Spatiotemporal Assessment of Soil Organic Carbon Change Using Machine-Learning in Arid Regions
Abstract
:1. Introduction
2. Materials and Methods
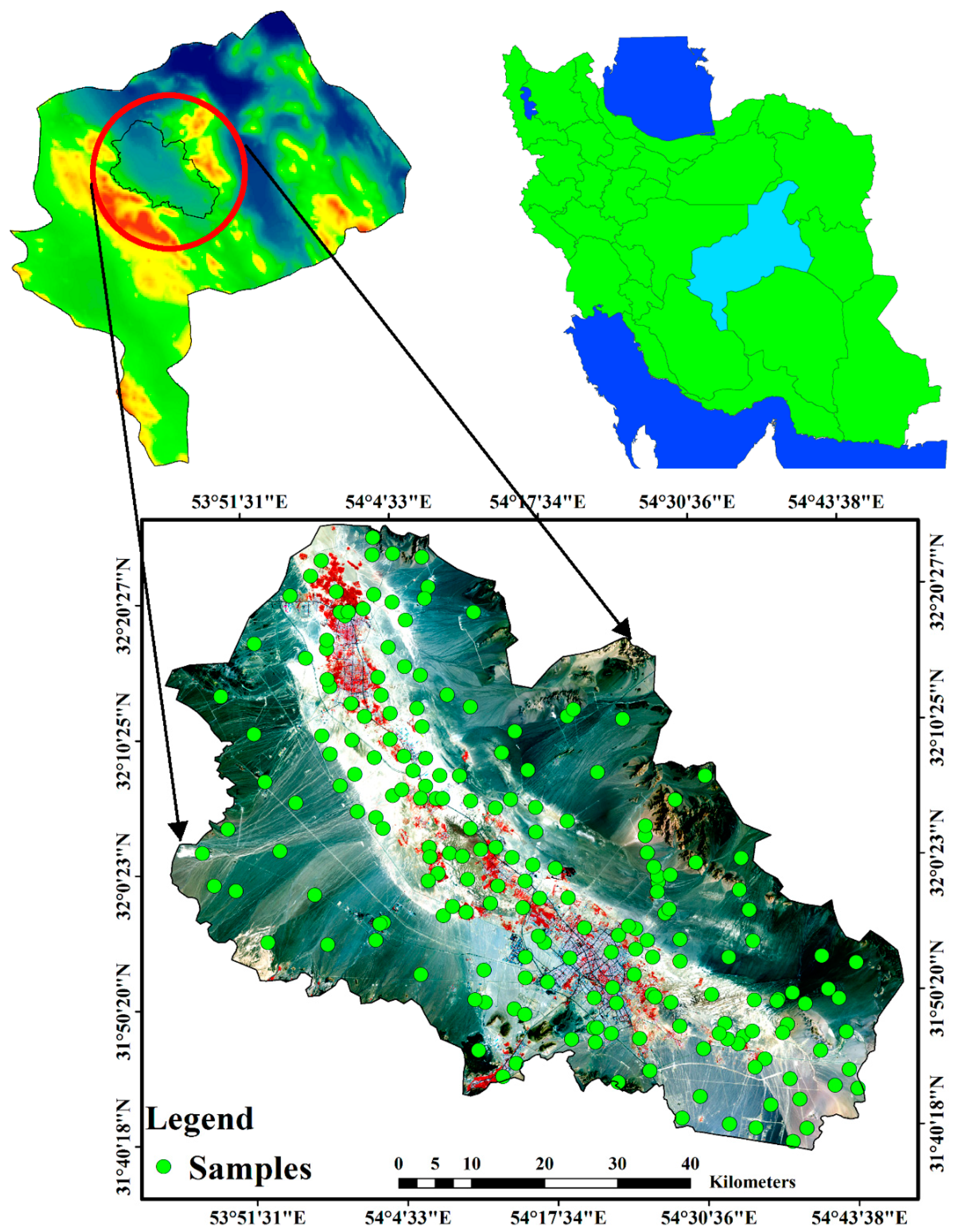
2.1. Study Area & Soil Sampling
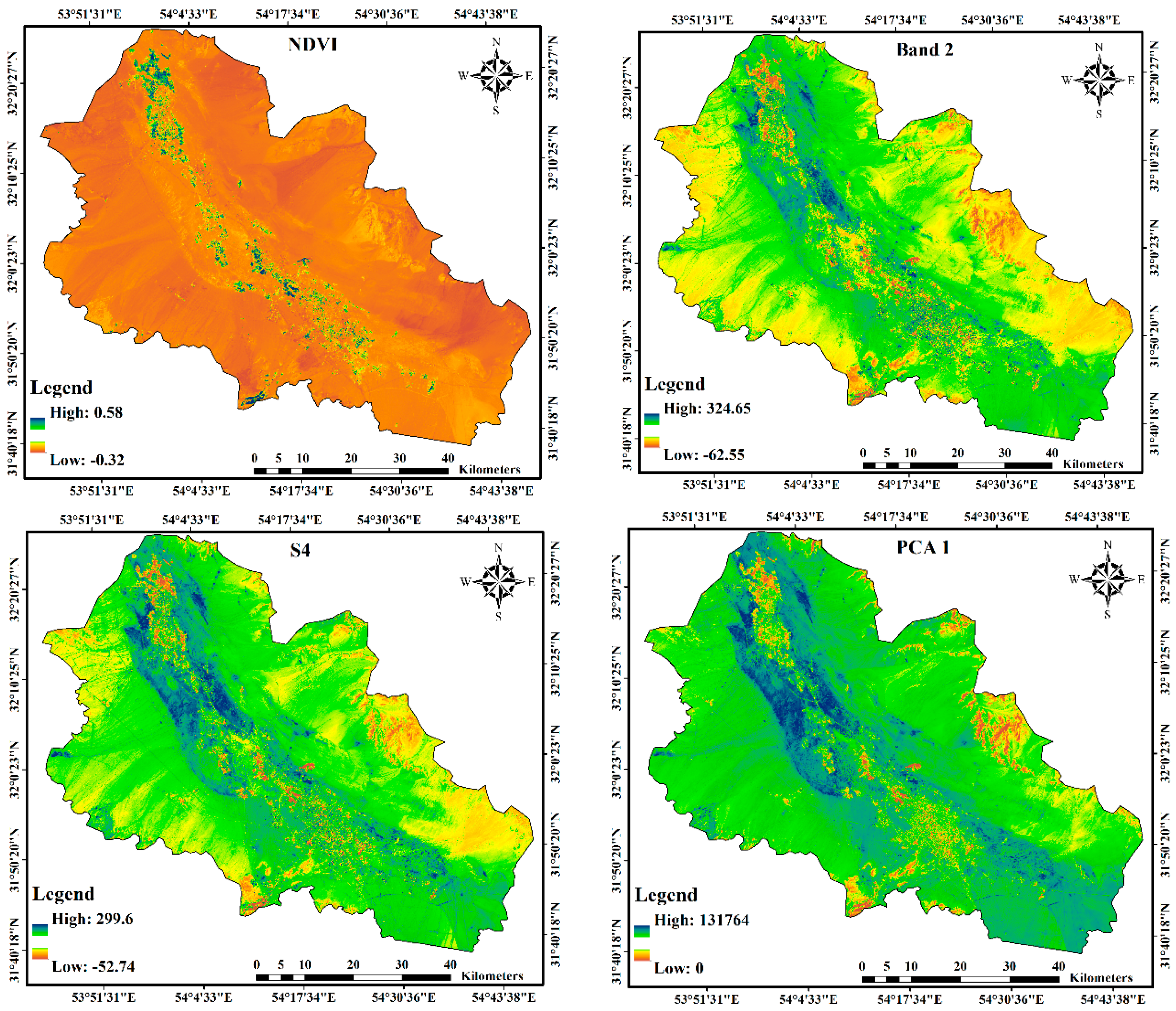
2.2. Environmental Predictors
2.3. Machine Learning
2.3.1. Random Forest (RF)
2.3.2. Support Vector Regression (SVR)
2.3.3. Artificial Neural Networks (ANN)
2.4. Accuracy and Uncertainty Assessment
3. Results and Discussion
3.1. Summary Statistics
3.2. Accuracy and Uncertainty Assessments
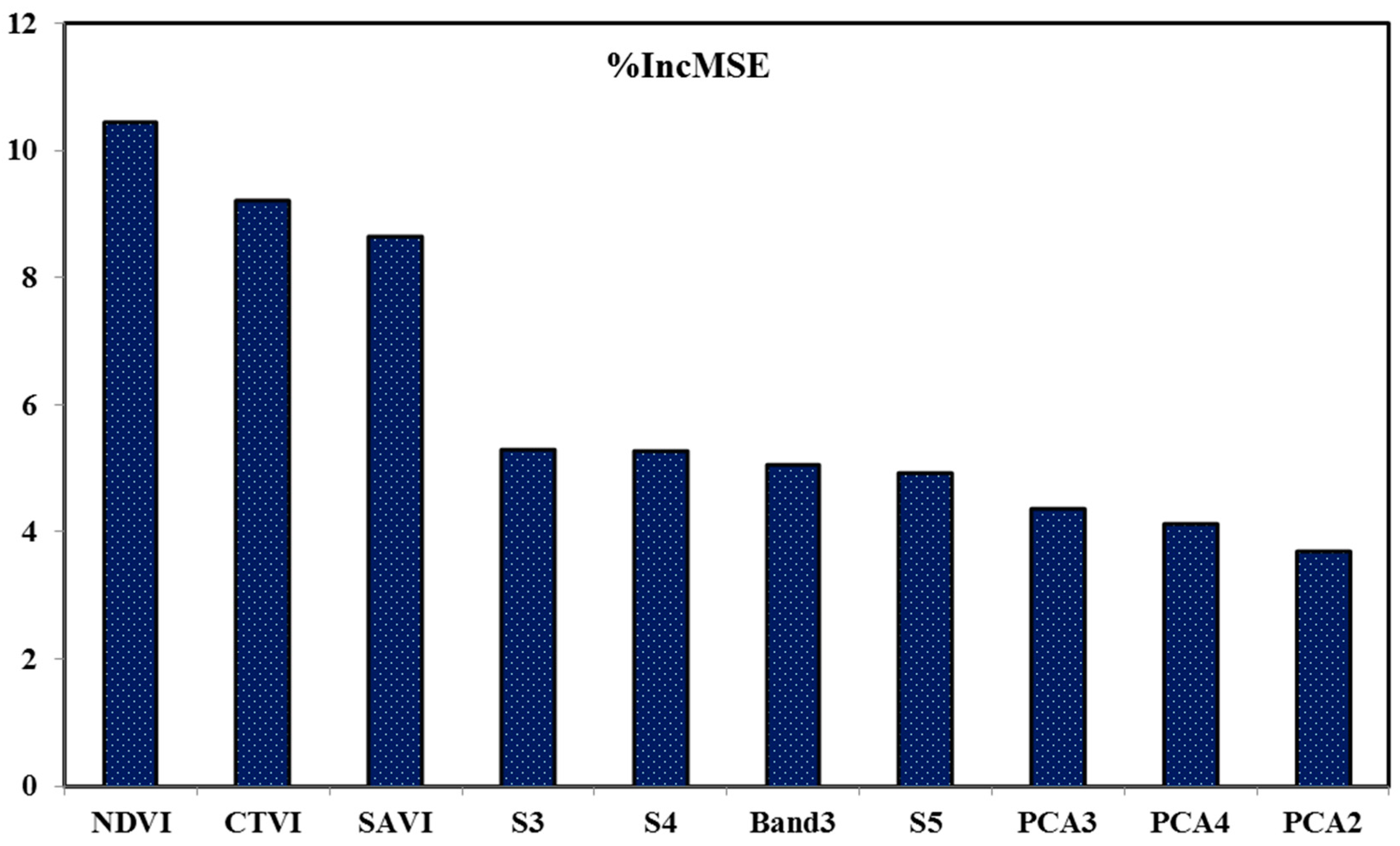
3.3. Variable Importance Analysis
3.4. Soil Organic Carbon Trends for 1986, 1999, 2010, and 2016
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Manlay, R.J.; Feller, C.; Swift, M. Historical evolution of soil organic matter concepts and their relationships with the fertility and sustainability of cropping systems. Agric. Ecosyst. Environ. 2007, 119, 217–233. [Google Scholar] [CrossRef]
- Ayoubi, S.; Karchegani, P.M.; Mosaddeghi, M.R.; Honarjoo, N. Soil aggregation and organic carbon as affected by topography and land use change in western Iran. Soil Tillage Res. 2012, 121, 18–26. [Google Scholar] [CrossRef]
- Malone, B.P.; Minasny, B.; McBratney, A.B. Using R for Digital Soil Mapping; Springer International Publishing: Cham, Switzerland, 2017. [Google Scholar]
- Guo, P.-T.; Li, M.-F.; Luo, W.; Tang, Q.-F.; Liu, Z.-W.; Lin, Z.-M. Digital mapping of soil organic matter for rubber plantation at regional scale: An application of random forest plus residuals kriging approach. Geoderma 2015, 237–238, 49–59. [Google Scholar] [CrossRef]
- Arrouays, D.; McBratney, A.; Minasny, B.; Hempel, J.; Heuvelink, G.; Macmillan, R.; Hartemink, A.; Lagacherie, P.; McKenzie, N. The GlobalSoilMap project specifications. GlobalSoilMap 2014, 494, 9–12. [Google Scholar] [CrossRef]
- Kempen, B.; Brus, D.; Stoorvogel, J. Three-dimensional mapping of soil organic matter content using soil type–specific depth functions. Geoderma 2011, 162, 107–123. [Google Scholar] [CrossRef] [Green Version]
