Evaluation of Methods for Measuring Fusarium-Damaged Kernels of Wheat
Abstract
:1. Introduction
2. Materials and Methods
2.1. Summary of Regional Trials
2.1.1. Florence, South Carolina 2019 and 2020 (FSC19, FSC20) and Winnsboro Louisiana 2020 (WLA)
2.1.2. Mt. Holly, Virginia (MtVA19)
2.2. Creation of Phenotypic Dataset
2.2.1. Harvest and Threshing
2.2.2. Manual Separation with Electric Counting (MANUAL)
2.2.3. Visual Estimates (VISUAL)
2.2.4. Near-Infrared Spectroscopy (NIR)
2.2.5. Vibe QM3 Grain Analyzer (VIBE)
2.2.6. Deoxynivalenol (DON) Analysis
2.3. Genotypic Data
2.4. Phenotypic Analysis
2.5. Genetic Correlations between FDK Platforms and DON
2.6. Genome-Wide Association Studies
2.7. Genomic Prediction
3. Results
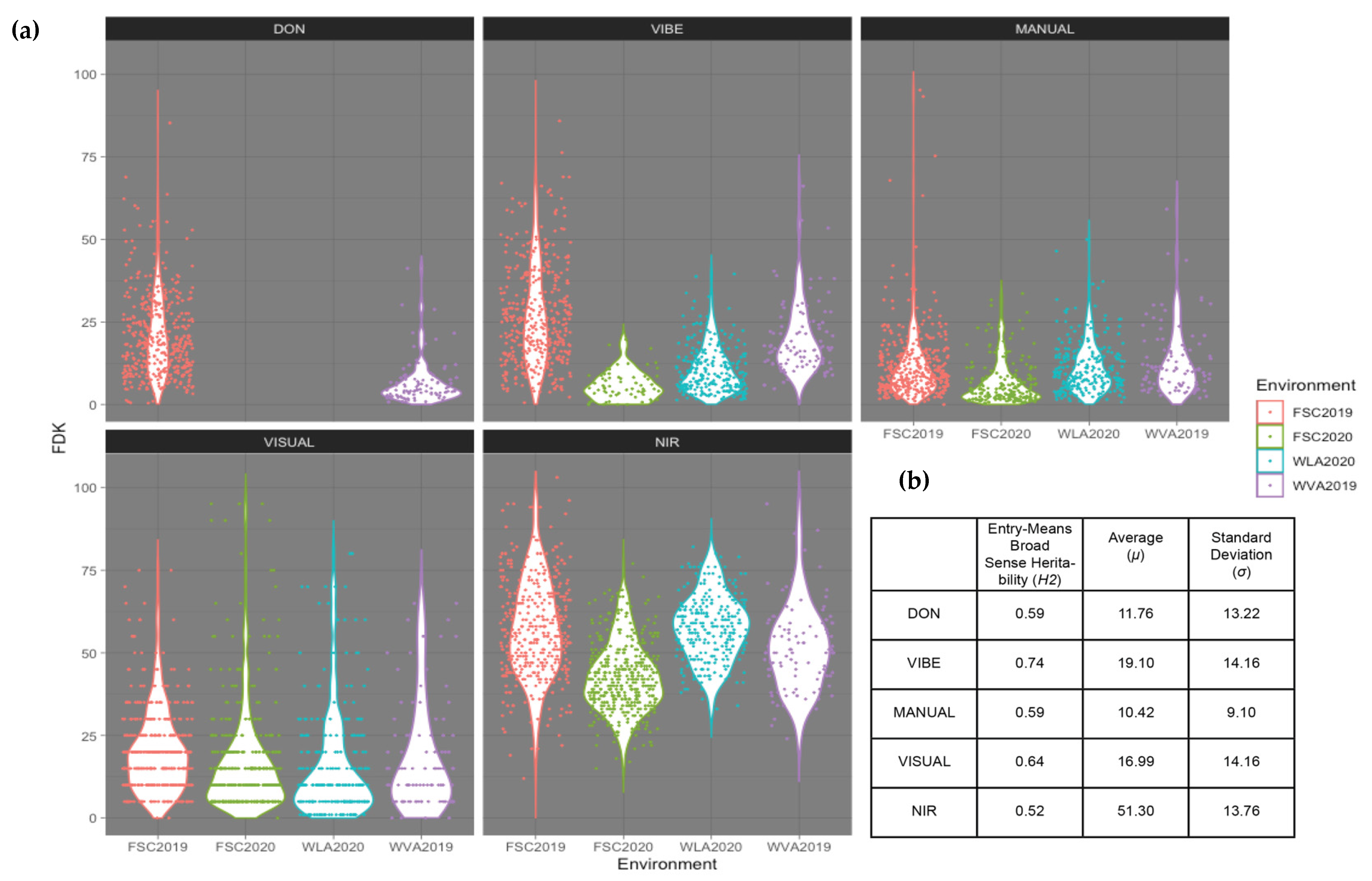
3.1. Variation among FDK Platforms and DON Content
3.2. Vibe Demonstrates Strongest Association with DON Content
3.2.1. Raw Phenotypic Data Correlation with DON
3.2.2. Genetic Correlations among Platforms
3.2.3. FDK Platforms Ability to Predict DON Resistance
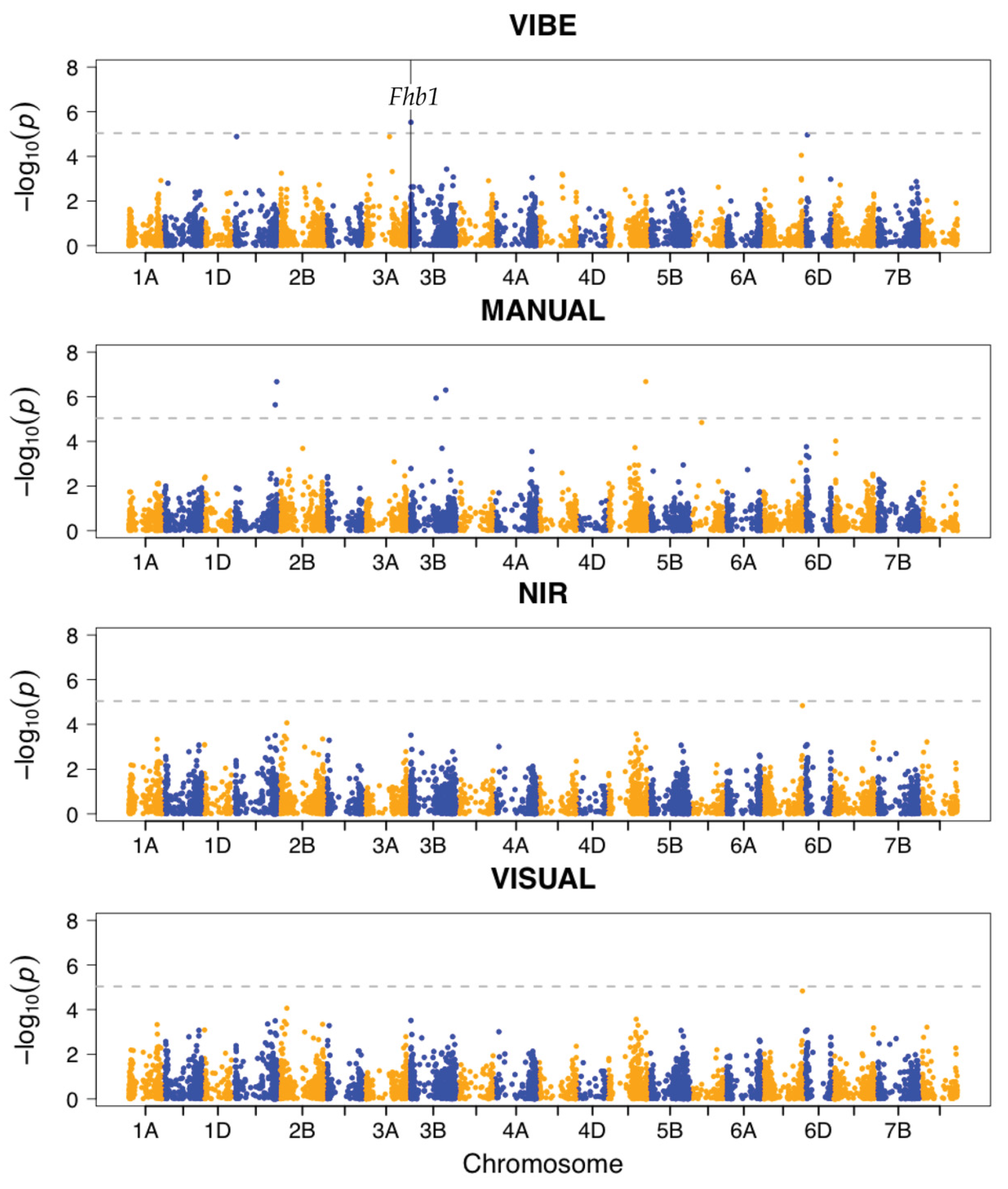
3.3. VIBE Detects Significant SNP Association in Fhb1
3.4. VISUAL and VIBE Show Highest Prediction Accuracy among FDK Platforms
4. Discussion
- Minimization of cost and the ability to uncover maximum FDK heritability.
