Assessing Intraspecific Variability and Diversity in African Yam Bean Landraces Using Agronomic Traits
Abstract
:1. Introduction
2. Materials and Methods
2.1. Germplasm, Research Sites and Experimental Design
2.2. Data Collection
2.3. Data Analysis
3. Results
3.1. Soil and Climatic Conditions under Field Evaluation
3.2. Variability among Accessions
3.3. Relationships among Traits
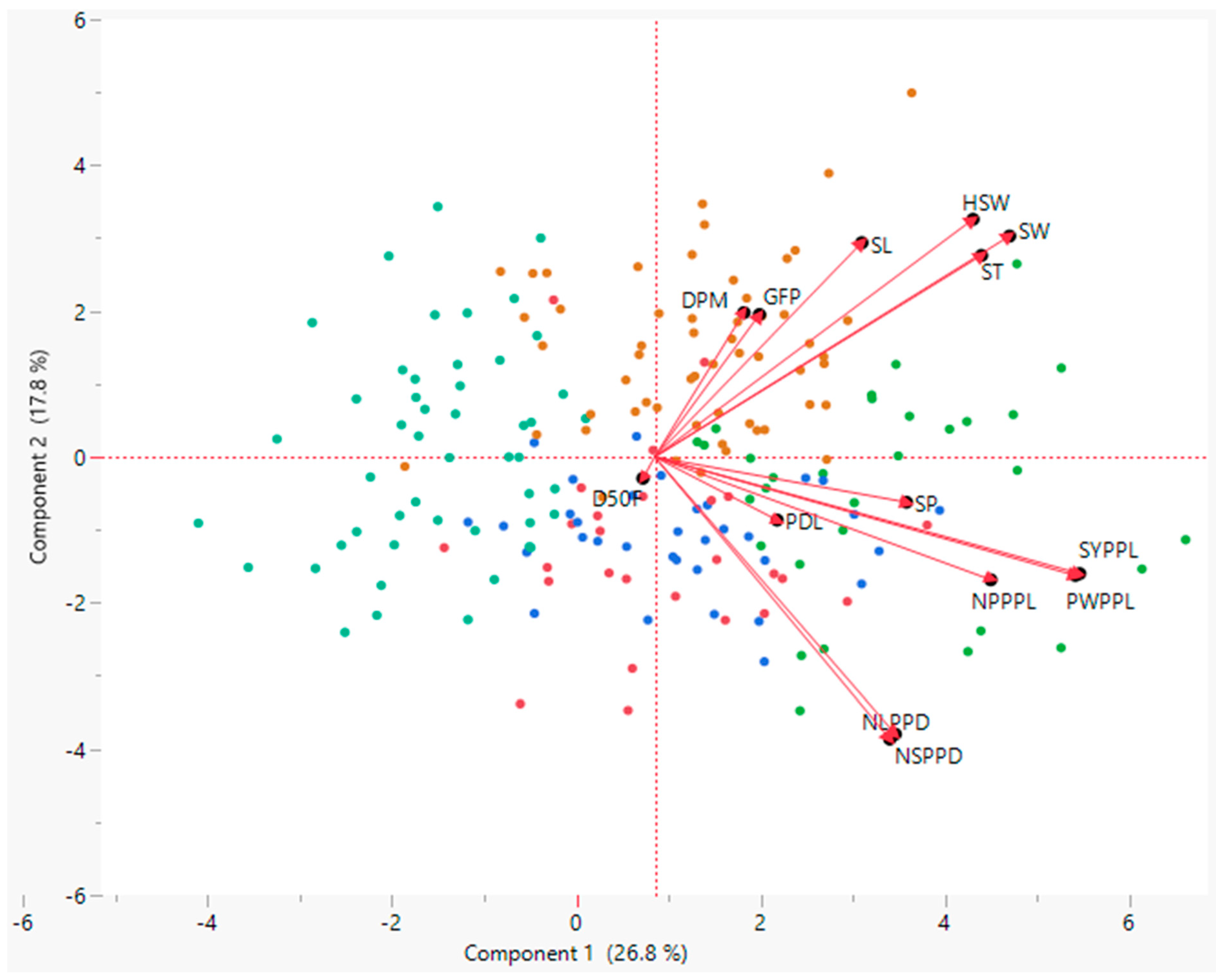
3.4. Principal Component Analysis (PCA)
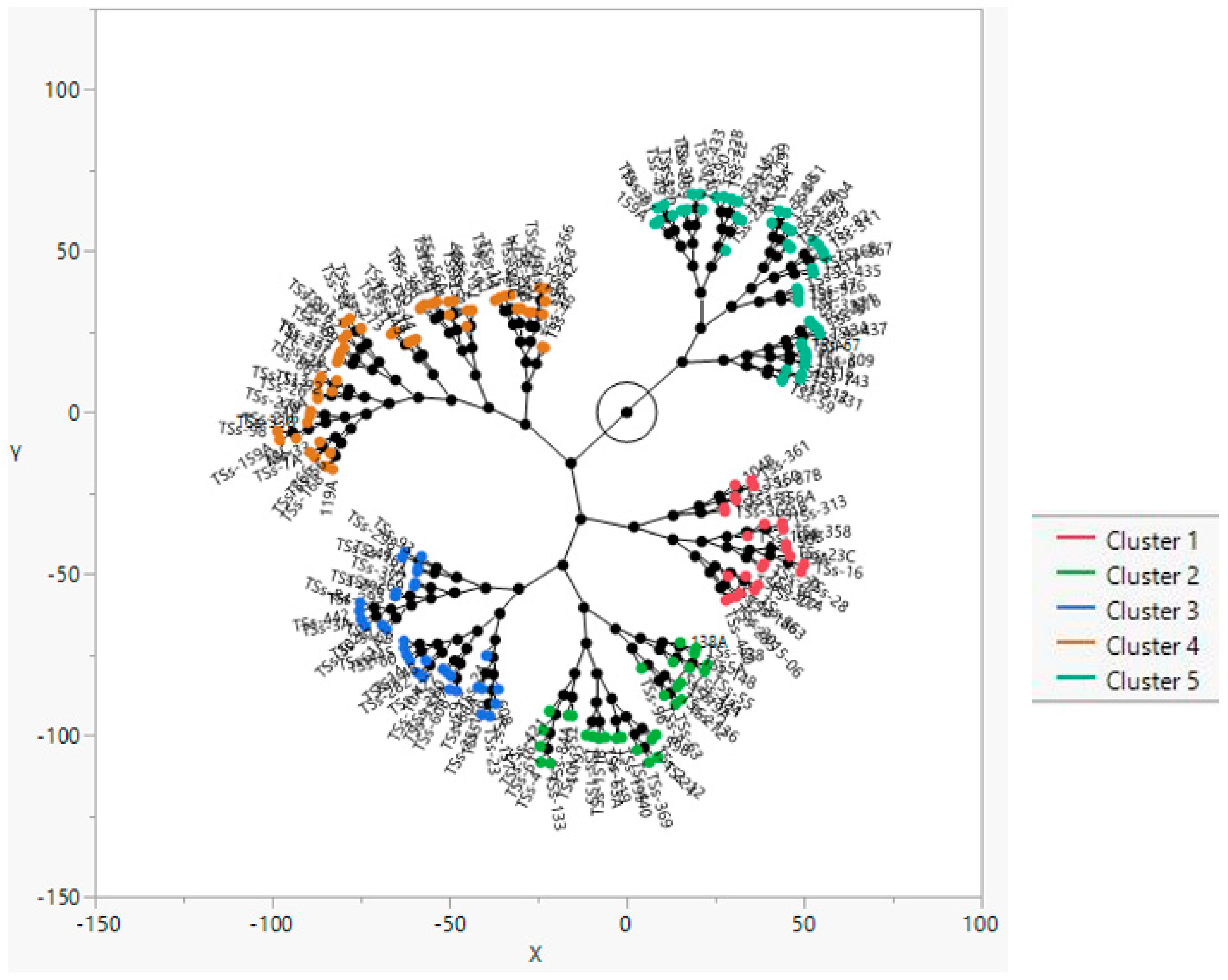
3.5. Cluster Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Xu, X.; Sharma, R.; Tondelli, A.; Russell, J.; Comadran, J.; Schnaithmann, F.; Pillen, K.; Kilian, B.; Cattivelli, L.; Thomas, W.T.B.; et al. Genome-Wide association analysis of grain yield-associated traits in a pan-European barley cultivar collection. Plant Genome 2018, 11, 170073. [Google Scholar] [CrossRef] [PubMed]
- Aremu, C.O.; Ojuederie, O.B.; Ayo-Vaughan, F.; Dahunsi, O.; Adekiya, A.O.; Olayanju, A.; Adebiyi, O.T.; Sunday, I.; Inegbedion, H.; Asaleye, A.J.; et al. Morphometric analysis and characterization of the nutritional quality in African yam bean accessions. Plant Physiol. Rep. 2019, 24, 446–459. [Google Scholar] [CrossRef]
- Okigbo, B.N. Introducing the yam bean: Stenostylis stenocarpa (Hochst ex A. Rich) harms. Proc. First IITA Grain 1973, 7, 224–237. [Google Scholar]
- Oganale, D. Nodulation and nitrogen fixation of African yam beam (Sphenostylis stenocarpa). In Proceedings of the African Crop Science Society Conference, Science and Technology Supporting Food Security in Africa, Cape Town, Africa, 28 September–1 October 2009. [Google Scholar]
- Omitogun, O.G.; Jackai, L.E.N.; Thottappilly, G. Isolation of insecticidal lectin-enrich extracts from African yam bean (Sphenostylis stenocarpa) and other legume species. Entomol. Exp. Appl. 1991, 90, 301–311. [Google Scholar] [CrossRef]
- Potter, D. Economic botany of Sphenostylis (Leguminosae). Econ. Bot. 1992, 46, 262–275. [Google Scholar] [CrossRef]
- Saka, J.O.; Ajibade, S.R.; Adeniyan, O.N.; Olowoyo, R.B.; Ogunbodede, B.A. Survey of underutilized grain Legume production systems in the Southwest Agricultural zone of Nigeria. J. Agric. Food Inf. 2004, 6, 93–107. [Google Scholar] [CrossRef]
- Okpara, D.A.; Omaliko, C.P.E. Effects of Staking, nitrogen and Phosphorus fertilizer rates on yield and yield components of African yam bean (Sphenostylis stenocarpa). Ghana J. Agric. Sci. 1995, 28, 23–28. [Google Scholar] [CrossRef]
- Fasoyiro, S.B.; Ajibade, S.R.; Omole, A.J.; Adeniyan, O.N.; Farinde, E.O. Proximate, minerals and antinutritional factors of some underutilized grain legumes in south-western Nigeria. Nutr. Food Sci. 2006, 36, 18–23. [Google Scholar] [CrossRef]
