crop breeding. Crop Sci. 2017, 57, 1070–1082. [Google Scholar] [CrossRef]
- Rubiales, D.; Fondevilla, S.; Chen, W.; Gentzbittel, L.; Higgins, T.J.V.; Castillejo, M.A.; Singh, K.B.; Rispail, N. Achievements and challenges in legume breeding for pest and disease resistance. CRC Crit. Rev. Plant Sci. 2015, 34, 195–236. [Google Scholar] [CrossRef] [Green Version]
- Schaefer, H.; Hechenleitner, P.; Santos-Guerra, A.; De Sequeira, M.M.; Pennington, R.T.; Kenicer, G.; Carine, M.A. Systematics, biogeography, and character evolution of the legume tribe Fabeae with special focus on the middle-atlantic island lineages. BMC Evol. Biol. 2012, 12, 250. [Google Scholar] [CrossRef] [Green Version]
- Heywood, V.; Casas, A.; Ford-Lloyd, B.; Kell, S.; Maxted, N. Conservation and sustainable use of crop wild relatives. Agric. Ecosyst. Environ. 2007, 121, 245–255. [Google Scholar] [CrossRef]
- Hajjar, R.; Hodgkin, T. The use of wild relatives in crop improvement: A survey of developments over the last 20 years. Euphytica 2007, 156, 1–13. [Google Scholar] [CrossRef]
- Yunus, A.G.; Jackson, M.T. The gene pools of the grasspea (Lathyrus sativus L.). Plant Breed. 1991, 106, 319–328. [Google Scholar] [CrossRef]
- Rubiales, D.; González-Bernal, M.J.; Carrilo-Perdomo, E.; Almeida, N.F.; Gonçalves, L.; Vaz Patto, M.C. Lathyrus sativus and L. cicera breeding: Characterization of Iberian germplasm, QTL mapping, interspecific hybridization and cultivar development. In Proceedings of the Joint Meeting of Fodder Crops and Amenity Grasses Section and Protein Crops Working Group of EUCARPIA-Oil and Protein Crops Section. Breeding Grasses and Protein Crops in the Era of Genomics, Vilnius, Lithuania, 11–14 September 2017; p. 35. [Google Scholar]
- Sillero, J.C.; Cubero, J.I.; Fernandéz-Aparicio, M.; Rubiales, D. Search for resistance to create broomrape (Orobanche crenata) in Lathyrus. Lathyrus Latthyrism Newsl. 2005, 4, 7–9. [Google Scholar]
- Fernández-Aparicio, M.; Flores, F.; Rubiales, D. Recognition of root exudates by seeds of broomrape (Orobanche and Phelipanche) species. Ann. Bot. 2009, 103, 423–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Aparicio, M.; Rubiales, D. Characterisation of resistance to crenate broomrape (Orobanche crenata Forsk.) in Lathyrus cicera L. Euphytica 2010, 173, 77–84. [Google Scholar] [CrossRef]
- Bohra, A.; Kilian, B.; Sivasankar, S.; Caccamo, M.; Mba, C.; McCouch, S.R.; Varshney, R.K. Reap the crop wild relatives for breeding future crops. Trends Biotechnol. 2022, 40, 412–431. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Kushwah, N.; Kumar, S.; Singh, P.; Rathore, M.; Singh, N.P.; Das, A. Genome editing using CRISPR/Cas systems in legumes. In Genome Editing in Plants: Principles and Applications, 1st ed.; Gupta, O.P., Karkute, S.G., Eds.; CRC Press: Boca Raton, FL, USA, 2021; pp. 89–107. [Google Scholar] [CrossRef]
- Wolter, F.; Schindele, P.; Puchta, H. Plant breeding at the speed of light: The power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biol. 2019, 19, 176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhowmik, P.; Konkin, D.; Polowick, P.; Hodgins, C.L.; Subedi, M.; Xiang, D.; Yu, B.; Patterson, N.; Rajagopalan, N.; Babic, V.; et al. CRISPR/Cas9 gene editing in legume crops: Opportunities and challenges. Legume Sci. 2021, 3, e96. [Google Scholar] [CrossRef]