- Zeraatpisheh, M.; Ayoubi, S.; Jafari, A.; Finke, P. Comparing the efficiency of digital and conventional soil mapping to predict soil types in a semi-arid region in Iran. Geomorphology 2017, 285, 186–204. [Google Scholar] [CrossRef]
- McBratney, A.; Santos, M.M.; Minasny, B. On digital soil mapping. Geoderma 2003, 117, 3–52. [Google Scholar] [CrossRef]
- Grunwald, S. Multi-criteria characterization of recent digital soil mapping and modeling approaches. Geoderma 2009, 152, 195–207. [Google Scholar] [CrossRef]
- Behrens, T.; Scholten, T. Chapter 25 A Comparison of Data-Mining Techniques in Predictive Soil Mapping. Dev. Soil Sci. 2006, 31, 353–617. [Google Scholar] [CrossRef]
- Zhu, A.X.; Liu, J.; Du, F.; Zhang, S.J.; Qin, C.-Z.; Burt, J.; Behrens, T.; Scholten, T. Predictive soil mapping with limited sample data. Eur. J. Soil Sci. 2015, 66, 535–547. [Google Scholar] [CrossRef]
- Stumpf, F.; Schmidt, K.; Goebes, P.; Behrens, T.; Schönbrodt-Stitt, S.; Wadoux, A.; Xiang, W.; Scholten, T. Uncertainty-guided sampling to improve digital soil maps. Catena 2017, 153, 30–38. [Google Scholar] [CrossRef]
- Taghizadeh, R.; Nabiollahi, K.; Kerry, R. Digital mapping of soil organic carbon at multiple depths using different data mining techniques in Baneh region, Iran. Geoderma 2016, 266, 98–110. [Google Scholar] [CrossRef]
- Taghizadeh, R.; Toomanian, N.; Khavaninzadeh, A.R.; Jafari, A.; Triantafilis, J. Predicting and mapping of soil particle-size fractions with adaptive neuro-fuzzy inference and ant colony optimization in central Iran. Eur. J. Soil Sci. 2016, 67, 707–725. [Google Scholar] [CrossRef]
- Taghizadeh, R.; Minasny, B.; Sarmadian, F.; Malone, B. Digital mapping of soil salinity in Ardakan region, central Iran. Geoderma 2014, 213, 15–28. [Google Scholar] [CrossRef]
- Liu, D.; Wang, Z.; Zhang, B.; Song, K.; Li, X.; Li, J.; Li, F.; Duan, H. Spatial distribution of soil organic carbon and analysis of related factors in croplands of the black soil region, Northeast China. Agric. Ecosyst. Environ. 2006, 113, 73–81. [Google Scholar] [CrossRef]