- Paliwal, R.; Abberton, M.; Faloye, B.; Olaniyi, O. Developing the role of legumes in West Africa under climate change. Curr. Opin. Plant Biol. 2020, 56, 242–258. [Google Scholar] [CrossRef]
- Akande, S.R. Germplasm characterization of African yam bean from southwest Nigeria. Acta Hortic. 2009, 806, 695–700. [Google Scholar] [CrossRef]
- Popoola, J.O.; Adegbite, A.E.; Obembe, O.O.; Adewale, B.D.; Odu, B.O. Morphological intraspecific variabilities in African yam bean (AYB) (Sphenostylis stenocarpa Ex. A. Rich) Harms. Sci. Res. Essay 2011, 6, 507–515. [Google Scholar]
- Adewale, B.D.; Dumet, D.J.; Vroh-Bi, I.; Kehinde, O.B.; Ojo, D.K.; Adegbite, A.E.; Franco, J. Morphological diversity analysis of African yam bean (Sphenostylis stenocarpa Hochst. ex A. Rich.) Harms and prospects for utilization in germplasm conservation and breeding. Genet. Resour. Crop Evol. 2012, 59, 927–936. [Google Scholar] [CrossRef]
- Aremu, C.O.; Ibirinde, D.B. Bio-diversity studies on accessions of African yam bean (Sphenostylis stenocarpa). Int. J. Agric. Res. 2012, 7, 78–85. [Google Scholar] [CrossRef]
- Ibirinde, D.O.; Aremu, C.O. Trait variability studies on African yam bean (Sphenostylis stenocarpa) grown in the Guinea Savannah Zone of Southwestern Nigeria. Int. J. Adv. Biol. Res. 2013, 3, 422–427. [Google Scholar]
- Aina, I.A.; Ilori, O.C.; Ekanem, O.U.; Oyatomi, A.O.; Potter, D.; Abberton, M.T. Morphological Characterization and Variability Analysis of African Yam Bean (Sphenostylis stenocarpa Hochst. ex. A. Rich) Harms. Int. J. Plant Res. 2020, 10, 45–52. [Google Scholar]
- Moyib, O.K.; Gbadegesin, M.A.; Aina, O.O.; Odunola, A.O. Genetic variation within a collection of Nigerian accessions of African yam bean (Sphenostylis stenocarpa) revealed by RAPD primers. Afr. J. Biotechnol. 2008, 7, 1839–1846. [Google Scholar] [CrossRef] [Green Version]
- Ojuederie, O.B.; Balogun, M.O.; Fawole, I.; Igwe, D.O.; Olowolafe, M.O. Assessment of the genetic diversity of African yam bean (Sphenostylis stenocarpa Hochst ex. A Rich. Harms) accessions using amplified fragment length polymorphism (AFLP) markers. Afr. J. Biotechnol. 2014, 13, 18. [Google Scholar]
- Adewale, B.D.; Vroh-Bi, I.; Dumet, D.J.; Nnadi, S.; Kehinde, O.B.; Ojo, D.K.; Adegbite, A.E.; Franco, J. Genetic diversity in African yam bean accessions based on AFLP markers: Towards a platform for germplasm improvement and utilization. Plant Genet. Resour. 2015, 13, 111. [Google Scholar] [CrossRef]