- Ochatt, S.; Conreux, C.; Moussa Mcolo, R.; Despierre, G.; Magnin-Robert, J.B.; Raffiot, B. Phytosulfokine-alpha, an enhancer of in vitro regeneration competence in recalcitrant legumes. Plant Cell. Tissue Organ Cult. 2018, 135, 189–201. [Google Scholar] [CrossRef] [Green Version]
- Zambre, M.; Chowdhury, B.; Kuo, Y.H.; Van Montagu, M.; Angenon, G.; Lambein, F. Prolific regeneration of fertile plants from green nodular callus induced from meristematic tissues in Lathyrus sativus L. (grass pea). Plant Sci. 2002, 163, 1107–1112. [Google Scholar] [CrossRef]
- Barik, D.P.; Mohapatra, U.; Chand, P.K. High frequency in vitro regeneration of Lathyrus sativus L. Biol. Plant. 2005, 49, 637–639. [Google Scholar] [CrossRef]
- Goldsmith, M.; Barad, S.; Knafo, M.; Savidor, A.; Ben-Dor, S.; Brandis, A.; Mehlman, T.; Peleg, Y.; Albeck, S.; Dym, O.; et al. Identification and characterization of the key enzyme in the biosynthesis of the neurotoxin β-ODAP in grass pea. J. Biol. Chem. 2022, accepted. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, M.; Barad, S.; Peleg, Y.; Albeck, S.; Dym, O.; Brandis, A.; Mehlman, T.; Reich, Z. The Identification and characterization of an oxalyl-CoA synthetase from grass pea (Lathyrus sativus L.). RSC Chem. Biol. 2022, 3, 320–333. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Watson, A.; Gonzalez-Navarro, O.E.; Ramirez-Gonzalez, R.H.; Yanes, L.; Mendoza-Suárez, M.; Simmonds, J.; Wells, R.; Rayner, T.; Green, P.; et al. Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat. Protoc. 2018, 13, 2944–2963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Li, P.; Zou, C.; Lu, Y.; Xie, C.; Zhang, X.; Prasanna, B.M.; Olsen, M.S. Enhancing genetic gain in the era of molecular breeding. J. Exp. Bot. 2017, 68, 2641–2666. [Google Scholar] [CrossRef]
- Yang, W.; Feng, H.; Zhang, X.; Zhang, J.; Doonan, J.H.; Batchelor, W.D.; Xiong, L.; Yan, J. Crop phenomics and high-throughput phenotyping: Past decades, current challenges, and future perspectives. Mol. Plant 2020, 1, 187–214. [Google Scholar] [CrossRef] [Green Version]
- Cabrera-Bosquet, L.; Crossa, J.; von Zitzewitz, J.; Serret, M.D.; Luis Araus, J. High-throughput phenotyping and genomic selection: The frontiers of crop breeding converge. J. Integr. Plant Biol. 2012, 54, 312–320. [Google Scholar] [CrossRef] [Green Version]
- Hickey, L.T.; Hafeez, A.N.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef] [Green Version]
- Croser, J.S.; Pazos-Navarro, M.; Bennett, R.G.; Tschirren, S.; Edwards, K.; Erskine, W.; Creasy, R.; Ribalta, F.M. Time to flowering of temperate pulses in vivo and generation turnover in vivo–in vitro of narrow-leaf lupin accelerated by low red to far-red ratio and high intensity in the far-red region. Plant Cell. Tissue Organ Cult. 2016, 127, 591–599. [Google Scholar] [CrossRef]
- Barpete, S.; Gupta, P.; Khawar, K.M.; Ozcan, S.; Kumar, S. In vitro approaches for shortening generation cycles and faster breeding of low β-N-oxalyl-L-α, β -diaminopropionic acid content of grass pea (Lathyrus sativus L.). Fresenius Environ. Bull. 2020, 29, 2698–2706. [Google Scholar]