- Maximization of the genetic relationship of the FDK trait and DON resistance.
- Accurate prediction of DON content resulting in effective use as a proxy phenotype.
- Ability to detect associations within major FHB resistance QTL known to be within sampling populations that control variation for FDK and DON: Fhb1, F1BJ, F1AN.
- Optimize the FDK trait to provide increased accuracy in genomic prediction models.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Balut, A.L.; Clark, A.J.; Brown-Guedira, G.; Souza, E.; Sanford, D.A.V. Validation of Fhb1 and QFhs.Nau-2DL in Several Soft Red Winter Wheat Populations. Crop Sci. 2013, 53, 934–945. [Google Scholar] [CrossRef] [Green Version]
- Cowger, C.; Weisz, R.; Arellano, C.; Murphy, P. Profitability of Integrated Management of Fusarium Head Blight in North Carolina Winter Wheat. Phytopathology 2016, 106, 814–823. [Google Scholar] [CrossRef] [Green Version]
- Carter, J.P.; Rezanoor, H.N.; Holden, D.; Desjardins, A.E.; Plattner, R.D.; Nicholson, P. Variation in Pathogenicity Associated with the Genetic Diversity of Fusarium Graminearum. Eur. J. Plant Pathol. 2002, 108, 573–583. [Google Scholar] [CrossRef]
- Aviles, A.C.; Harrison, S.A.; Arceneaux, K.J.; Brown-Guidera, G.; Mason, R.E.; Baisakh, N. Identification of QTLs for Resistance to Fusarium Head Blight Using a Doubled Haploid Population Derived from Southeastern United States Soft Red Winter Wheat Varieties AGS 2060 and AGS 2035. Genes 2020, 11, 699. [Google Scholar] [CrossRef] [PubMed]
- Buerstmayr, M.; Steiner, B.; Buerstmayr, H. Breeding for Fusarium Head Blight Resistance in Wheat—Progress and Challenges. Plant Breed. 2020, 139, 429–454. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Ban, T.; Anderson, J.A. QTL Mapping and Marker-Assisted Selection for Fusarium Head Blight Resistance in Wheat: A Review. Plant Breed. 2009, 128, 1–26. [Google Scholar] [CrossRef]
- Mesterhazy, A. Updating the Breeding Philosophy of Wheat to Fusarium Head Blight (FHB): Resistance Components, QTL Identification, and Phenotyping—A Review. Plants 2020, 9, 1702. [Google Scholar] [CrossRef] [PubMed]
- Mesterhazy, A. Types and Components of Resistance to Fusarium Head Blight of Wheat. Plant Breed. 1995, 114, 377–386. [Google Scholar] [CrossRef]
- Hales, B.; Steed, A.; Giovannelli, V.; Burt, C.; Lemmens, M.; Molnár-Láng, M.; Nicholson, P. Type II Fusarium Head Blight Susceptibility Conferred by Region on Wheat Chromosome 4D. J. Exp. Bot. 2020, 71, 4703–4714. [Google Scholar] [CrossRef]
- Schroeder, H.W.; Christensen, J.J. Factors Affecting Resistance of Wheat to Scab Caused by Gibberella Zeae. Phytopathology 1963, 53, 831–838. [Google Scholar]
- Khaeim, H.M.; Clark, A.; Pearson, T.; Sanford, D.D.V. Methods of Assessing Fusarium Damage to Wheat Kernels. Al-Qadisiyah J. Agric. Sci. QJAS 2019, 9, 297–308. [Google Scholar] [CrossRef]
- Góral, T.; Wiśniewska, H.; Ochodzki, P.; Nielsen, L.; Walentyn-Góral, D.; Stępień, Ł. Relationship between Fusarium Head Blight, Kernel Damage, Concentration of Fusarium Biomass, and Fusarium Toxins in Grain of Winter Wheat Inoculated with Fusarium Culmorum. Toxins 2018, 11, 2. [Google Scholar] [CrossRef] [Green Version]
- Yi, X.; Cheng, J.; Jiang, Z.; Hu, W.; Bie, T.; Gao, D.; Li, D.; Wu, R.; Li, Y.; Chen, S.; et al. Genetic Analysis of Fusarium Head Blight Resistance in CIMMYT Bread Wheat Line C615 Using Traditional and Conditional QTL Mapping. Front. Plant Sci. 2018, 9, 573. [Google Scholar] [CrossRef]