- Shitta, N.S.; Abberton, M.T.; Adesoye, A.I.; Adewale, D.B.; Oyatomi, O. Analysis of genetic diversity of African yam bean using SSR markers derived from cowpea. Plant Genet. Resour. 2016, 14, 50–56. [Google Scholar] [CrossRef]
- Kalisz, S.; Kramer, E.M. Variation and constraint in plant evolution and development. Heredity 2008, 100, 171–177. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, D.N.; Tripathi, S.R.; Tripathi, M.P.; Khatri, N.; Bastola, B.R. Genetic Variability and Correlation Coefficients of Major Traits in Early Maturing Rice under Rainfed Lowland Environments of Nepal. Adv. Agric. 2019, 2019, 5975901. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Sharma, S.D.; Sharma, N.C.; Devi, M. The path coefficient analysis of yield components for leaf nutrient concentrations in mango (Mangifera indica L.) under rainfed agroclimatic conditions of north-west Himalaya. Sci. Hortic. 2015, 190, 31–35. [Google Scholar] [CrossRef]
- Sesay, S.; Ojo, D.K.; Ariyo, O.J.; Meseka, S.K.; Fayeun, L.S.; Omikunle, A.O.; Oyetunde, A.O. Correlation and path coefficient analysis of top-cross and three-way cross hybrid maize populations. Afr. J. Agric. Res. 2017, 12, 780–789. [Google Scholar]
- Adewale, D.B.; Dumet, D.J. Descriptors for African Yam Bean, Sphenostylis stenocarpa (Hochst ex. A. Rich.) Harms; Genetic Resources Center, International Institute of Tropical Agriculture (IITA): Ibadan, Nigeria, 2011; pp. 1–13. [Google Scholar]
- SAS Institute Inc. Base SAS® 9.4 Procedures Guide: Statistical Procedures; SAS Institute Inc.: Cary, NC, USA, 2017. [Google Scholar]
- Robinson, H.F.; Comstock, R.E.; Harvey, P.H. Estimates of Heritability and the Degree of Dominance in Corn. Agron. J. 1949, 41, 353–359. [Google Scholar] [CrossRef]
- Singh, R.K.; Chaudhary, B.D. Biometrical Methods in Quantitative Genetic Analysis; Kalyani Publishers: New Delhi, India, 1977; pp. 57–58. [Google Scholar]
- Sivasubramanian, S.; Menon, M. Heterosis and inbreeding depression in rice. Adv. Agron. 1973, 47, 85–140. [Google Scholar]
- Alvarado, G.L.; Vargas, M.; Pacheco, M.; Rodríguez, Á.F.; Burgueño, J.; Crossa, J. META-R (Multi Environment Trail Analysis with R for Windows) Version 6.04. CIMMYT Research Data & Software Repository Network, V23, 2015. Available online: https://hdl.handle.net/11529/10201 (accessed on 5 September 2021).
- Ward, J.H., Jr. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Adewale, B.D.; Aremu, C.O.; Amazue, U.E. Intra-Specific variability and diversity analysis of African yam bean by seed size parameters. J. Agric. Biol. Sci. 2012, 7, 454–461. [Google Scholar]