- Chai, X.; Shen, C.; Yuan, X.; Huang, Y. Spatial prediction of soil organic matter in the presence of different external trends with REML-EBLUP. Geoderma 2008, 148, 159–166. [Google Scholar] [CrossRef]
- Sumfleth, K.; Duttmann, R. Prediction of soil property distribution in paddy soil landscapes using terrain data and satellite information as indicators. Ecol. Indic. 2008, 8, 485–501. [Google Scholar] [CrossRef]
- Pahlavan-Rad, M.R.; Dahmardeh, K.; Brungard, C. Predicting soil organic carbon concentrations in a low relief landscape, eastern Iran. Geoderma Reg. 2018, 15, e00195. [Google Scholar] [CrossRef]
- Zeraatpisheh, M.; Jafari, A.; Bodaghabadi, M.B.; Ayoubi, S.; Taghizadeh-Mehrjardi, R.; Toomanian, N.; Kerry, R.; Xu, M. Conventional and digital soil mapping in Iran: Past, present, and future. Catena 2019, 188, 104424. [Google Scholar] [CrossRef]
- McBratney, A.B.; Stockmann, U.; Angers, D.A.; Minasny, B.; Field, D.J. Challenges for Soil Organic Carbon Research. In Soil Carbon; Springer: Cham, Switzerland, 2014; pp. 3–16. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, G.-L.; Sun, Y.-J.; Zhao, Y.-G.; Li, D.-C. Mapping the Three-Dimensional Distribution of Soil Organic Matter across a Subtropical Hilly Landscape. Soil Sci. Soc. Am. J. 2013, 77, 1241–1253. [Google Scholar] [CrossRef]
- Fathizad, H.; Ardakani, M.A.H.; Sodaiezadeh, H.; Kerry, R.; Taghizadeh-Mehrjardi, R. Investigation of the spatial and temporal variation of soil salinity using random forests in the central desert of Iran. Geoderma 2020, 365, 114233. [Google Scholar] [CrossRef]
- Taghizadeh-Mehrjardi, R.; Fathizad, H.; Ardakani, M.A.H.; Sodaiezadeh, H.; Kerry, R.; Heung, B.; Scholten, T. Spatio-Temporal Analysis of Heavy Metals in Arid Soils at the Catchment Scale Using Digital Soil Assessment and a Random Forest Model. Remote Sens. 2021, 13, 1698. [Google Scholar] [CrossRef]
- Minasny, B.; McBratney, A. A conditioned Latin hypercube method for sampling in the presence of ancillary information. Comput. Geosci. 2006, 32, 1378–1388. [Google Scholar] [CrossRef]
- Walkley, A.; Black, I.A. An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef] [Green Version]
