Combining Variable Selection and Multiple Linear Regression for Soil Organic Matter and Total Nitrogen Estimation by DRIFT-MIR Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Soil Sampling
2.2. Spectral Measurement and SOM and TN Content Analysis
2.3. Spectral Pre-Processing
2.4. Variable Selection Methods
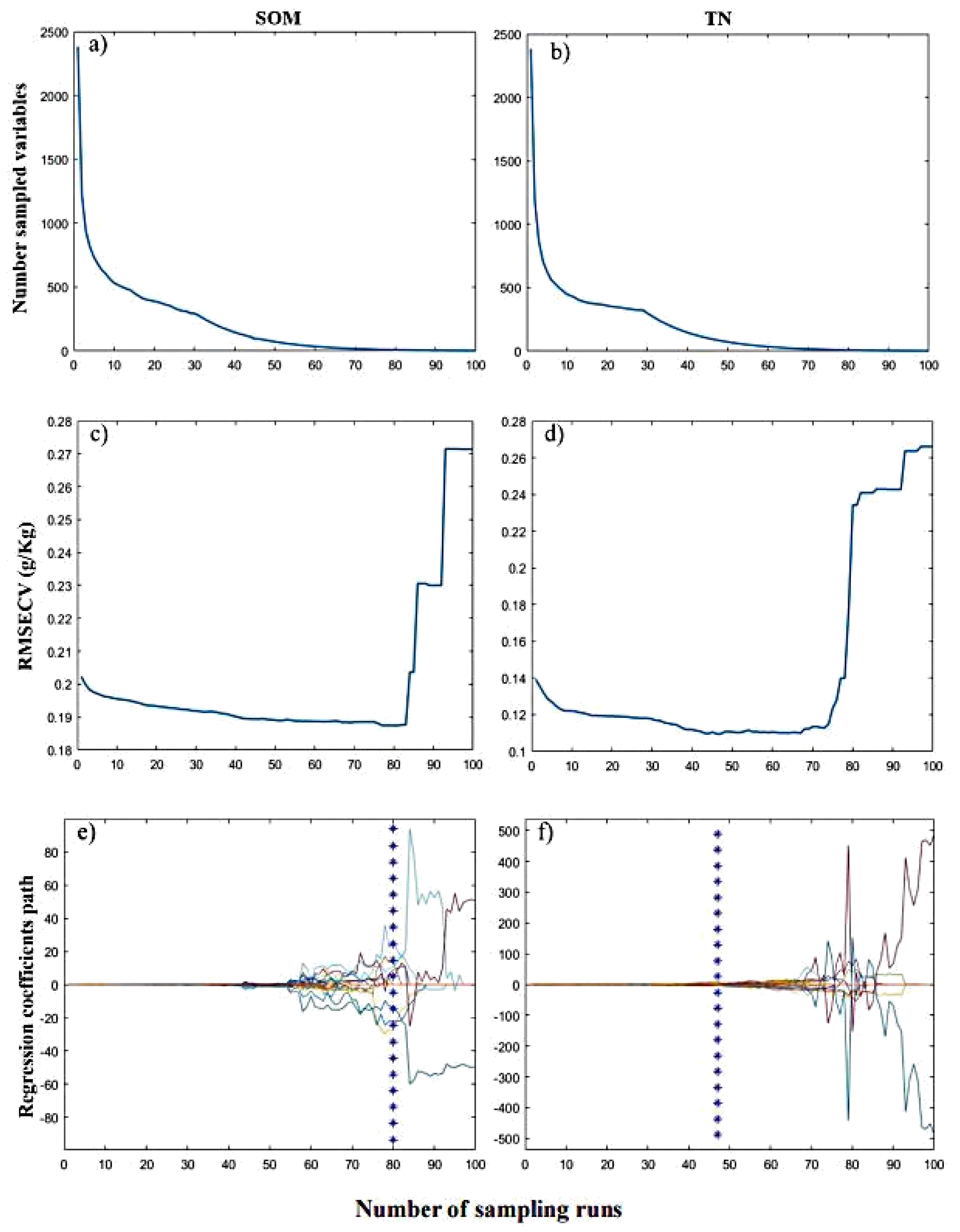
2.4.1. Stability Competitive Adaptive Reweighting Algorithm (sCARS)
2.4.2. Bootstrapping Soft Shrinkage (BOSS)
2.5. Model Calibration
2.6. Model Evaluation
3. Results
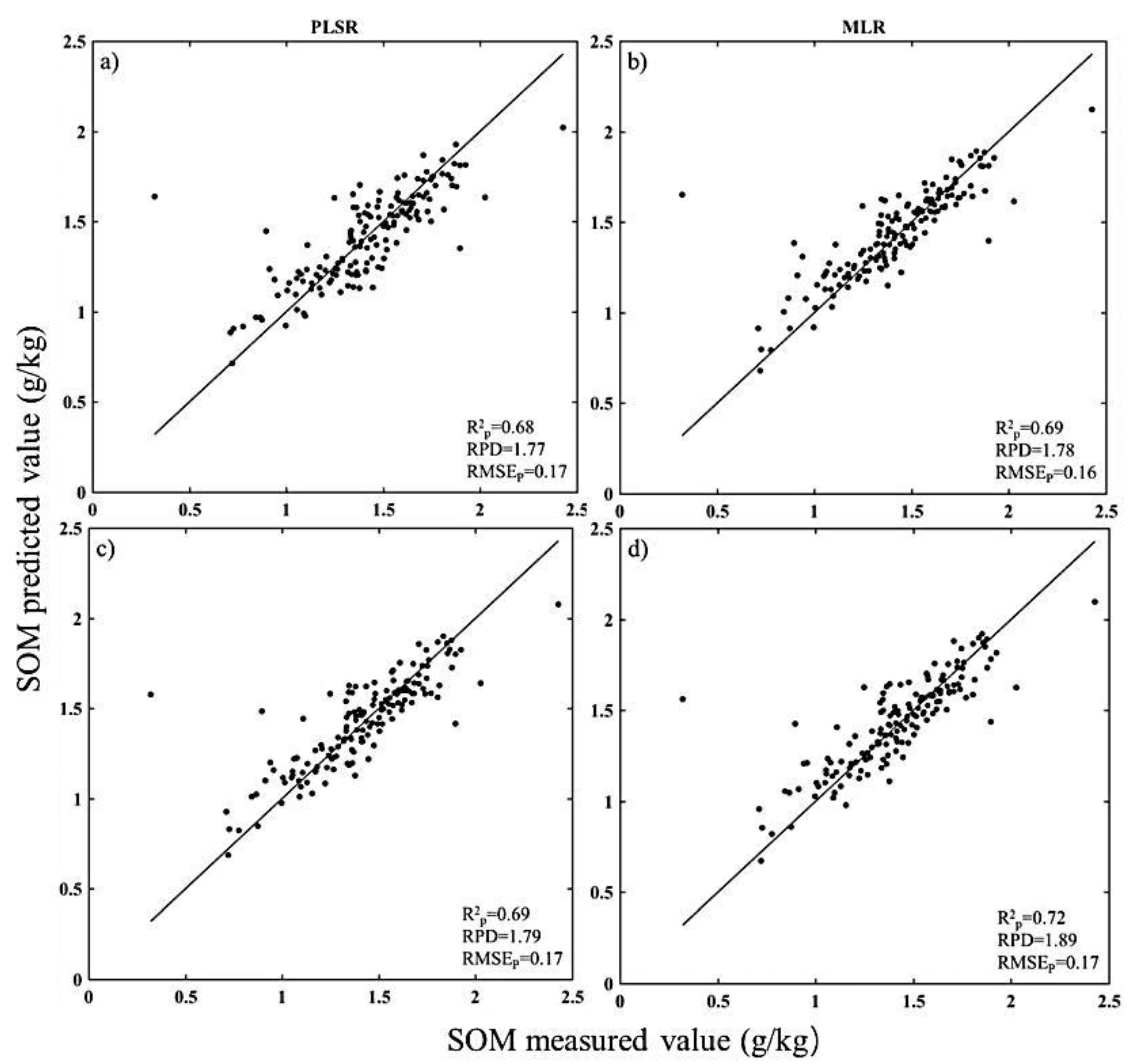
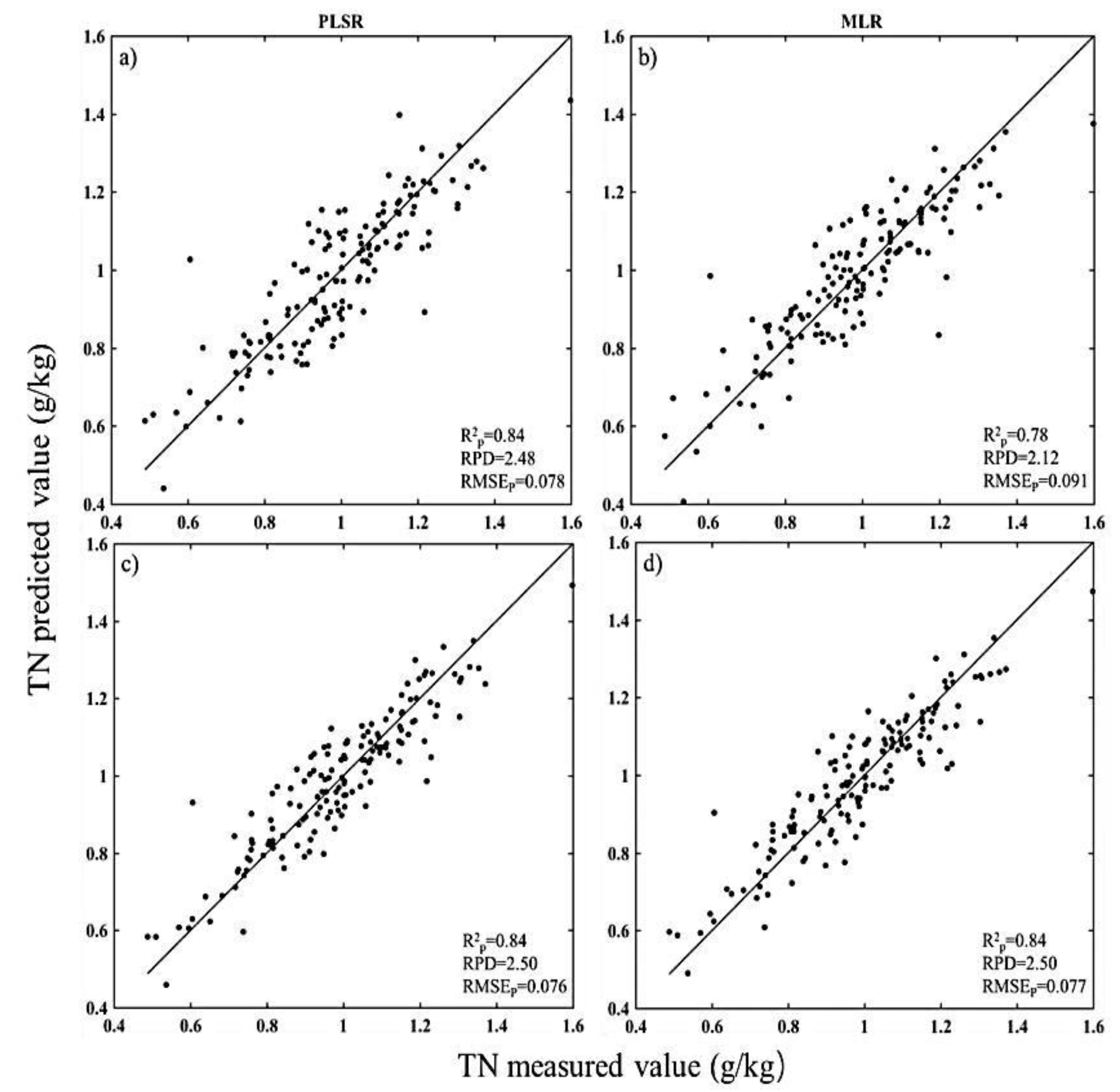
3.1. Estimation Accuracies and Model Parsimony
3.2. Key Wavenumbers
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ARS | Adaptive reweighted sampling |
| BOSS | Bootstrapping soft shrinkage |
| BSS | Bootstrap sampling |
| CARS | Competitive adaptive reweighted sampling |
| EDF | Exponentially decreasing function |
| ELM | Extreme learning machine |
| FT-IR | Fourier transform infrared spectrometer |
| GA | Genetic algorithm |
| LV | Latent variables |
| MCS | Monte Carlo sampling |
| MCUVE | Monte Carlo uninformative variables elimination |
| MIR | Mid-infrared spectroscopy |
| MIR | Cation exchange capacity |
| MLR | Multiple linear regression |
| MPA | Model population analysis |
| MSC | Multiplicative scatter correction |
| NIR | Near-infrared spectroscopy |
| PLSR | Partial least-squares regression |
| Rp2 | Coefficient determination of prediction |
| RC | Regression coefficients |
| RMSE | Root-mean-square error |
| RMSECV | Root-mean-square error of cross-validation |
| RMSEp | Root-mean-square error of prediction |
| RPD | Residual prediction deviation |
| sCARS | Stability competitive adaptive reweighted sampling |
| SNV | Standard normalized variate |
| SOM | Soil organic matter |
| SPA | Successive projections algorithm |
| TN | Total nitrogen |
| Vis-NIR | Visible and near-infrared spectroscopy |
| WBS | Weighted bootstrap sampling |
References
- Ahmadi, A.; Emami, M.; Daccache, A.; He, L.Y. Soil properties prediction for precision agriculture using visible and near-infrared spectroscopy: A systematic review and Meta-analysis. Agronomy 2021, 11, 433. [Google Scholar] [CrossRef]