- Gunupuru, L.R.; Perochon, A.; Doohan, F.M. Deoxynivalenol Resistance as a Component of FHB Resistance. Trop. Plant Pathol. 2017, 42, 175–183. [Google Scholar] [CrossRef] [Green Version]
- Paul, P.A.; Lipps, P.E.; Madden, L.V. Relationship between Visual Estimates of Fusarium Head Blight Intensity and Deoxynivalenol Accumulation in Harvested Wheat Grain: A Meta-Analysis. Phytopathology 2005, 95, 1225–1236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaire, R.; Brown-Guedira, G.; Dong, Y.; Ohm, H.; Mohammadi, M. Genome-Wide Association Studies for Fusarium Head Blight Resistance and Its Trade-Off with Grain Yield in Soft Red Winter Wheat. Plant Dis. 2021, 105, 2435–2444. [Google Scholar] [CrossRef]
- Cowger, C.; Arellano, C. Fusarium Graminearum Infection and Deoxynivalenol Concentrations During Development of Wheat Spikes. Phytopathology 2013, 103, 460–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, S.; Lyerly, J.H.; Maloney, P.V.; Brown-Guedira, G.; Cowger, C.; Costa, J.M.; Dong, Y.; Murphy, J.P. Mapping of Fusarium Head Blight Resistance Quantitative Trait Loci in Winter Wheat Cultivar NC-Neuse. Crop Sci. 2016, 56, 1473–1483. [Google Scholar] [CrossRef]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-Wide Association Studies. Nat. Rev. Methods Primers 2021, 1, 59. [Google Scholar] [CrossRef]
- Li, Y. Development of New Tools and Germplasms for Improvement of Wheat Resistance to Fusarium Head Blight. Ph.D. Thesis, Kansas State University, Manhattan, KS, USA, 2019. [Google Scholar]
- Maloney, P.V.; Petersen, S.; Navarro, R.A.; Marshall, D.; McKendry, A.L.; Costa, J.M.; Murphy, J.P. Digital Image Analysis Method for Estimation of Fusarium-Damaged Kernels in Wheat. Crop Sci. 2014, 54, 2077–2083. [Google Scholar] [CrossRef]
- MacRae, C.A.; Vasan, R.S. Next-Generation Genome-Wide Association Studies. Circ. Cardiovasc. Genet. 2011, 4, 334–336. [Google Scholar] [CrossRef] [PubMed]
- Arruda, M.P.; Brown, P.; Brown-Guedira, G.; Krill, A.M.; Thurber, C.; Merrill, K.R.; Foresman, B.J.; Kolb, F.L. Genome-Wide Association Mapping of Fusarium Head Blight Resistance in Wheat Using Genotyping-by-Sequencing. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, N.R.; Wright, E.; Malla, S.; Singh, L.; Sanford, D.V.; Clark, A.; Harrison, S.; Murphy, J.P.; Costa, J.; Chao, S.; et al. Identification and Validation of Fusarium Head Blight Resistance QTL in the U.S. Soft Red Winter Wheat Cultivar ‘Jamestown’. Crop Sci. 2020, 60, 2919–2930. [Google Scholar] [CrossRef]
- Agostinelli, A.M.; Clark, A.J.; Brown-Guedira, G.; Sanford, D.A.V. Optimizing Phenotypic and Genotypic Selection for Fusarium Head Blight Resistance in Wheat. Euphytica 2012, 186, 115–126. [Google Scholar] [CrossRef]
- Rutkoski, J.; Benson, J.; Jia, Y.; Brown-Guedira, G.; Jannink, J.; Sorrells, M. Evaluation of Genomic Prediction Methods for Fusarium Head Blight Resistance in Wheat. Plant Genome 2012, 5, 51. [Google Scholar] [CrossRef] [Green Version]
- Larkin, D.L.; Mason, R.E.; Moon, D.E.; Holder, A.L.; Ward, B.P.; Brown-Guedira, G. Predicting Fusarium Head Blight Resistance for Advanced Trials in a Soft Red Winter Wheat Breeding Program with Genomic Selection. Front. Plant Sci. 2021, 12, 715314. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Boyle, K.; Brûlé-Babel, A.L.; Fedak, G.; Gao, P.; Djama, Z.R.; Polley, B.; Cuthbert, R.D.; Randhawa, H.S.; Jiang, F.; et al. Genetic Characterization of Multiple Components Contributing to Fusarium Head Blight Resistance of FL62R1, a Canadian Bread Wheat Developed Using Systemic Breeding. Front. Plant Sci. 2020, 11, 580833. [Google Scholar] [CrossRef] [PubMed]