- Adewale, B.D.; Kehinde, O.B.; Aremu, C.O.; Popoola, J.O.; Dumet, D.J. Seed metrics for genetic and shape determination in African yam bean. Afr. J. Plant Sci. 2010, 4, 107–115. [Google Scholar]
- Ojuederie, O.B.; Balogun, M.O.; Akande, S.R.; Korie, S.; Omodele, T. Intraspecific variability in agro-morphological traits of African yam bean Sphenostylis stenocarpa (Hochst ex. A. Rich) Harms. J. Crop Sci. Biotechnol. 2015, 18, 53–62. [Google Scholar] [CrossRef]
- Ibirinde, D.O.; Aremu, C.O.; Balogun, K.; Oladokun, L. Assessment of Seed and Tuber Production Potential in Varieties of Sphenostylis stenocarpa (Africa Yam Bean). Agric. Sci. 2019, 10, 870–881. [Google Scholar]
- Aremu, C.; Abberton, M.; Adebiyi, T.; Asaleye, A.J.; Inegbedion, H.; Abolusoro, S.; Adekiya, A.; Aboyeji, C.; Dunsin, O. Selecting yield and nutritional traits in Sphenostylis stenocarpa landraces for food improvement. Recent Pat. Food Nutr. Agric. 2020, 11, 69–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez, K.A.; Gomez, A.A. Statistical Procedures for Agricultural Research; John Wiley and Sons: Hoboken, NJ, USA, 1984. [Google Scholar]
- Alake, C.O.; Porbeni, B.J. Genetic variation and intraspecific diversity for agro-morphological and nutritional traits in African yam bean. J. Crop Improv. 2020, 34, 139–163. [Google Scholar] [CrossRef]
- Nwofia, G.E.; Awaraka, R.O.; Agbo, C.U. Genetic Variability and Trait Association Studies in African Yam Bean (Sphenostylis sternocarpa) Hochst ex A. Rich. Am.-Eurasian J. Agric. Environ. Sci. 2013, 13, 1547–1553. [Google Scholar]
- Bose, L.K.; Das, S.; Pradhan, S.K.; Subudhi, H.; Singh, S.; Singh, O. Genetic variability of quality characters and grain yield in lowland rice genotypes of Eastern India. Korean J. Breed. Sci. 2007, 39, 1–6. [Google Scholar]
- Adesoye, A.; Ukwueze, C. Genetic and environmental variation of seed yield and yield components in African yam bean (Sphenostylis stenocarpa (Hochst. Ex. A. Rich.)). Adv. Life Sci. Technol. 2015, 38, 7–16. [Google Scholar]
- Osuagwu, A.N.; Chukwurah, P.N.; Ekpo, I.A.; Akpakpan, E.E.; Agbor, R.B. Variation, correlation and path coefficient analyses in seed yield and related characters in local accessions of African Yam Bean (Sphenostylis stenocarpa) from Southern Nigeria. Afr. J. Agric. Res. 2014, 9, 211–215. [Google Scholar]
- Das, S.; Das, S.S.; Chakraborty, I.; Roy, N.; Nath, M.K.; Sarma, D. Principal component analysis in plant breeding. Biomol. Rep. Int. e-Newsl. 2017, 3, 1–3. [Google Scholar]
- Guei, R.G.; Sanni, K.A.; Abamu, F.J.; Fawole, I. Genetic diversity of rice (Oryza sativa L.). Agron. Afr. 2005, 5, 17–28. [Google Scholar]