- Reda, R.; Saffaj, T.; Ilham, B.; Saidi, O.; Issam, K.; Brahim, L.; El Hadrami, E.M. A comparative study between a new method and other machine learning algorithms for soil organic carbon and total nitrogen prediction using near infrared spectroscopy. Chemometr. Intell. Lab. Syst. 2019, 195, 103873. [Google Scholar] [CrossRef]
- Gebbers, R.; Adamchuk, V.I. Precision Agriculture and Food Security. Science 2010, 327, 828–831. [Google Scholar] [CrossRef]
- Xu, D.; Zhao, R.; Li, S.; Chen, S.; Jiang, Q.; Zhou, L.; Shi, Z. Multi-sensor fusion for the determination of several soil properties in the Yangtze River Delta, China. Eur. J. Soil Sci. 2019, 70, 162–173. [Google Scholar] [CrossRef] [Green Version]
- Reeves, J.B. Near- versus mid-infrared diffuse reflectance spectroscopy for soil analysis emphasizing carbon and laboratory versus on-site analysis: Where are we and what needs to be done? Geoderma 2010, 158, 3–14. [Google Scholar] [CrossRef]
- Bellon-Maurel, V.; Mcbratney, A. Near-infrared (NIR) and mid-infrared (MIR) spectroscopic techniques for assessing the amount of carbon stock in soils-Critical review and research perspectives. Soil Biol. Biochem. 2011, 43, 1398–1410. [Google Scholar] [CrossRef]
- Soriano-Disla, J.M.; Janik, L.J.; Viscarra, R.; Macdonald, L.M.; McLaughlin, M.J. The performance of visible, near-, and mid-infrared reflectance spectroscopy for prediction of soil physical, chemical, and biological properties. Appl. Spectrosc. Rev. 2014, 49, 139–186. [Google Scholar] [CrossRef]
- Barra, I.; Haefele, S.M.; Sakrabani, R.; Kebede, F. Soil spectroscopy with the use of chemometrics, machine learning and pre-processing techniques in soil diagnosis: Recent advances—A review. TrAC Trends Anal. Chem. 2021, 135, 116166. [Google Scholar] [CrossRef]
- Guo, P.; Li, T.; Gao, H.; Chen, X.W.; Cui, Y.F.; Huang, Y.R. Evaluating calibration and spectral variable selection methods for predicting three soil nutrients using Vis-NIR spectroscopy. Remote Sens. 2021, 13, 4000. [Google Scholar] [CrossRef]
- Lazaar, A.; Mouazen, A.M.; EL Hammouti, K.; Fullen, M.; Pradhan, B.; Memon, M.S.; Andich, K.; Monir, A. The application of proximal visible and near-infrared spectroscopy to estimate soil organic matter on the Triffa Plain of Morocco. Int. Soil Water Conserv. Res. 2020, 8, 195–204. [Google Scholar] [CrossRef]