- Huete, A. Extension of soil spectra to the satellite: Atmosphere, geometric, and sensor considerations. Photo Interprétation 1996, 34, 101–118. [Google Scholar]
- Leblon, B. Soil and vegetation optical properties. Faculty of Forestry and Environmental Management University of New Brunswick, Fredericton (NB) Canada. Int. J. For. Eng. 1997, 13, 57. [Google Scholar]
- Crippen, R. Calculating the vegetation index faster. Remote Sens. Environ. 1990, 34, 71–73. [Google Scholar] [CrossRef]
- Foody, G.M.; Cutler, M.; McMorrow, J.; Pelz, D.; Tangki, H.; Boyd, D.; Douglas, I. Mapping the biomass of Bornean tropical rain forest from remotely sensed data. Glob. Ecol. Biogeogr. 2001, 10, 379–387. [Google Scholar] [CrossRef]
- Nield, S.J.; Boettinger, J.L.; Ramsey, R.D. Digitally Mapping Gypsic and Natric Soil Areas Using Landsat ETM Data. Soil Sci. Soc. Am. J. 2007, 71, 245–252. [Google Scholar] [CrossRef] [Green Version]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Arzani, H.; King, G.W. Application of Remote Sensing (Landsat TM Data) for Vegetation Parameters Measurement in Western Division of NSW; International Grassland Congress: Hohhot, China, 2008. [Google Scholar]
- Jordan, C.F. Derivation of Leaf-Area Index from Quality of Light on the Forest Floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Pettorelli, N.; Vik, J.O.; Mysterud, A.; Gaillard, J.-M.; Tucker, C.J.; Stenseth, N.C. Using the satellite-derived NDVI to assess ecological responses to environmental change. Trends Ecol. Evol. 2005, 20, 503–510, Erratum in Trends Ecol. Evol. 2006, 21, 11. [Google Scholar] [CrossRef]
- Kullberg, E.G.; DeJonge, K.C.; Chávez, J.L. Evaluation of thermal remote sensing indices to estimate crop evapotranspiration coefficients. Agric. Water Manag. 2017, 179, 64–73. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.M.; Rastoskuev, V.V.; Sato, Y.; Shiozawa, S. Assessment of hydrosaline land degradation by using a simple approach of remote sensing indicators. Agric. Water Manag. 2005, 77, 96–109. [Google Scholar] [CrossRef]
- Wilson, E.H.; Sader, S.A. Detection of forest harvest type using multiple dates of Landsat TM imagery. Remote Sens. Environ. 2002, 80, 385–396. [Google Scholar] [CrossRef]
