Selection and Evaluation of Reference Genes for Quantitative Real-Time PCR in Tomato (Solanum lycopersicum L.) Inoculated with Oidium neolycopersici
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant and Fungal Materials
2.2. Inoculation with O. neolycopersici
2.3. RNA Isolation and cDNA Synthesis
2.4. Candidate Reference Genes Selection
2.5. Specificity Detection of Candidate Reference Genes Primers and qRT-PCR
2.6. Data Analysis and Stability Evaluation of Candidate Reference Genes
2.7. Reference Gene Validation
3. Results
3.1. RNA Quality Detection and Reference Gene Primer Specificity Analysis
3.2. Expression Level Analysis of Candidate Reference Genes
3.3. Expression Stability of Candidate Reference Genes in Tomato after Powdery Mildew Infection
3.3.1. geNorm Analysis
3.3.2. NormFinder Analysis
3.3.3. BestKeeper Analysis
3.3.4. The Comparative ∆CT Method Analysis
3.4. Comprehensive Analysis of the Candidate Reference Genes of Tomato under Powdery Mildew Stress
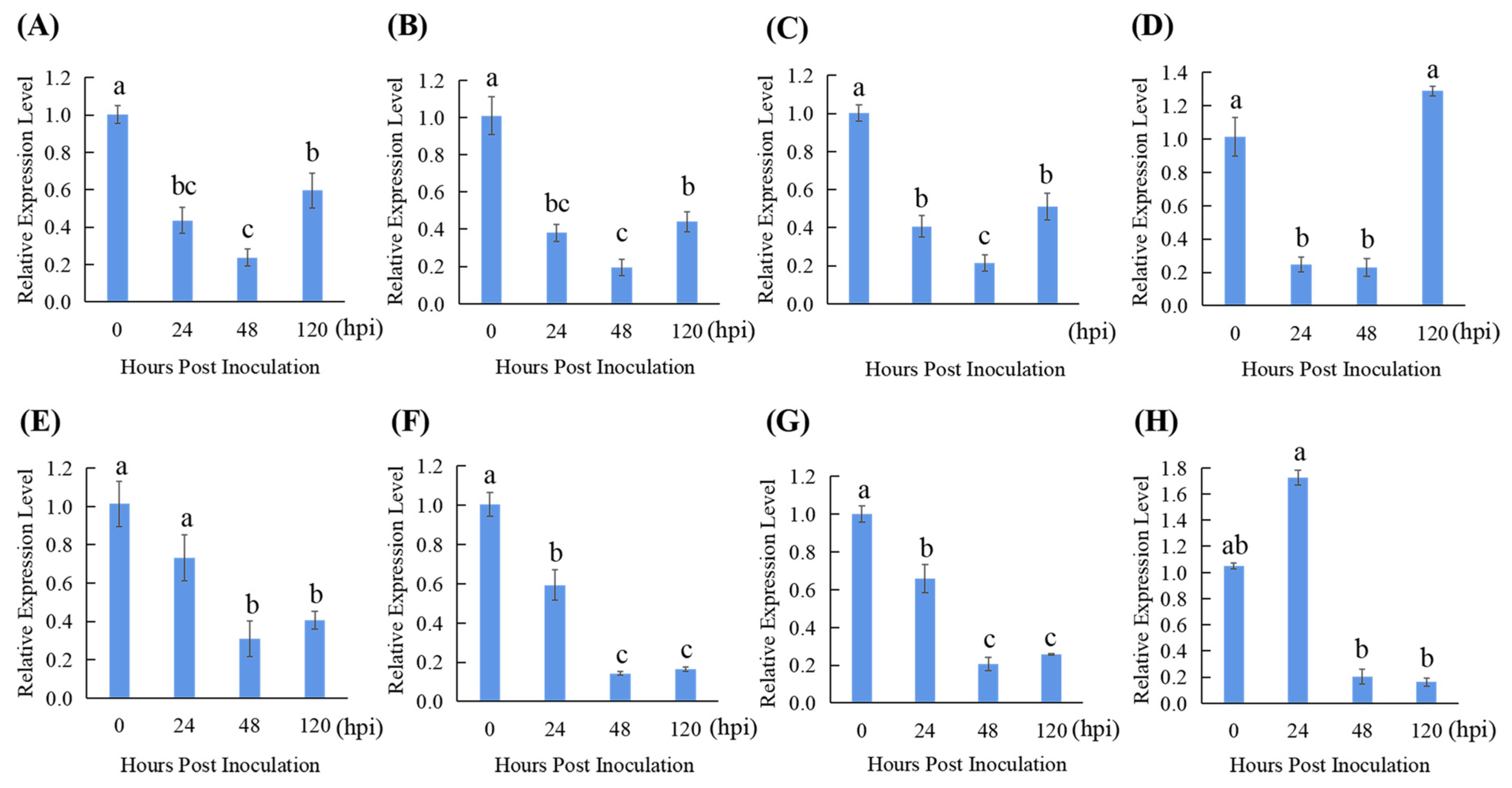
3.5. Reference Gene Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bergougnoux, V. The history of tomato: From domestication to biopharming. Biotechnol. Adv. 2014, 32, 170–189. [Google Scholar] [CrossRef]
- Doudna Jennifer, A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Jones, H.; Whipps, J.M.; Gurr, S.J. The tomato powdery mildew fungus Oidium neolycopersici. Mol. Plant Pathol. 2001, 2, 303–309. [Google Scholar] [CrossRef]
- Lebeda, A.; Mieslerová, B.; Jankovics, T.; Kiss, L.; Van, E.J. First detection of tomato powdery mildew caused by Oidium neolycopersici in South Africa. S. Afr. J. Bot. 2015, 99, 153–157. [Google Scholar] [CrossRef]
- Mamgain, A.; Biswas, M.K.; Dey, N. Potential role of chemical elicitors in induced systemic resistance for the effective management of Alternaria blight in mustard. J. Pharmacogn. Phytochem. 2019, 8, 2246–2250. [Google Scholar]
- Riaz, S.; Boursiquot, J.M.; Dangl, G.S.; Lacombe, T.; Laucou, V.; Tenscher, A.C.; Walker, M.A. Identification of mildew resistance in wild and cultivated Central Asian grape germplasm. BMC Plant Biol. 2013, 13, 149–169. [Google Scholar] [CrossRef]
- Zou, S.; Wang, H.; Li, Y.; Kong, Z.; Tang, D. The NB-LRR gene Pm60 confers powdery mildew resistance in wheat. New Phytol. 2017, 218, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Sun, X.; Liu, X.; Li, C.; He, L.; Chen, S.; Su, J. Selection of reliable reference genes for gene expression studies on Rhododendron molle G. Don. Front. Plant Sci. 2016, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; Preter, K.D.; Pattyn, F.; Poppe, B.; Roy, N.V.; Paepe, A.D.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef]
- Lu, Y.; Liao, J.; Guo, Z.; Zheng, H.; Lu, L.; Cai, Z.; Zeng, Z.; Ying, Z.; Chen, M. Selection and evaluation of potential reference genes for quantitative real-time PCR in Agaricus blazei based on transcriptome sequencing data. BioMed. Res. Int. 2021, 2021, 6661842. [Google Scholar] [CrossRef]
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Chen, M.; Dong, B.; Wang, N.; Yu, Q.; Wang, X.; Xuan, L.; Wang, Y.; Zhang, S.; Shen, Y. Transcriptomic analysis of flower bud differentiation in Magnolia sinostellata. Genes 2018, 9, 212–226. [Google Scholar] [CrossRef]