- de Santana, F.B.; de Souza, A.M.; Poppi, R.J. Green methodology for soil organic matter analysis using a national near infrared spectral library in tandem with learning machine. Sci. Total Environ. 2019, 658, 895–900. [Google Scholar] [CrossRef] [PubMed]
- Terra, F.S.; Dematte, J.A.M.; Viscarra Rossel, R.A. Spectral libraries for quantitative analyses of tropical Brazilian soils: Comparing vis–NIR and mid-IR reflectance data. Geoderma 2015, 255, 81–93. [Google Scholar] [CrossRef]
- Ji, W.J.; Li, S.; Chen, S.C.; Shi, Z.; Viscarra Rossel, R.A.; Mouazen, A.M. Prediction of soil attributes using the Chinese soil spectral library and standardized spectra recorded at field conditions. Soil Till. Res. 2016, 155, 492–500. [Google Scholar] [CrossRef]
- Xu, S.X.; Zhao, Y.C.; Wang, M.Y.; Shi, X.Z. Comparison of multivariate methods for estimating selected soil properties from intact soil cores of paddy fields by Vis–NIR spectroscopy. Geoderma 2018, 310, 29–43. [Google Scholar] [CrossRef]
- Morellos, A.; Pantazi, X.E.; Moshou, D.; Alexandridis, T.; Whetton, R.; Tziotzios, G.; Wiebensohn, J.; Bill, R.; Mouazen, A.M. Machine learning based prediction of soil total nitrogen, organic carbon and moisture content by using VIS-NIR spectroscopy. Biosyst. Eng. 2016, 152, 104–116. [Google Scholar] [CrossRef] [Green Version]
- Conforti, M.2; Matteucci, G.; Buttafuoco, G. Using laboratory Vis-NIR spectroscopy for monitoring some forest soil properties. J. Soils Sediments 2018, 18, 1009–1019. [Google Scholar] [CrossRef]
- Askari, M.S.; O’Rourke, S.M.; Holden, N.M. Evaluation of soil quality for agricultural production using visible-near-infrared spectroscopy. Geoderma 2015, 243, 80–91. [Google Scholar] [CrossRef]
- Tümsavaş, Z. Application of visible and near infrared reflectance spectroscopy to predict total nitrogen in soil. J. Environ. Biol. 2017, 38, 1101–1106. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, M.Z.; Zheng, L.H.; Qin, Q.M.; Lee, W.S. Spectral features extraction for estimation of soil total nitrogen content based on modified ant colony optimization algorithm. Geoderma 2019, 333, 23–34. [Google Scholar] [CrossRef]
- McCarty, G.W.; Reeves III, J.B.; Reeves, V.B.; Follett, R.F.; Kimble, J.M. Mid-infrared and near-infrared diffuse reflectance spectroscopy for soil carbon management. Soil Sci. Soc. Am. J. 2002, 66, 640–646. [Google Scholar] [CrossRef]