| Properties | E1 | E2 | E3 | E4 | E5 | E6 |
|---|---|---|---|---|---|---|
| pH (1:1) | 5.9 | 6.8 | 5.8 | 5.4 | 4.8 | 5.2 |
| Bray P (mg/kg) | 9 | 5 | 12 | 10 | 2 | 3 |
| Organic Carbon (g/kg) | 4.3 | 4.2 | 3.8 | 8.7 | 5.1 | 4.8 |
| N (g/kg) | 0.6 | 1.3 | 0.2 | 0.6 | 1.1 | 2.8 |
| Particle size (g/kg) | ||||||
| Sand | 750 | 650 | 850 | 770 | 810 | 850 |
| Silt | 60 | 80 | 60 | 140 | 60 | 90 |
| Clay | 190 | 270 | 90 | 90 | 130 | 60 |
| Textural class | SL | SCL | LS | SL | SL | LS |
| Coordinates | 7°29′12.89″ N, 3°54′07.38″ E, 237.07 m altitude | 7°29′07.95″ N, 3°54′03.79″ E, 211.6 m altitude | 12°08′21.97″ N, 8°40′05.55″ E, 427.8 m altitude | 12°08′23.59″ N, 8°40′11.03″ E, 425.5 m altitude | 6°40′09.40″ N, 6°20′28.08″ E, 334.4 m altitude | 6°40′09.40″ N, 6°20′28.08″ E, 334.4 m altitude |
| Month | Year | Rainfall (mm) | Solar Radiation (MJ/m2/day) | Temp. Min (°C) | Temp. Max (°C) | Relative Humidity (%) | Month | Year | Rainfall (mm) | Solar Radiation (MJ/m2/day) | Temp. Min (°C) | Temp. Max (°C) | Relative Humidity (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E1 | E2 | ||||||||||||
| 4–31 August | 2018 | 91.55 | 13.2 | 22.23 | 28.33 | 84.91 | 6–31 August | 2019 | 236.85 | 14.916 | 22.27 | 28.95 | 82.35 |
| September | 2018 | 251.56 | 14.81 | 22.44 | 29.39 | 82.66 | September | 2019 | 305.3 | 15.73 | 22.29 | 28.96 | 81.14 |
| October | 2018 | 280 | 18.15 | 21.92 | 30.9 | 78.08 | October | 2019 | 299.95 | 16.92 | 22.05 | 29.23 | 79.06 |
| November | 2018 | 16.9 | 18.94 | 23.39 | 32.04 | 69.28 | November | 2019 | 32.4 | 18.18 | 23.3 | 32.3 | 70.05 |
| December | 2018 | 0 | 18.77 | 20.54 | 33.83 | 51.48 | December | 2019 | 9 | 18.6 | 21.51 | 33.84 | 60.39 |
| January | 2019 | 7.1 | 14.28 | 22.1 | 35.02 | 57.86 | January | 2020 | 0 | 18.93 | 19.81 | 34.71 | 46.69 |
| Average | 107.85 | 16.36 | 22.1 | 31.59 | 70.71 | Average | 147.25 | 17.21 | 21.87 | 31.33 | 69.95 | ||
| E3 | E4 | ||||||||||||
| 20–31 July | 2018 | 63.72 | 19.18 | 22.04 | 31.59 | 88.33 | 17–31 July | 2019 | 114.21 | 20.44 | 22.19 | 31.15 | 71.34 |
| August | 2018 | 249.86 | 17.85 | 21.45 | 30.28 | 92.26 | August | 2019 | 244.25 | 18.99 | 21.16 | 29.21 | 79.22 |
| September | 2018 | 85.56 | 19.98 | 21.72 | 31.94 | 89.09 | September | 2019 | 72.8 | 20.38 | 21.96 | 31.79 | 71.63 |
| October | 2019 | 10.64 | 20.34 | 21.53 | 31.8 | 67.83 | October | 2019 | 53.87 | 20.34 | 21.53 | 31.8 | 67.83 |
| November | 2018 | 0 | 19.96 | 16.17 | 34.54 | 54.88 | November | 2019 | 21.5 | 20.78 | 18.03 | 34.08 | 41.21 |
| December | 2018 | 0 | 16.99 | 14.12 | 30.19 | 42.2 | December | 2019 | 0 | 20.63 | 11.87 | 30.42 | 26.88 |