- Die, J.V.; Román, B.; Nadal, S.; González-Verdejo, C.I. Evaluation of candidate reference genes for expression studies in Pisum sativum under different experimental conditions. Planta 2010, 232, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Coker, J.S.; Davies, E. Selection of Candidate Housekeeping Controls in Tomato Plants Using EST Data. Biotechniques 2003, 35, 740–748. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Fan, C.; Li, H.; Zhang, Q.; Fu, Y. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol. Biol. 2009, 10, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, P.; Wrzesińska, B.; Obrępalska-Stęplowska, A. Assessment of reference gene stability influenced by extremely divergent disease symptoms in Solanum Lycopersicum L. J. Virol. Methods 2013, 194, 161–168. [Google Scholar] [CrossRef]
- Zheng, Z.; Nonomura, T.; Appiano, M.; Pavan, S.; Matsuda, Y.; Toyoda, H.; Wolters, A.A.; Visser, R.G.F.; Bai, Y. Loss of function in Mlo orthologs reduces susceptibility of pepper and tomato to powdery mildew disease caused by Leveillula taurica. PLoS ONE 2013, 8, e70723. [Google Scholar] [CrossRef]
- Jarošová, J.; Kundu, J.K. Validation of reference genes as internal control for studying viral infections in cereals by quantitative real-time RT-PCR. BMC Plant Biol. 2010, 10, 146–154. [Google Scholar] [CrossRef]
- Scholtz, J.J.; Visser, B. Reference gene selection for qPCR gene expression analysis of rust-infected wheat. Physiol. Mol. Plant Pathol. 2013, 81, 22–25. [Google Scholar] [CrossRef]
- Foss, D.L.; Baarsch, M.J.; Murtaugh, M.P. Regulation of hypoxanthine phosphoribosyltransferase, glyceraldehyde-3-phosphate dehydrogenase and β-actin mRNA expression in porcine immune cells and tissues. Anim. Biotechnol. 1998, 9, 67–78. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Zakrajsek, B.A. Effect of experimental treatment on housekeeping gene expression: Validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Methods 2000, 46, 69–81. [Google Scholar] [CrossRef]
- Du, M.; Zhou, M.; Deng, L.; Li, C.; Li, C. Current status and prospects on tomato molecular breeding—From gene cloning to cultivar improvement. Acta Hortic. Sin. 2017, 44, 581–600. (In Chinese) [Google Scholar] [CrossRef]
- Eissa, N.; Kermarrec, L.; Hussein, H.; Bernstein, C.N.; Ghia, J.E. Appropriateness of reference genes for normalizing messenger RNA in mouse 2,4-dinitrobenzene sulfonic acid (DNBS)-induced colitis using quantitative real time PCR. Sci. Rep. 2017, 7, 42427. [Google Scholar] [CrossRef]
- Albuquerque, G.M.R.; Fonseca, F.C.A.; Boiteux, L.S.; Borges, R.C.F.; Miller, R.N.G.; Lopes, C.A.; Souza, E.B.; Fonseca, M.E.N. Stability analysis of reference genes for RT-qPCR assays involving compatible and incompatible Ralstonia solanacearum-tomato ‘Hawaii 7996’ interactions. Sci. Rep. 2021, 11, 18719. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Wang, X.; Kong, W.; Wang, L.; Bai, S.; Li, X.; Guo, M.; Cheng, G. Identification of pathogen causing tomato powdery mildew in Yinchuan. J. Hunan Agric. Univ. (Nat. Sci.) 2022, 48, 196–200. (In Chinese) [Google Scholar] [CrossRef]