- McCarty, G.W.; Reeves, J.B., III. Comparison of near infrared and mid infrared diffuse reflectance spectroscopy for field-scale measurement of soil fertility parameters. Soil Sci. 2006, 171, 94–102. [Google Scholar] [CrossRef] [Green Version]
- Xie, H.T.; Yang, X.M.; Drury, C.F.; Yang, J.Y.; Zhang, X.D. Predicting soil organic carbon and total nitrogen using mid- and near-infrared spectra for Brookston clay loam soil in Southwestern Ontario, Canada. Can. J Soil Sci. 2011, 91, 53–63. [Google Scholar] [CrossRef]
- Vohland, M.; Ludwig, M.; Thiele-Bruhn, S.; Ludwig, B. Determination of soil properties with visible to near- and mid-infrared spectroscopy: Effects of spectral variable selection. Geoderma 2014, 223–225, 88–96. [Google Scholar] [CrossRef]
- Knox, N.M.; Grunwald, S.; McDowell, M.L.; Bruland, G.L.; Myers, D.B. Harris W G. Modelling soil carbon fractions with visible near-infrared (VNIR) and mid-infrared (MIR) spectroscopy. Geoderma 2015, 239–240, 229–239. [Google Scholar] [CrossRef]
- Haghi, R.K.; Perez-Fernandez, E.; Robertson, A.H.J. Prediction of various soil properties for a national spatial dataset of Scottish soils based on four different chemometric approaches: A comparison of near infrared and mid-infrared spectroscopy. Geoderma 2021, 396, 115071. [Google Scholar] [CrossRef]
- dos Santos, U.J.; de Melo Dematte, J.A.; Cezar Menezes, R.S.; Dotto, A.C.; Barbosa Guimaraes, C.C.; Rodrigues Alves, B.J.; Primo, D.C.; de Sa Barretto Sampaio, E.V. Predicting carbon and nitrogen by visible near-infrared (Vis-NIR) and mid-infrared (MIR) spectroscopy in soils of Northeast Brazil. Geoderma Reg. 2020, 23, e00333. [Google Scholar] [CrossRef]
- Johnson, J.M.; Vandamme, E.; Senthilkumar, K.; Sila, A.; Shepherd, K.D.; Saito, K. Near-infrared, mid-infrared or combined diffuse reflectance spectroscopy for assessing soil fertility in rice fields in sub-Saharan Africa. Geoderma 2019, 354, 113840. [Google Scholar] [CrossRef]
- Seybold, C.A.; Ferguson, R.; Wysocki, D.; Bailey, B.; Anderson, J.; Nester, B.; Schoeneberger, P.; Wills, S.; Libohova, Z.; Hoover, D.; et al. Application of Mid-Infrared Spectroscopy in Soil Survey. Soil Sci. Soc. Am. J. 2019, 83, 1746–1759. [Google Scholar] [CrossRef]
- Xiong, Y.R.; Zhang, R.Q.; Zhang, F.Y.; Yang, W.Y.; Kang, Q.D.; Chen, W.C.; Du, Y.P. A spectra partition algorithm based on spectral clustering for interval variable selection. Infrared Phys. Technol. 2020, 105, 103259. [Google Scholar] [CrossRef]