| 1–19 January | 2019 | 0 | 19.73 | 13.8 | 19.72 | 18.71 | 1–16 January | 2020 | 0 | 19.38 | 13.03 | 29.28 | 28.58 |
| Average | 58.45 | 19.15 | 18.69 | 30.01 | 64.76 | Average | 72.38 | 20.13 | 18.54 | 31.1 | 55.24 | ||
| E5 | E6 | ||||||||||||
| 25–31 August | 2018 | 91.49 | 17.47 | 21.911 | 28.33 | 88.9 | June | 2019 | 315.76 | 16.55 | 22.9 | 29.07 | 88.36 |
| September | 2018 | 325.42 | 16.43 | 22.37 | 28.01 | 90.35 | July | 2019 | 241.36 | 16.38 | 22.32 | 28.21 | 89.2 |
| October | 2018 | 139.97 | 18.45 | 22.57 | 28.58 | 89.53 | August | 2019 | 386.17 | 15.65 | 21.99 | 27.77 | 90.09 |
| November | 2018 | 76.72 | 19.58 | 22.54 | 30.14 | 83.15 | September | 2019 | 391.53 | 16.51 | 22.46 | 27.81 | 91.14 |
| December | 2018 | 0.01 | 19.03 | 19.27 | 29.72 | 66.92 | October | 2019 | 417.39 | 16.63 | 22.19 | 27.94 | 90.08 |
| January | 2019 | 14.89 | 17.78 | 20.52 | 30.4 | 72.76 | November | 2019 | 62.6 | 19.03 | 22.75 | 30.05 | 84.45 |
| 1–24 February | 2019 | 15.94 | 18.57 | 21.5 | 30.42 | 72 | Average | 302.47 | 16.79 | 22.44 | 28.48 | 88.89 | |
| Average | 94.92 | 18.19 | 21.53 | 29.37 | 80.52 | ||||||||
| SOV | DF | D50F | DPM | GFP | NPPPL | PWPPL | SP | PDL |
|---|---|---|---|---|---|---|---|---|
| ACC | 195 | 111.12 *** | 248.33 * | 300.56 ** | 0.11 ** | 774.96 * | 1.8 * | 19.61 *** |
| ENV (LOC * YR) | 5 | 105,468 *** | 410,740 *** | 252,643 *** | 18.17 *** | 178,126 *** | 156.3 *** | 3725.93 *** |
| REP (ENV) | 12 | 463.82 *** | 1422.3 *** | 1174.56 *** | 1.07 *** | 19,861 *** | 8.26 *** | 62.84 *** |
| BLK (REP * ENV) | 234 | 62.96 *** | 134.33 *** | 158.49 *** | 0.07 *** | 765.32 *** | 0.92 ** | 6.63 * |
| ACC * ENV | 975 | 64.13 *** | 209.3 *** | 236.84 *** | 0.09 *** | 644.24 *** | 1.52 *** | 11.74 *** |
| Error | 2157 | 35.84 | 75.46 | 97.98 | 0.04 | 336.72 | 0.73 | 5.54 |
| CV (%) | 6.72 | 5.68 | 15.45 | 21.56 | 56.43 | 13.12 | 11.37 | |
| SOV | DF | NLPPD | NSPPD | HSW | SYPPL | SL | SW | ST |
| ACC | 195 | 0.02 *** | 0.02 ** | 52.18 *** | 225.74 * | 1.26 *** | 0.55 *** | 0.88 *** |
| ENV (LOC * YR) | 5 | 8.71 *** | 9.56 *** | 2582.22 *** | 56,656 ** | 59.79 *** | 57.48 *** | 63.52 *** |
| REP (ENV) | 12 | 0.05 *** | 0.05 *** | 66.67 *** | 6967.85 *** | 0.54 ** | 0.37 ** | 0.52 ** |
| BLK (REP * ENV) | 234 | 0.01 | 0.01 * | 17.14 * | 232.52 *** | 0.22 | 0.14 | 0.2 |
| ACC * ENV | 975 | 0.02 *** | 0.02 *** | 25.69 *** | 181.26 *** | 0.42 *** | 0.28 *** | 0.38 *** |
| Error | 2157 | 0.01 | 0.01 | 14.1 | 96.36 | 0.22 | 0.14 | 0.21 |
| CV (%) | 7.98 | 8.56 | 18.24 | 64.62 | 5.89 | 6.06 | 7.76 |
| Traits | Mean | σ2e | σ2p | σ2g | σ2ge | GVC | PCV | H2 (%) |