- Zheng, Z.; Appiano, M.; Pavan, S.; Bracuto, V.; Ricciardi, L.; Visser, R.G.F.; Wolters, A.A.; Bai, Y. Genome-wide study of the tomato SlMLO gene family and its functional characterization in response to the powdery mildew fungus Oidium neolycopersici. Front. Plant Sci. 2016, 7, 380. [Google Scholar] [CrossRef]
- Mascia, T.; Santovito, E.; Gallitelli, D.; Cillo, F. Evaluation of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in infected tomato plants. Mol. Plant Pathol. 2010, 11, 805–816. [Google Scholar] [CrossRef]
- Zhang, N.; Wei, Z.; Du, L.; Qing, L.; Sun, X. Selection of reference genes in tobacco and tomato stressed by TMV or PVX. J. Southwest Univ. (Nat. Sci. Ed. ) 2016, 38, 13–18. (In Chinese) [Google Scholar] [CrossRef]
- Expósito-Rodríguez, M.; Borges, A.A.; Borges-Pérez, A.; Pérez, J.A. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 2008, 8, 131–142. [Google Scholar] [CrossRef]
- Lacerda, A.L.M.; Fonseca, L.N.; Blawid, R.; Boiteux, L.S.; Ribeiro, S.G.; Brasileiro, A.C.M. Reference gene selection for qPCR analysis in tomato-bipartite begomovirus interaction and validation in additional tomato-virus pathosystems. PLoS ONE 2015, 10, e0136820. [Google Scholar] [CrossRef]
- Jose, S.; Abbey, J.; Jaakola, L.; Percival, D. Selection and validation of reliable reference genes for gene expression studies from Monilinia vaccinii-corymbosi infected wild blueberry phenotypes. Sci. Rep. 2020, 10, 11688. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Rntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Horvath, D.P.; Chao, W.S.; Yang, Y.; Wang, X.; Xiao, B. Identification and evaluation of reliable reference genes for quantitative real-time PCR analysis in tea plant (Camellia Sinensis (L.) O. Kuntze). Int. J. Mol. Sci. 2014, 15, 22155–22172. [Google Scholar] [CrossRef]
- Wang, J.; Liu, L.; Tang, Q.; Sun, K.; Zeng, L.; Wu, Z. Evaluation and selection of suitable qRT-PCR reference genes for light responses in Tea Plant (Camellia Sinensis). Sci. Hortic. 2020, 289, 110488. [Google Scholar] [CrossRef]
- Li, R.; Cui, K.; Xie, Q.; Xie, S.; Chen, X.; Zhuo, L.; Cao, A.; Shen, H.; Jin, X.; Wang, F.; et al. Selection of the reference genes for quantitative gene expression by RT-qPCR in the Desert Plant Stipagrostis pennata. Sci. Rep. 2021, 11, 21711. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, B.; Wang, X.; Wei, X. Screening of stable internal reference gene of quinoa under hormone treatment and abiotic stress. Physiol. Mol. Biol. Plants 2021, 27, 2459–2470. [Google Scholar] [CrossRef]
- Yin, Z.; Xie, F.; Michalak, K.; Zhang, B.; Zimnoch-Guzowska, E. Reference gene selection for miRNA and mRNA normalization in potato in response to potato virus Y. Mol. Cell. Probes 2021, 55, 101691. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Xue, Y.; Zhou, H.; Li, S.; Zhang, Z.; Hou, R.; Ding, Y.; Hu, K. Evaluation of reference gene suitability for quantitative expression analysis by quantitative polymerase chain reaction in the mandibular condyle of sheep. Mol. Med. Rep. 2015, 12, 5633–5640. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Y.; Zhang, H. Genome-wide analysis of the mildew resistance locus o (MLO) gene family in tomato (Solanum Lycopersicum L.). Plant Omics J. 2014, 7, 87–93. [Google Scholar]