- Lao, C.C.; Chen, J.Y.; Zhang, Z.T.; Chen, Y.W.; Ma, Y.; Chen, H.R.; Gu, X.B.; Ning, J.F.; Jin, J.M.; Li, X.W. Predicting the contents of soil salt and major water-soluble ions with fractional-order derivative spectral indices and variable selection. Comput. Electron. Agric. 2021, 182, 106031. [Google Scholar] [CrossRef]
- Kawamura, K.; Nishigaki, T.; Tsujimoto, Y.; Andriamananjara, A.; Rabenaribo, M.; Asai, H.; Rakotoson, T.; Razafimbelo, T. Exploring relevant wavelength regions for estimating soil total carbon contents of rice fields in Madagascar from Vis-NIR spectra with sequential application of backward interval PLS. Plant Prod. Sci. 2021, 24, 1–14. [Google Scholar] [CrossRef]
- Vohland, M.; Ludwig, M.; Harbich, M.; Emmerling, C.; Thiele-Bruhn, S. Using variable selection and wavelets to exploit the full potential of visible-near infrared spectra for predicting soil properties. J. Near Infrared Spectrosc. 2016, 24, 255–269. [Google Scholar] [CrossRef]
- Cheng, H.; Wang, J.; Du, Y.K. Combining multivariate method and spectral variable selection for soil total nitrogen estimation by Vis-NIR spectroscopy. Arch. Agron. Soil Sci. 2020, 67, 1665–1678. [Google Scholar] [CrossRef]
- Zheng, K.Y.; Li, Q.Q.; Wang, J.J.; Geng, J.P.; Cao, P.; Sui, T.; Wang, X.; Du, Y.P. Stability competitive adaptive reweighted sampling (SCARS) and its applications to multivariate calibration of NIR spectra. Chemometr. Intell. Lab. Syst. 2012, 112, 48–54. [Google Scholar] [CrossRef]
- Yan, H.; Song, X.Z.; Tian, K.D.; Gao, J.X.; Li, Q.Q.; Xiong, Y.M.; Min, S.G. A modification of the bootstrapping soft shrinkage approach for spectral variable selection in the issue of over-fitting, model accuracy and variable selection credibility. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2019, 210, 362–371. [Google Scholar] [CrossRef]
- Deng, B.C.; Yun, Y.H.; Cao, D.S.; Yin, Y.L.; Wang, W.T.; Lu, H.M.; Luo, Q.Y.; Liang, Y.Z. A bootstrapping soft shrinkage approach for variable selection in chemical modeling. Anal. Chim. Acta 2016, 908, 63–74. [Google Scholar] [CrossRef]
- Zhang, F.; Tang, X.J.; Tong, A.X.; Wang, B.; Wang, J.W. Bootstrapping soft shrinkage variable selection method based on the combination of frequency and regression coefficient. Chin. J. Sci. Instrum. 2020, 41, 64–70. [Google Scholar]