|---|---|---|---|---|---|---|---|---|
| D50F | 89.11 | 36.09 | 6.61 | 2.97 | 9.76 | 1.9 | 2.9 | 45.0 |
| DPM | 153.03 | 75.67 | 14.76 | 2.53 | 48.17 | 1.0 | 2.5 | 17.1 |
| GFP | 64.05 | 98.48 | 17.87 | 4.15 | 49.52 | 3.2 | 6.6 | 23.2 |
| NPPPL | 9.23 | 25.46 | 4.02 | 0.87 | 10.42 | 10.1 | 21.7 | 21.6 |
| PWPPL | 32.52 | 334.79 | 45.72 | 7.86 | 115.58 | 8.6 | 20.8 | 17.2 |
| SP | 43.80 | 112.38 | 16.2 | 2.72 | 43.43 | 3.8 | 9.2 | 16.8 |
| PDL | 20.69 | 5.54 | 1.18 | 0.50 | 2.27 | 3.4 | 5.3 | 42.0 |
| NLPPD | 12.35 | 4.96 | 0.79 | 0.22 | 1.74 | 3.8 | 7.2 | 28.1 |
| NSPPD | 11.42 | 4.74 | 0.79 | 0.25 | 1.65 | 4.4 | 7.8 | 30.8 |
| HSW | 20.58 | 14.24 | 3.07 | 1.58 | 4.17 | 6.1 | 8.5 | 51.6 |
| SYPPL | 15.19 | 95.7 | 13.62 | 2.97 | 32.01 | 11.3 | 24.3 | 21.8 |
| SL | 7.98 | 0.22 | 0.08 | 0.05 | 0.08 | 2.8 | 3.5 | 66.4 |
| SW | 6.19 | 0.14 | 0.03 | 0.02 | 0.05 | 2.1 | 2.9 | 50.0 |
| ST | 5.97 | 0.21 | 0.05 | 0.03 | 0.06 | 3.0 | 3.9 | 57.8 |
| Traits | D50F | DPM | GFP | NPPPL | PWPPL | SP | PDL | NLPPD | NSPPD | HSW | SL | SW | ST | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DPM | rg | 0.35 ** | ||||||||||||
| rp | 0.21 ** | |||||||||||||
| GFP | rg | −0.61 ** | 0.53 ** | |||||||||||
| rp | −0.42 ** | 0.78 ** | ||||||||||||
| NPPPL | rg | −0.57 ** | 0.01 | 0.53 ** | ||||||||||
| rp | −0.2 ** | 0.16 * | 0.27 ** | |||||||||||
| PWPPL | rg | −0.15 * | 0.32 ** | 0.40 ** | 0.71 ** | |||||||||
| rp | −0.09 | 0.14 * | 0.19 ** | 0.83 ** | ||||||||||
| SP | rg | 0.29 ** | 0.40 ** | 0.12 | 0.78 ** | 0.70 ** | ||||||||
| rp | 0.08 | 0.06 | 0.01 | 0.21 ** | 0.21 ** | |||||||||
| PDL | rg | 0.35 ** | 0.21 ** | −0.14 | −0.33 ** | −0.23 ** | −0.83 ** | |||||||
| rp | 0.12 | −0.003 | −0.08 | 0.02 | 0.12 | −0.16 * | ||||||||
| NLPPD | rg | 0.48 ** | 0.21 ** | −0.2 | 0.12 | 0.19 ** | 0.31 ** | 0.01 | ||||||
| rp | 0.14 | 0.02 | −0.06 | 0.18 ** | 0.26 ** | 0.21 ** | 0.39 ** | |||||||
| NSPPD | rg | 0.31 ** | 0.09 | −0.14 | 0.01 | 0.11 | 0.27 ** | −0.08 | 0.97 ** | |||||
| rp | 0.08 | 0.02 | −0.02 | 0.18 * | 0.27 ** | 0.23 ** | 0.33 ** | 0.94 ** | ||||||
| HSW | rg | 0.09 | 0.12 | 0.05 | −0.18 * | 0.26 ** | 0.28 ** | 0.11 | −0.27 ** | −0.28 ** | ||||
| rp | 0.05 | 0.11 | 0.08 | 0.05 | 0.21 ** | 0.26 ** | 0.12 | −0.06 | −0.07 | |||||
| SL | rg | 0.03 | 0.15 * | 0.09 | −0.16 * | 0.07 | −0.21 ** | 0.42 ** | −0.25 ** | −0.36 ** | 0.73 ** | |||
| rp | 0.05 | 0.1 | 0.06 | −0.01 | 0.09 | 0.02 | 0.30 ** | −0.07 | −0.13 | 0.6 ** | ||||
| SW | rg | 0.01 | 0.07 | 0.07 | −0.16 * | 0.25 ** | 0.43 ** | 0.04 | 0.06 | −0.01 | 0.86 ** | 0.50 ** | ||