- Saha, I. Are Rice Markets in Bangladesh Efficiently Pricing Rice Based on Quality? An Empirical Assessment. Master’s Thesis, University of Arkansas, Fayetteville, AR, USA, 2021. [Google Scholar]
- Tacke, B.K.; Casper, H.H. Determination of Deoxynivalenol in Wheat, Barley, and Malt by Column Cleanup and Gas Chromatography with Electron Capture Detection. J. AOAC Int. 1996, 79, 472–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for Association Mapping of Complex Traits in Diverse Samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Covarrubias-Pazaran, G. Genome-Assisted Prediction of Quantitative Traits Using the R Package Sommer. PLoS ONE 2016, 11, e0156744. [Google Scholar] [CrossRef] [Green Version]
- VanRaden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [Green Version]
- Waldron, B.L.; Moreno-Sevilla, B.; Anderson, J.A.; Stack, R.W.; Frohberg, R.C. RFLP Mapping of QTL for Fusarium Head Blight Resistance in Wheat. Crop Sci. 1999, 39, 805–811. [Google Scholar] [CrossRef]
- Su, Z.; Bernardo, A.; Tian, B.; Chen, H.; Wang, S.; Ma, H.; Cai, S.; Liu, D.; Zhang, D.; Li, T.; et al. A Deletion Mutation in TaHRC Confers Fhb1 Resistance to Fusarium Head Blight in Wheat. Nat. Genet. 2019, 51, 1099–1105. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Bates, D.; Maechler, M.; Walker, S.; Bolker, B.; Christensen, R.H.B. Lme4. 2021. Available online: https://www.jstatsoft.org/article/view/v082i13 (accessed on 10 December 2021).
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using Lme4. J. Stat. Softw. 2014, 67, 1–48. [Google Scholar]
- Lenth, R.V. Emmeans: Estimated Marginal Means, Aka Least-Squares Means. 2021. Available online: https://rdrr.io/cran/emmeans/ (accessed on 10 December 2021).
- Kruijer, W. Heritability: Marker-Based Estimation of Heritability Using Individual Plant or Plot Data. Genetics 2019, 199, 1–20. [Google Scholar]
- Montesinos-López, O.A.; Montesinos-López, A.; Luna-Vázquez, F.J.; Toledo, F.H.; Pérez-Rodríguez, P.; Lillemo, M.; Crossa, J. An R Package for Bayesian Analysis of Multi-Environment and Multi-Trait Multi-Environment Data for Genome-Gased Prediction. G3 Genes Genomes Genet. 2019, 9, 1355–1369. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A Package for the next Level of Genome-Wide Association Studies with Both Individuals and Markers in the Millions. Gigascience 2018, 8, 154. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genom. Proteom. Bioinform. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, Z.; Zhang, Z.; Li, H.; Liu, D.; Zhang, Q.; Bradbury, P.J.; Buckler, E.S.; Zhang, Z. Expanding the BLUP Alphabet for Genomic Prediction Adaptable to the Genetic Architectures of Complex Traits. Heredity 2018, 121, 648–662. [Google Scholar] [CrossRef] [PubMed]
- Endelman, J.B. Ridge Regression and Other Kernels for Genomic Selection with R Package RrBLUP. Plant Genome 2011, 4, 250. [Google Scholar] [CrossRef] [Green Version]
- Peterson, B.G.; Carl, P. PerformanceAnalytics. 2020. Available online: https://cran.r-project.org/web/packages/PerformanceAnalytics/index.html (accessed on 10 December 2021).