- Hutengs, C.; Ludwig, B.; Jung, A.; Eisele, A.; Vohland, M. Comparison of portable and bench-top spectrometers for mid-infrared diffuse reflectance measurements of soils. Sensors 2018, 18, 993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terhoeven-Urselmans, T.; Vagen, T.G.; Spaargaren, O.; Shepherd, K.D. Prediction of soil fertility properties from a globally distributed soil mid-Infrared spectral library. Soil Sci. Soc. Am. J. 2010, 74, 1792–1799. [Google Scholar] [CrossRef] [Green Version]
- Viscarra Rossel, R.A.; Behrens, T. Using data mining to model and interpret soil diffuse reflectance spectra. Geoderma 2010, 158, 46–54. [Google Scholar] [CrossRef]
- Al Maliki, A.; Bruce, D.; Owens, G. Prediction of lead concentration in soil using reflectance spectroscopy. Environ. Sci. Technol. 2014, 1–2, 8–15. [Google Scholar] [CrossRef]
- Deiss, L.; Margenot, A.J.; Culman, S.W.; Demyan, M.S. Tuning support vector machines regression models improves prediction accuracy of soil properties in MIR spectroscopy. Geoderma 2020, 365, 114227. [Google Scholar] [CrossRef]
- Le Guillou, F.; Wetterlind, W.; Rossel, R.A.V.; Hicks, W.; Grundy, M.; Tuomi, S. How does grinding affect the mid-infrared spectra of soil and their multivariate calibrations to texture and organic carbon? Soil Res. 2015, 53, 913–921. [Google Scholar] [CrossRef]
- Janik, L.J.; Soriano-Disla, J.M.; Forrester, S.T.; McLaughlin, M.J. Effects of soil composition and preparation on the prediction of particle size distribution using mid-infrared spectroscopy and partial least-squares regression. Soil Res. 2016, 54, 889–904. [Google Scholar] [CrossRef]
- Agricultural Chemistry Committee of China. Conventional Methods of Soil and Agricultural Chemistry Analysis; Science Press: Beijing, China, 1983. (In Chinese) [Google Scholar]
- Nelson, D.; Sommers, L. Total nitrogen analysis of soil and plant tissues. JAOAC 1980, 63, 770–780. [Google Scholar] [CrossRef]
- Wang, D.M.; Ji, J.M.; Gao, H.Z. The effect of MSC spectral pretreatment regions on near infrared spectroscopy calibration results. Spectrosc. Spectr. Anal. 2014, 34, 2387–2390. [Google Scholar]
- Genkawa, T.; Shinzawa, H.; Kato, H.; Ishikawa, D.; Murayama, K.; Komiyama, M.; Ozaki, Y. Baseline correction of diffuse reflection near-infrared spectra using searching region standard normal variate (SRSNV). Appl. Spectrosc. 2015, 69, 1432–1441. [Google Scholar] [CrossRef]