- Major, D.J.; Baret, F.; Guyot, G. A ratio vegetation index adjusted for soil brightness. Int. J. Remote Sens. 1990, 11, 727–740. [Google Scholar] [CrossRef]
- Bannari, A.; Guedon, A.M.; El-Harti, A.; Cherkaoui, F.Z.; El-Ghmari, A. Characterization of Slightly and Moderately Saline and Sodic Soils in Irrigated Agricultural Land using Simulated Data of Advanced Land Imaging (EO-1) Sensor. Commun. Soil Sci. Plant Anal. 2008, 39, 2795–2811. [Google Scholar] [CrossRef]
- Taghizadeh-Mehrjardi, R.; Schmidt, K.; Toomanian, N.; Heung, B.; Behrens, T.; Mosavi, A.; Band, S.B.; Amirian-Chakan, A.; Fathabadi, A.; Scholten, T. Improving the spatial prediction of soil salinity in arid regions using wavelet transformation and support vector regression models. Geoderma 2021, 383, 114793. [Google Scholar] [CrossRef]
- Douaoui, A.E.K.; Nicolas, H.; Walter, C. Detecting salinity hazards within a semiarid context by means of combining soil and remote-sensing data. Geoderma 2006, 134, 217–230. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Nabiollahi, K.; Taghizadeh-Mehrjardi, R.; Shahabi, A.; Heung, B.; Amirian-Chakan, A.; Davari, M.; Scholten, T. Assessing agricultural salt-affected land using digital soil mapping and hybridized random forests. Geoderma 2021, 385, 114858. [Google Scholar] [CrossRef]
- Taghizadeh-Mehrjardi, R.; Hamzehpour, N.; Hassanzadeh, M.; Heung, B.; Goydaragh, M.G.; Schmidt, K.; Scholten, T. Enhancing the accuracy of machine learning models using the super learner technique in digital soil mapping. Geoderma 2021, 399, 115108. [Google Scholar] [CrossRef]
- Vapnik, V.N.; Lerner, A. Pattern recognition using generalized portrait method. Autom. Remote Control. 1963, 24, 774–780. [Google Scholar]
- Smola, A.J.; Schölkopf, B. A tutorial on support vector regression. Stat. Comput. 2004, 14, 199–222. [Google Scholar] [CrossRef] [Green Version]
- Ripley, B.D. Pattern Recognition and Neural Networks; Cambridge University Press: Cambridge, NY, USA, 1966. [Google Scholar]