- Plans, M.; Simó, J.; Casañas, F.; Sabaté, J.; Rodriguez-Saona, L. Characterization of common beans (Phaseolus vulgaris L.) by infrared spectroscopy: Comparison of MIR, FT-NIR and dispersive NIR using portable and benchtop instruments. Food Res. Int. 2013, 54, 1643–1651. [Google Scholar] [CrossRef]
- Carbonaro, M.; Nucara, A. Secondary structure of food proteins by Fourier Transform spectroscopy in the Mid-Infrared region. Amino Acids 2010, 38, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, D.S.; Galão, O.F.; Pallone, J.A.L.; Poppi, R.J. Comparison and application of Near-Infrared (NIR) and Mid-Infrared (MIR) Spectroscopy for determination of quality parameters in soybean samples. Food Control 2014, 35, 227–232. [Google Scholar] [CrossRef]
- Mendonza, F.A.; Cichy, K.A.; Sprague, C.; Goffnett, A.; Lu, R.; Kelly, J.D. Prediction of canned black bean texture (Phaseolus vulgaris L.) from intact dry seeds using visible/ near infrared spectroscopy and hyperspectral imaging data. J. Sci. Food Agric. 2018, 98, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Plans, M.; Simó, J.; Casañas, F.; Sabaté, J. Near-infrared spectroscopy analysis of seed coats of common beans (Phaseolus vulgaris L.): A potential tool for breeding and quality evaluation. J. Agric. Food Chem. 2012, 60, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Plans, M.; Simó, J.; Casañas, F.; del Castillo, R.R.; Rodriguez-Saona, L.E.; Sabaté, J. Estimating sensory properties of common beans (Phaseolus vulgaris L.) by near infrared spectroscopy. Food Res. Int. 2014, 56, 55–62. [Google Scholar] [CrossRef]
- Wafula, E.N.; Wainaina, I.N.; Buvé, C.; Kinyanjui, P.K.; Saeys, W.; Sila, D.N.; Hendrickx, M.E.G. Prediction of cooking times of freshly harvested common beans and their susceptibility to develop the hard-to-cook defect using near infrared spectroscopy. J. Food Eng. 2021, 298, 110495. [Google Scholar] [CrossRef]
- Wafula, E.N.; Onduso, M.; Wainaina, I.N.; Buvé, C.; Kinyanjui, P.K.; Githiri, S.M.; Saeys, W.; Sila, D.N.; Hendrickx, M. Antinutrient to mineral molar ratios of raw common beans and their rapid prediction using near-infrared spectroscopy. Food Chem. 2022, 368, 130773. [Google Scholar] [CrossRef]
- Wang, J.; Liu, H.; Ren, G. Near-infrared spectroscopy (NIRS) evaluation and regional analysis of chinese faba bean (Vicia faba L.). Crop J. 2014, 2, 28–37. [Google Scholar] [CrossRef] [Green Version]
- Erny, G.L.; Brito, E.; Pereira, A.B.; Bento-Silva, A.; Vaz Patto, M.C.; Bronze, M.R. Projection to latent correlative structures, a dimension reduction strategy for spectral-based classification. RSC Adv. 2021, 11, 29124–29129. [Google Scholar] [CrossRef]
- Bento-Silva, A.; Gonçalves, L.; Mecha, E.; Pereira, F.; Patto, M.C.V.; Bronze, M.D.R. An improved HILIC HPLC-MS/MS method for the determination of β-ODAP and its α isomer in Lathyrus sativus. Molecules 2019, 24, 3043. [Google Scholar] [CrossRef] [Green Version]