- Rheenen, W.; van Peyrot, W.J.; Schork, A.J.; Lee, S.H.; Wray, N.R. Genetic Correlations of Polygenic Disease Traits: From Theory to Practice. Nat. Rev. Genet. 2019, 20, 567–581. [Google Scholar] [CrossRef] [PubMed]
- Gaire, R.; Ohm, H.; Brown-Guedira, G.; Mohammadi, M. Identification of Regions under Selection and Loci Controlling Agronomic Traits in a Soft Red Winter Wheat Population. Plant Genome 2020, 13, e20031. [Google Scholar] [CrossRef] [PubMed]
- Petersen, S.; Lyerly, J.H.; McKendry, A.L.; Islam, M.S.; Brown-Guedira, G.; Cowger, C.; Dong, Y.; Murphy, J.P. Validation of Fusarium Head Blight Resistance QTL in US Winter Wheat. Crop Sci. 2017, 57, 1–12. [Google Scholar] [CrossRef]
- Alisaac, E.; Behmann, J.; Rathgeb, A.; Karlovsky, P.; Dehne, H.-W.; Mahlein, A.-K. Assessment of Fusarium Infection and Mycotoxin Contamination of Wheat Kernels and Flour Using Hyperspectral Imaging. Toxins 2019, 11, 556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Femenias, A.; Gatius, F.; Ramos, A.J.; Sanchis, V.; Marín, S. Standardisation of near Infrared Hyperspectral Imaging for Quantification and Classification of DON Contaminated Wheat Samples. Food Control 2020, 111, 107074. [Google Scholar] [CrossRef]
- Beyer, M.; Pogoda, F.; Ronellenfitsch, F.K.; Hoffmann, L.; Udelhoven, T. Estimating Deoxynivalenol Contents of Wheat Samples Containing Different Levels of Fusarium-Damaged Kernels by Diffuse Reflectance Spectrometry and Partial Least Square Regression. Int. J. Food Microbiol. 2010, 142, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Tanabata, T.; Shibaya, T.; Hori, K.; Ebana, K.; Yano, M. SmartGrain: High-Throughput Phenotyping Software for Measuring Seed Shape through Image Analysis. Plant Physiol. 2012, 160, 1871–1880. [Google Scholar] [CrossRef] [Green Version]





| Trial | Year | Location | Field Replicates | Entries |
|---|---|---|---|---|
| USSN | 2019 | Florence, SC | 2 | 47 |
| USSN | 2019 | Mt. Holly, VA | 2 | 50 |
| USSN | 2020 | Florence, SC | 2 | 48 |
| USSRWWN | 2019 | Florence, SC | 2 | 40 |
| USSRWWN | 2020 | Florence, SC | 2 | 38 |
| USSRWWN | 2020 | Winnsboro, LA | 2 | 38 |
| GAWN | 2019 | Florence, SC | 2 | 54 |
| GAWN | 2020 | Florence, SC | 1 | 50 |
| GAWN | 2020 | Winnsboro, LA | 2 | 50 |
| SunWheat | 2019 | Florence, SC | 2 | 91 |
| SunWheat | 2020 | Florence, SC | 1 | 94 |
| SunWheat | 2020 | Winnsboro, LA | 1 | 94 |
| UBWT | 2020 | Florence, SC | 1 | 45 |
| OVT | 2020 | Florence, SC | 1 | 71 |
| Total samples: | 1266 |
| Predetermined Standards: | 0% | 35% | 50% | 85% | 100% | Pearson’s Correlation | Standard Error |
|---|---|---|---|---|---|---|---|
| QC Test 1—3 August 2021 | 5.68 | 37.95 | 51.41 | 86.07 | 96.5 | 0.9994 | 1.55 |