| rp | 0.01 | 0.12 | 0.11 | 0.1 | 0.24 ** | 0.23 ** | 0.11 | 0.11 | 0.08 | 0.68 ** | 0.54 ** | |||
| ST | rg | 0.09 | 0.09 | 0.05 | −0.20 ** | 0.28 ** | 0.50 ** | −0.02 | 0.14 | 0.15 * | 0.70 ** | 0.19 ** | 0.82 ** | |
| rp | 0.01 | 0.11 | 0.11 | 0.05 | 0.22 ** | 0.26 ** | 0.02 | 0.08 | 0.09 | 0.61 ** | 0.3 ** | 0.80 ** | ||
| SYPPL | rg | 0.28 ** | 0.45 ** | 0.15 * | 0.53 ** | 0.89 ** | 0.76 ** | −0.44 ** | 0.26 ** | 0.2 ** | 0.29 ** | −0.06 | 0.36 ** | 0.41 ** |
| rp | 0.03 | 0.15 * | 0.12 | 0.70 ** | 0.91 ** | 0.44 ** | 0.02 | 0.27 ** | 0.28 ** | 0.26 ** | 0.05 | 0.26 ** | 0.28 ** |
| Variables | PC1 | PC2 | PC3 | PC4 | PC5 | PC6 |
|---|---|---|---|---|---|---|
| D50F | −0.01 | −0.03 | 0.25 | 0.13 | 0.60 | 0.59 |
| DPM | 0.09 | 0.22 | −0.37 | 0.51 | 0.36 | 0.12 |
| GFP | 0.10 | 0.22 | −0.49 | 0.41 | −0.02 | −0.26 |
| NPPPL | 0.33 | −0.19 | −0.36 | −0.13 | −0.19 | 0.17 |
| PWPPL | 0.41 | −0.18 | −0.24 | −0.13 | −0.14 | 0.27 |
| SP | 0.24 | −0.07 | 0.04 | −0.28 | 0.43 | −0.26 |
| PDL | 0.12 | −0.10 | 0.27 | 0.47 | −0.36 | 0.28 |
| NLPPD | 0.23 | −0.42 | 0.24 | 0.30 | 0.05 | −0.24 |
| NSPPD | 0.23 | −0.43 | 0.21 | 0.27 | 0.06 | −0.30 |
| HSW | 0.31 | 0.36 | 0.20 | −0.08 | −0.04 | 0.01 |
| SYPPL | 0.41 | −0.18 | −0.19 | −0.21 | 0.07 | 0.22 |
| SL | 0.20 | 0.32 | 0.25 | 0.07 | −0.31 | 0.22 |
| SW | 0.35 | 0.33 | 0.20 | 0.00 | 0.01 | −0.15 |
| ST | 0.32 | 0.30 | 0.15 | −0.03 | 0.16 | −0.24 |
| Eigenvalue | 3.75 | 2.49 | 2.12 | 1.46 | 1.23 | 1.08 |
| Proportion % | 26.8 | 17.8 | 15.2 | 10.4 | 8.7 | 7.7 |
| Cumulative % | 26.8 | 44.6 | 59.8 | 70.2 | 78.9 | 86.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olomitutu, O.E.; Abe, A.; Oyatomi, O.A.; Paliwal, R.; Abberton, M.T. Assessing Intraspecific Variability and Diversity in African Yam Bean Landraces Using Agronomic Traits. Agronomy 2022, 12, 884. https://doi.org/10.3390/agronomy12040884
Olomitutu OE, Abe A, Oyatomi OA, Paliwal R, Abberton MT. Assessing Intraspecific Variability and Diversity in African Yam Bean Landraces Using Agronomic Traits. Agronomy. 2022; 12(4):884. https://doi.org/10.3390/agronomy12040884
Chicago/Turabian StyleOlomitutu, Oluwaseyi E., Ayodeji Abe, Olaniyi A. Oyatomi, Rajneesh Paliwal, and Michael T. Abberton. 2022. "Assessing Intraspecific Variability and Diversity in African Yam Bean Landraces Using Agronomic Traits" Agronomy 12, no. 4: 884. https://doi.org/10.3390/agronomy12040884
APA StyleOlomitutu, O. E., Abe, A., Oyatomi, O. A., Paliwal, R., & Abberton, M. T. (2022). Assessing Intraspecific Variability and Diversity in African Yam Bean Landraces Using Agronomic Traits. Agronomy, 12(4), 884. https://doi.org/10.3390/agronomy12040884