- Santos, C.; Polanco, C.; Rubiales, D.; Vaz Patto, M.C. The MLO1 powdery mildew susceptibility gene in Lathyrus species: The power of high-density linkage maps in comparative mapping and synteny analysis. Plant Genome 2021, 14, e20090. [Google Scholar] [CrossRef] [PubMed]
- Emmrich, P.M.F.; Sarkar, A.; Njaci, I.; Kaithakottil, G.G.; Ellis, N.; Moore, C.; Edwards, A.; Heavens, D.; Waite, D.; Cheema, J.; et al. A draft genome of grass pea (Lathyrus sativus), a resilient diploid legume. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kreplak, J.; Madoui, M.A.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019, 51, 1411–1422. [Google Scholar] [CrossRef] [PubMed]
- Bugdol, M.; Jedynak, P. Integrated Management Systems, 1st ed.; Springer: Cham, Switzerland, 2015; pp. 1–194. [Google Scholar] [CrossRef]
- Lulie, B. Intercropping practice as an alternative pathway for sustainable agriculture: A review. Acad. Res. J. Agric. Sci. Res. 2017, 5, 440–452. [Google Scholar] [CrossRef]
- Sarwar, C.D.M.; Malek, M.A.; Sarker, A.; Hassan, M.S. Genetic resources of grass pea (Lathyrus sativus L.) in Bangladesh. In Lathyrus Genetic Resouces in Asia, Proceedings of the a Regional Workshop, Indira Gandhi Agricultural University, Raipur, India, 27–29 December 1995; IPGRI Office for South Asia: New Delhi, India, 1995; p. 13. [Google Scholar]
- Das, N.R. Lathyrus sativus in rainfed multiple cropping systems in West Bengal, India—A review. Lathyrus Lathyrism Newsl. 2000, 1, 25–27. [Google Scholar]
- Atis, I.; Acikalin, S. Yield, quality and competition properties of grass pea and wheat grown as pure and binary mixture in different plant densities. Turk. J. Field Crop. 2020, 25, 18–25. [Google Scholar] [CrossRef]
- Rhaman, M.; Awal, M.; Shelley, I. Interception and use of solar radiation in mustard/ grass pea intercropping. Int. J. Plant Soil Sci. 2016, 12, 1–13. [Google Scholar] [CrossRef]
- Rao, S.C.; Northup, B.K. Planting date affects production and quality of grass pea forage. Crop Sci. 2008, 48, 1629–1635. [Google Scholar] [CrossRef]
- Calderón, F.J.; Vigil, M.F.; Nielsen, D.C.; Benjamin, J.G.; Poss, D.J. Water use and yields of no-till managed dryland grasspea and yellow pea under different planting configurations. Field Crops Res. 2012, 125, 179–185. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonçalves, L.; Rubiales, D.; Bronze, M.R.; Vaz Patto, M.C. Grass Pea (Lathyrus sativus L.)—A Sustainable and Resilient Answer to Climate Challenges. Agronomy 2022, 12, 1324. https://doi.org/10.3390/agronomy12061324
Gonçalves L, Rubiales D, Bronze MR, Vaz Patto MC. Grass Pea (Lathyrus sativus L.)—A Sustainable and Resilient Answer to Climate Challenges. Agronomy. 2022; 12(6):1324. https://doi.org/10.3390/agronomy12061324
Chicago/Turabian StyleGonçalves, Letice, Diego Rubiales, Maria R. Bronze, and Maria C. Vaz Patto. 2022. "Grass Pea (Lathyrus sativus L.)—A Sustainable and Resilient Answer to Climate Challenges" Agronomy 12, no. 6: 1324. https://doi.org/10.3390/agronomy12061324
APA StyleGonçalves, L., Rubiales, D., Bronze, M. R., & Vaz Patto, M. C. (2022). Grass Pea (Lathyrus sativus L.)—A Sustainable and Resilient Answer to Climate Challenges. Agronomy, 12(6), 1324. https://doi.org/10.3390/agronomy12061324