- Tajik, S.; Ayoubi, S.; Zeraatpisheh, M. Digital mapping of soil organic carbon using ensemble learning model in Mollisols of Hyrcanian forests, northern Iran. Geoderma Reg. 2020, 20, e00256. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Q.; Adhikari, K.; Jia, S.; Jin, X.; Liu, H. Spatial-Temporal Changes of Soil Organic Carbon Content in Wafangdian, China. Sustainability 2016, 8, 1154. [Google Scholar] [CrossRef] [Green Version]
- Lamichhane, S.; Kumar, L.; Wilson, B. Digital soil mapping algorithms and covariates for soil organic carbon mapping and their implications: A review. Geoderma 2019, 352, 395–413. [Google Scholar] [CrossRef]
- Gholizadeh, A.; Žižala, D.; Saberioon, M.; Borůvka, L. Soil organic carbon and texture retrieving and mapping using proximal, airborne and Sentinel-2 spectral imaging. Remote Sens. Environ. 2018, 218, 89–103. [Google Scholar] [CrossRef]
- Falahatkar, S.; Hosseini, S.M.; Ayoubi, S.; Salmanmahiny, A. Predicting soil organic carbon density using auxiliary environmental variables in northern Iran. Arch. Agron. Soil Sci. 2015, 62, 375–393. [Google Scholar] [CrossRef]
- Hong, S.Y.; Sudduth, K.A.; Kitchen, N.R.; Drummond, S.T.; Palm, H.L.; Wiebold, W.J. Estimating within-field variations in soil properties from airborne hyperspectral images. In Proceedings of the Pecora 15/Land Satellite Information IV/ISPRS Commission I/FIEOS 2002, Denver, CO, USA, 10–15 November 2002. [Google Scholar]
- Gomez, C.; Rossel, R.V.; McBratney, A. Soil organic carbon prediction by hyperspectral remote sensing and field vis-NIR spectroscopy: An Australian case study. Geoderma 2008, 146, 403–411. [Google Scholar] [CrossRef]
- Zhao, M.-S.; Zhang, G.-L.; Wu, Y.-J.; Li, D.-C.; Zhao, Y.-G. Driving forces of soil organic matter change in Jiangsu Province of China. Soil Use Manag. 2015, 31, 440–449. [Google Scholar] [CrossRef]



| Covariates | Definition | Reference |
|---|---|---|
| Difference vegetation index (DVI) | NIR–Red | [27] |
| Enhanced vegetation index (EVI) | G × (NIR–Red)/(NIR + c1 × Red–c2 × Blue + L) | [28] |
| Global vegetation index (GVI) | −0.29 × (G) − 0.56 × (Red) + 0.6 (IR) + 0.49 (NIR) | [29] |