- Kennard, R.W.; Stone, L.A. Computer aided design of experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Zou, H.M.; Li, X.C.; Shang, X.; Miao, C.H.; Huang, C.; Lu, J.H. Hyperspectral estimation of soil organic matter based on particle swarm optimization neural network. Sci. Surv. Mapp. 2019, 44, 146–150, 170. [Google Scholar]
- Briedis, C.; Baldock, J.; Sa, J.C.D.; dos Santos, J.B.; Milori, D.M.B.P. Strategies to improve the prediction of bulk soil and fraction organic carbon in Brazilian samples by using an Australian national mid-infrared spectral library. Geoderma 2020, 373, 114401. [Google Scholar] [CrossRef]
- Chang, C.W.; Laird, D.A.; Mausbach, M.J.; Hurburgh, C.R. Near-infrared reflectance spectroscopy-principal components regression analyses of soil properties. Soil Sci. Soc. Am. J. 2001, 65, 480–490. [Google Scholar] [CrossRef] [Green Version]
- Matějková, Š.; Šimon, T. Application of FTIR spectroscopy for evaluation of hydrophobic/hydrophilic organic components in arable soil. Plant Soil Environ. 2012, 58, 192–195. [Google Scholar] [CrossRef] [Green Version]
- Li, G.W.; Gao, X.H.; Xiao, N.W.; Xiao, Y.F. Estimation of soil organic matter content based on characteristic variable selection and regression methods. Acta Opt. Sin. 2019, 39, 361–371. [Google Scholar]
- Zheng, M.D.; Xiong, H.G.; Qiao, J.F.; Liu, J.C. Hyperspectral based estimation model about organic matter in desert soil at different levels of human disturbance. Arid Land Geogr. 2018, 41, 384–392. [Google Scholar]
- Jia, S.Y.; Tang, X.; Yang, X.L.; Li, G.; Zhang, J.M. Visible and near infrared spectroscopy combined with recursive variable selection to quantitatively determine soil total nitrogen and organic matter. Spectrosc. Spectr. Anal. 2014, 34, 2070–2075. [Google Scholar]

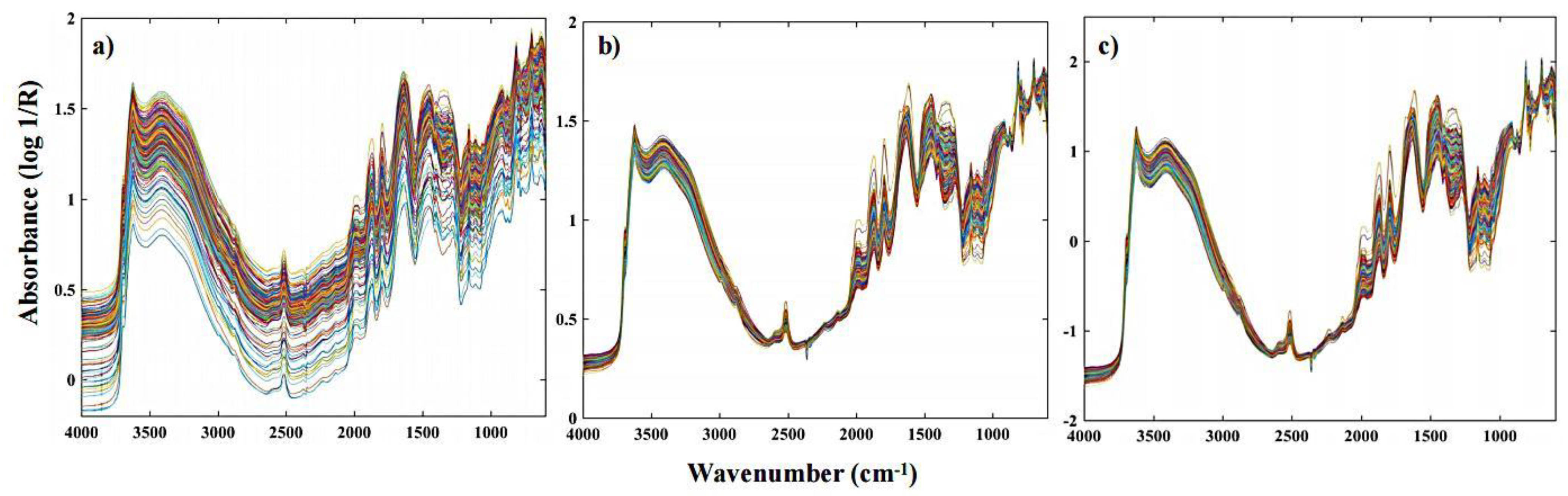

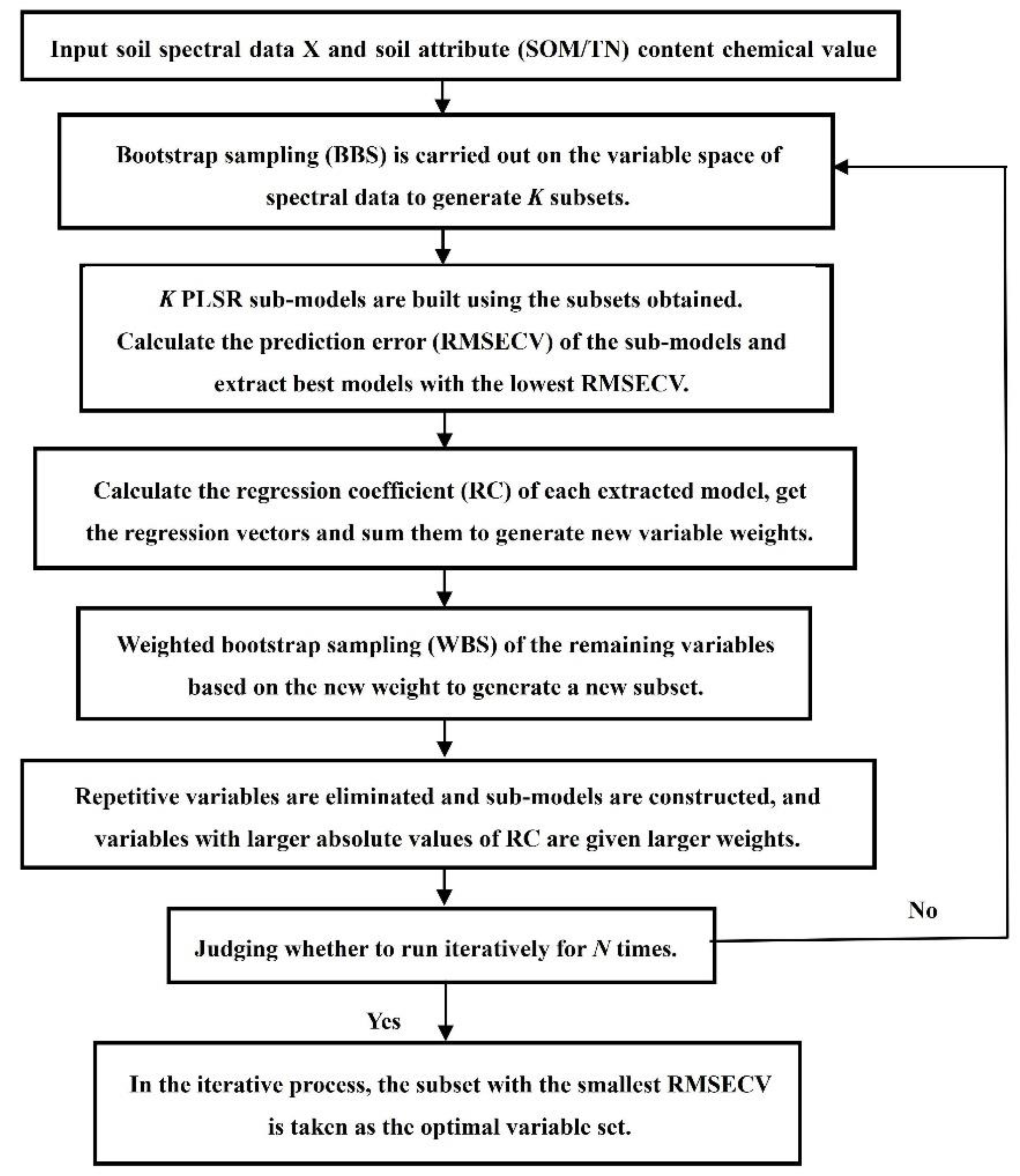
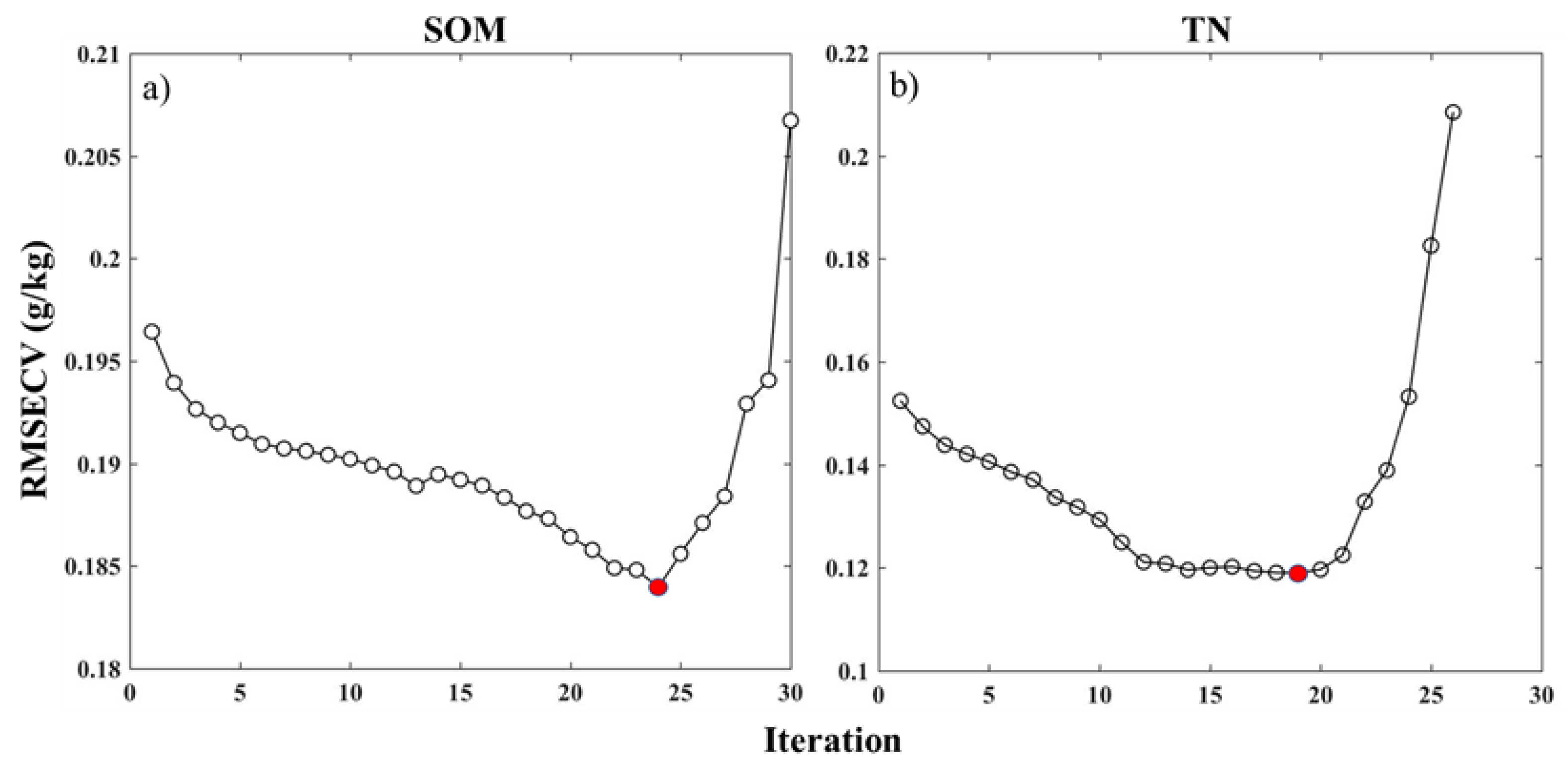
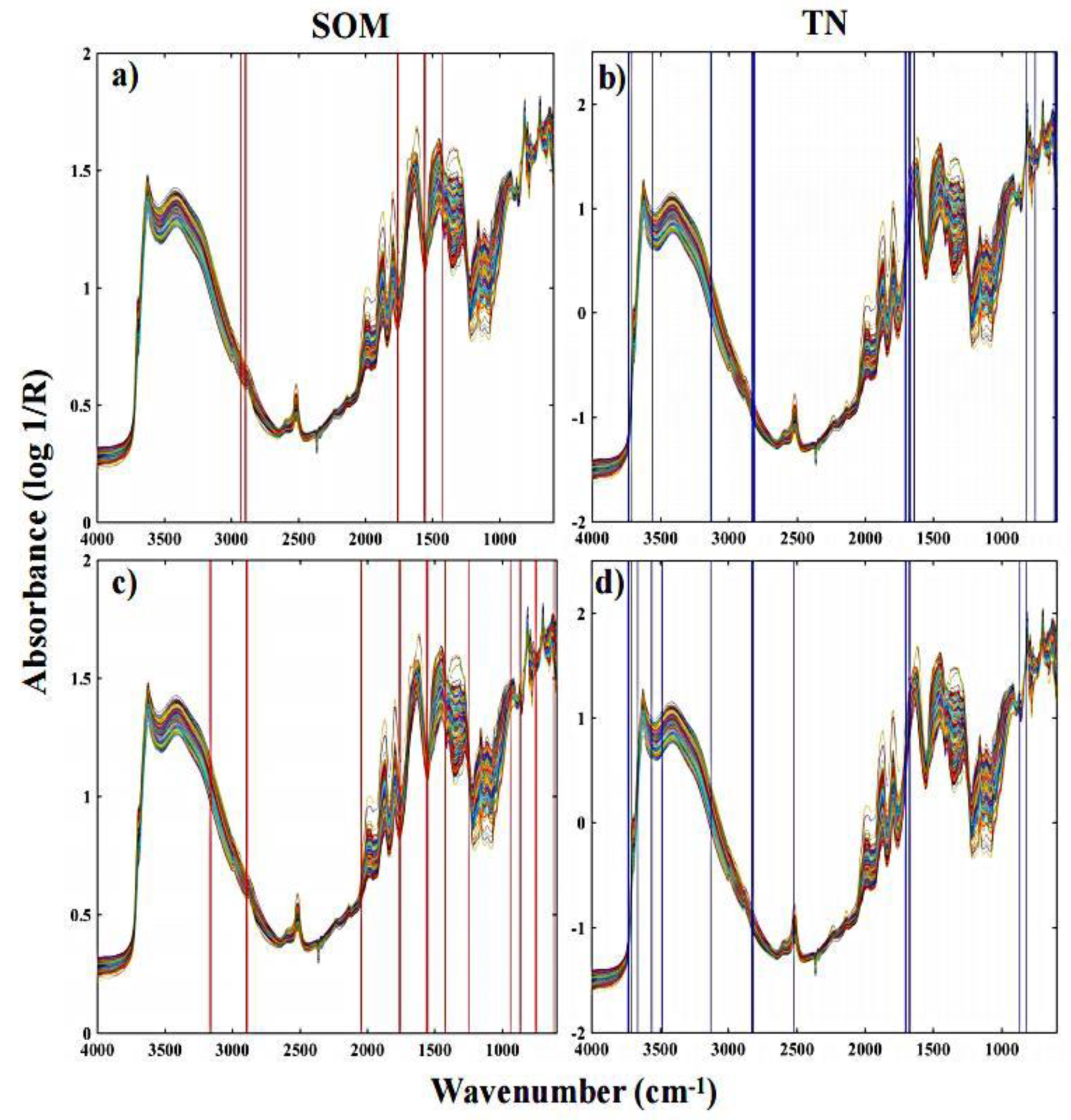
| Soil Property | Mean | Median | SD | Minimum | Maximum |
|---|---|---|---|---|---|
| SOM (g/kg) | 13.80 | 13.90 | 0.36 | 3.20 | 32.70 |
| TN (g/kg) | 0.96 | 0.96 | 0.26 | 0.21 | 2.43 |
| Soil Property | Model | Variable Number | Time (min) | Validation Set | ||