| QC Test 2—4 August 2021 | 7.23 | 38.68 | 50 | 84.58 | 96.59 | 0.9995 | 1.47 |
| QC Test 3—5 August 2021 | 7.92 | 40.18 | 53.45 | 86.39 | 96.76 | 0.9995 | 1.46 |
| QC Test 4—6 August 2021 | 6.41 | 40.38 | 52.75 | 85.88 | 97.09 | 0.9995 | 1.45 |
| QC Test 5—7 August 2021 | 5.1 | 35.39 | 50.64 | 85.8 | 97.75 | 0.9994 | 1.63 |
| QC Test 6—8 August 2021 | 5.02 | 38.52 | 51.22 | 86.62 | 97.94 | 0.9996 | 1.38 |
| QC Test 7—9 August 2021 | 5.89 | 37.89 | 51.05 | 85.15 | 97 | 0.9998 | 0.99 |
| QC Test 8—10 August 2021 | 5.83 | 37.71 | 50.44 | 85.25 | 97.07 | 0.9997 | 1.21 |
| QC Test 9—11 August 2021 | 3.47 | 39.29 | 51.62 | 84.83 | 96.92 | 0.9994 | 1.64 |
| Average | 5.84 | 38.44 | 51.40 | 85.62 | 97.07 | 0.9997 | 1.42 |
| Average Error of Obs vs. Pred | 5.84 | 3.44 | 1.40 | 0.62 | −2.93 |
| Platform | Equipment (Ascending Order by Cost) | Time | Broad-Sense Heritability (H2) | Genetic Correlation with Don Resistance | Ability to Predict Don Content (Raw Phenotype) | Ability to Predict Don Content (GEBV) | GWAS | Average Prediction Accuracy (R) |
|---|---|---|---|---|---|---|---|---|
| VISUAL | Predetermined Standards | 0:45 | 0.64 | 0.69 | 0.31 | 0.71 | N/A | 0.594 |
| MANUAL | Electric Seed Counter | 14:45 | 0.59 | 0.82 | 0.27 | 0.75 | N/A | 0.552 |
| VIBE | Vibe QM3 | 1:49 | 0.74 | 0.87 | 0.63 | 0.76 | Fhb1 | 0.588 |
| NIR | Near-Infrared Spectrometer | 1:45 | 0.52 | 0.57 | 0.31 | 0.48 | N/A | 0.404 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ackerman, A.J.; Holmes, R.; Gaskins, E.; Jordan, K.E.; Hicks, D.S.; Fitzgerald, J.; Griffey, C.A.; Mason, R.E.; Harrison, S.A.; Murphy, J.P.; et al. Evaluation of Methods for Measuring Fusarium-Damaged Kernels of Wheat. Agronomy 2022, 12, 532. https://doi.org/10.3390/agronomy12020532
Ackerman AJ, Holmes R, Gaskins E, Jordan KE, Hicks DS, Fitzgerald J, Griffey CA, Mason RE, Harrison SA, Murphy JP, et al. Evaluation of Methods for Measuring Fusarium-Damaged Kernels of Wheat. Agronomy. 2022; 12(2):532. https://doi.org/10.3390/agronomy12020532
Chicago/Turabian StyleAckerman, Arlyn J., Ryan Holmes, Ezekiel Gaskins, Kathleen E. Jordan, Dawn S. Hicks, Joshua Fitzgerald, Carl A. Griffey, Richard Esten Mason, Stephen A. Harrison, Joseph Paul Murphy, and et al. 2022. "Evaluation of Methods for Measuring Fusarium-Damaged Kernels of Wheat" Agronomy 12, no. 2: 532. https://doi.org/10.3390/agronomy12020532
APA StyleAckerman, A. J., Holmes, R., Gaskins, E., Jordan, K. E., Hicks, D. S., Fitzgerald, J., Griffey, C. A., Mason, R. E., Harrison, S. A., Murphy, J. P., Cowger, C., & Boyles, R. E. (2022). Evaluation of Methods for Measuring Fusarium-Damaged Kernels of Wheat. Agronomy, 12(2), 532. https://doi.org/10.3390/agronomy12020532