| Infrared percentage vegetation index (IPVI) | NIR/(NIR + Red) | [30] |
| Normalized difference vegetation index (NDVI) | (Red–NIR)/(Red + NIR) | [31] |
| Blue | Reflectance value of Landsat satellite band | Landsat satellite |
| Green | Reflectance value of Landsat satellite band | Landsat satellite |
| Red | Reflectance value of Landsat satellite band | Landsat satellite |
| Near-infrared (NIR) | Reflectance value of Landsat satellite band | Landsat satellite |
| Shortwave infrared (SWIR) | Reflectance value of Landsat satellite band | Landsat satellite |
| Principal components of Landsat bands | PC1, PC2, PC3, and PC4 | [32] |
| Normalized-NDVI | (NIR–(TM1 + Green))/(NIR + (TM1 + Green)) | [31] |
| Optimized soil-adjusted vegetation index (OSAVI) | (NIR–Red)/(NIR + Red + 0.16) | [33] |
| PD 311 | Red–TM1 | [34] |
| PD 312 | (Red–Blue)/(Red + TM1) | [34] |
| PD 321 | Red–Green | [34] |
| PD 322 | (Red–Green)/(Red + Green) | [34] |
| Ratio-Based | NIR/(Blue + Green) | [31] |
| Ratio vegetation index (RVI) | (NIR/Red) | [35] |
| Soil-adjusted vegetation index (SAVI) | [NIR–Red)/(NIR + Red + L)] × (1 + L) | [28] |
| Stress-related | (TM1 × Green)/Red | [31] |
| Transformed vegetation index (TVI) | (SWIR–Red)/(SWIR + Red) | [36] |
| VIT01 | Red/Thermal | [37] |
| VTI02 | Thermal/(Red + SWIR) | [37] |
| VIT03 | Thermal/Red | [37] |
| VIT04 | Thermal/(SWIR + Green) | [37] |
| Brightness index | BI = ((Red × Red) + (NIR × NIR))^0.5 | [38] |
| Normalized difference moisture index (NDMI) | NDMI = (NIR − SWIR)/(NIR + SWIR) | [39] |
| Normalized difference snow index (NDSI) | NDSI = (Red − NIR)/(Red + NIR) | [40] |
| Salinity index1 (S1) | S1 = Blue/Red | [41] |
| Salinity index2 (S2) | S2 = (Blue − Red)/(Blue + Red) | [42] |
| Salinity index3 (S3) | S3 = (Green × Red)/Blue | [42] |
| Salinity index4 (S4) | S4 = (Blue × Red)/Green | [42] |
| Salinity index5 (S5) | S5 = (Red × NIR)/Green | [42] |
| Salinity index6 (S6) | S6 = (Blue × Red)^0.5 | [38] |
| Salinity index7 (S7) | S7 = (Green × Red)^0.5 | [43] |
| Salinity index8 (S8) | S8 = (Blue2 × Green2 × Red2)^0.5 | [43] |