|---|---|---|---|---|---|---|
| RP2 | RPDp | RMSEP (g/kg) | ||||
| SOM | None-PLSR | 2382 | 2 | 0.68 | 1.77 | 0.17 |
| BOSS-PLSR | 14 | 200 | 0.68 | 1.77 | 0.17 | |
| BOSS-MLR | 14 | 3 | 0.69 | 1.78 | 0.16 | |
| sCARS-PLSR | 31 | 20 | 0.69 | 1.79 | 0.17 | |
| sCARS-MLR | 31 | 3 | 0.72 | 1.89 | 0.10 | |
| TN | None-PLSR | 2382 | 2 | 0.81 | 2.29 | 0.084 |
| BOSS-PLSR | 44 | 240 | 0.84 | 2.48 | 0.078 | |
| BOSS-MLR | 44 | 3 | 0.78 | 2.12 | 0.091 | |
| sCARS-PLSR | 27 | 20 | 0.84 | 2.50 | 0.076 | |
| sCARS-MLR | 27 | 3 | 0.84 | 2.50 | 0.077 | |
| Soil Property | Variable Selection Method | Variable Number | Characteristic Variable (cm−1) |
|---|---|---|---|
| SOM | BOSS | 14 | 2930,2928, 2894, 2893, 2891, 2890, 1760–1755, 1563, 1555, 1554, 1427 |
| sCARS | 31 | 3168, 3156, 2894–2888, 2045–2043, 1761–1755, 1561, 1560, 1555–1551, 1422, 1421, 1247, 938, 867–864, 754, 745, 617 | |
| TN | BOSS | 44 | 3736, 3733, 3728, 3707, 3560, 3130–3126, 2827–2818, 2810, 2808, 1707, 1705–1700, 1698, 1678–1665, 1641–1638, 817, 754, 617, 605, 604 |
| sCARS | 27 | 3733, 3728, 3727, 3707, 3661, 3560, 3558, 3484, 3483, 3127, 2825–2818, 2518, 1704–1700, 1675–1671, 867, 817 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Wang, J.; Zhang, J.; Liu, T.; Acquah, G.E.; Yuan, H. Combining Variable Selection and Multiple Linear Regression for Soil Organic Matter and Total Nitrogen Estimation by DRIFT-MIR Spectroscopy. Agronomy 2022, 12, 638. https://doi.org/10.3390/agronomy12030638
Li H, Wang J, Zhang J, Liu T, Acquah GE, Yuan H. Combining Variable Selection and Multiple Linear Regression for Soil Organic Matter and Total Nitrogen Estimation by DRIFT-MIR Spectroscopy. Agronomy. 2022; 12(3):638. https://doi.org/10.3390/agronomy12030638
Chicago/Turabian StyleLi, Hong, Junwei Wang, Jixiong Zhang, Tongqing Liu, Gifty E. Acquah, and Huimin Yuan. 2022. "Combining Variable Selection and Multiple Linear Regression for Soil Organic Matter and Total Nitrogen Estimation by DRIFT-MIR Spectroscopy" Agronomy 12, no. 3: 638. https://doi.org/10.3390/agronomy12030638
APA StyleLi, H., Wang, J., Zhang, J., Liu, T., Acquah, G. E., & Yuan, H. (2022). Combining Variable Selection and Multiple Linear Regression for Soil Organic Matter and Total Nitrogen Estimation by DRIFT-MIR Spectroscopy. Agronomy, 12(3), 638. https://doi.org/10.3390/agronomy12030638