| Index | PCC | Index | PCC |
|---|---|---|---|
| CTVI | 0.37 ** | S6 | −0.09 |
| DVI | 0.38 ** | S7 | −0.07 |
| EVI | −0.37 ** | S8 | 0.27 ** |
| GVI | 0.33 ** | SAVI | 0.37 ** |
| IPVI | −0.11 | Stress-related | −0.04 |
| NDSI | −0.37 ** | TTVI | 0.37 ** |
| NDVI | 0.37 ** | TVI | 0.37 ** |
| NIR | 0.36 ** | VIT01 | −0.06 |
| OSAVI | 0.37 ** | VIT02 | −0.14 * |
| PD311 | −0.09 | VIT03 | 0.12 |
| PD312 | −0.12 | VIT04 | −0.08 |
| PD321 | −0.15 * | PCA1 | −0.16 * |
| PD322 | −0.20 ** | PCA2 | 0.03 |
| Ratio Based | −0.08 | PCA3 | −0.31 ** |
| RVI | −0.37 ** | PCA4 | 0.26 ** |
| S1 | 0.13 | Band1 | −0.11 |
| S2 | 0.12 | Band2 | −0.10 |
| S3 | −0.08 | Band3 | −0.08 |
| S4 | −0.14 * | Band4 | −0.10 |
| S5 | 0.07 |
| Model | R2 | RMSE | MAE | MPI | PICP |
|---|---|---|---|---|---|
| RF | 0.536 | 0.082 | 0.058 | 0.14 | 81 |
| MLP | 0.501 | 0.142 | 0.110 | 0.19 | 75 |
| SVR | 0.407 | 0.181 | 0.134 | 0.21 | 67 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fathizad, H.; Taghizadeh-Mehrjardi, R.; Hakimzadeh Ardakani, M.A.; Zeraatpisheh, M.; Heung, B.; Scholten, T. Spatiotemporal Assessment of Soil Organic Carbon Change Using Machine-Learning in Arid Regions. Agronomy 2022, 12, 628. https://doi.org/10.3390/agronomy12030628
Fathizad H, Taghizadeh-Mehrjardi R, Hakimzadeh Ardakani MA, Zeraatpisheh M, Heung B, Scholten T. Spatiotemporal Assessment of Soil Organic Carbon Change Using Machine-Learning in Arid Regions. Agronomy. 2022; 12(3):628. https://doi.org/10.3390/agronomy12030628
Chicago/Turabian StyleFathizad, Hassan, Ruhollah Taghizadeh-Mehrjardi, Mohammad Ali Hakimzadeh Ardakani, Mojtaba Zeraatpisheh, Brandon Heung, and Thomas Scholten. 2022. "Spatiotemporal Assessment of Soil Organic Carbon Change Using Machine-Learning in Arid Regions" Agronomy 12, no. 3: 628. https://doi.org/10.3390/agronomy12030628
APA StyleFathizad, H., Taghizadeh-Mehrjardi, R., Hakimzadeh Ardakani, M. A., Zeraatpisheh, M., Heung, B., & Scholten, T. (2022). Spatiotemporal Assessment of Soil Organic Carbon Change Using Machine-Learning in Arid Regions. Agronomy, 12(3), 628. https://doi.org/10.3390/agronomy12030628