- Kim, D.; Jin, B.; Je, B.I.; Choi, Y.; Kim, B.S.; Jung, H.J.; Nou, I.S.; Park, Y. Development of DNA markers for Slmlo1.1, a new mutant allele of the powdery mildew resistance gene SlMlo1 in tomato (Solanum Lycopersicum). Genome 2018, 61, 703–712. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Løvdal, T.; Lillo, C. Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal. Biochem. 2009, 387, 238–242. [Google Scholar] [CrossRef]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Wang, Y.; Zhao, L.; Zhou, R.; Li, Y.; Zhao, T.; Yu, W. Selection of tomato reference genes for qRT-PCR. Jiangsu J. Agric. Sci. 2017, 33, 389–396. (In Chinese) [Google Scholar] [CrossRef]
- Moradi, N.; Rahimian, H.; Dehestani, A.; Babaeizad, V. Cucumber response to Sphaerotheca fuliginea: Differences in antioxidant enzymes activity and pathogenesis-related gene expression in susceptible and resistant genotypes. J. Plant Mol. Breed. 2016, 4, 33–40. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, K.; Pei, D.; Yu, D.; Zhang, J.; Li, X.; Chen, G.; Yang, H.; Zhou, W.; Li, C. ShORR-1, a novel tomato gene, confers enhanced host resistance to Oidium neolycopersici. Front. Plant Sci. 2019, 10, 1400. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cui, J.; Zhang, C.; Yang, X.; Pan, H.; Du, H.; Ahmad, A.; Wu, T.; Yao, C. Genome-wide identification and analysis of the MLO gene family for candidate powdery mildew susceptibility factors in Momordica charantia. Sci. Hortic. 2022, 283, 110119. [Google Scholar] [CrossRef]
- Eissa, H.F.; Hassanien, S.E.; Ramadan, A.M.; El-Shamy, M.M.; Saleh, O.M.; Shokry, A.M.; Abdelsattar, M.; Morsy, Y.B.; El-Maghraby, M.A.; Alameldin, H.F.; et al. Developing Transgenic wheat to encounter rusts and powdery mildew by overexpressing barley Chi26 gene for fungal resistance. Plant Methods 2017, 13, 41–53. [Google Scholar] [CrossRef]
- Qin, B.; Wang, M.; He, H.; Xiao, H.; Zhang, Y.; Wang, L. Identification and characterization of a potential candidate Mlo gene conferring susceptibility to powdery mildew in rubber tree. Phytopathology 2019, 109, 1236–1245. [Google Scholar] [CrossRef] [PubMed]
- Bracuto, V.; Appiano, M.; Ricciardi, L.; Göl, D.; Visser, R.G.F.; Bai, Y.; Pavan, S. Functional characterization of the powdery mildew susceptibility gene SmMLO1 in eggplant (Solanum Melongena L.). Transgenic Res. 2017, 26, 323–330. [Google Scholar] [CrossRef]
- Jiang, P.; Chen, Y.; Wilde, H.D. Reduction of MLO1 expression in petunia increases resistance to powdery mildew. Sci. Hortic. 2016, 201, 225–229. [Google Scholar] [CrossRef]
- Reddy, R.A.; Dubey, H.; Vijayan, K.; Ponnuvel, K.M.; Mishra, R.K.; Suresh, K. Genome wide characterization revealed MnMLO2 and MnMLO6A as candidate genes involved in powdery mildew susceptibility in mulberry. Mol. Biol. Rep. 2020, 47, 2889–2900. [Google Scholar] [CrossRef]
- Reilly, A.; Gibriel, H.A.Y.; Karki, S.J.; Twamley, A.; Finnan, J.; Kildea, S.; Feechan, A. Genome-wide identification of the oat MLO family and identification of a candidate AsMLO associated with powdery mildew susceptibility. bioRxiv 2021, 1–31. [Google Scholar] [CrossRef]
- Huibers, R.P.; Loonen, A.E.H.M.; Gao, D.; Ackerveken, G.V.; Visser, R.G.F.; Bai, Y. Powdery mildew resistance in tomato by impairment of SlPMR4 and SlDMR1. PLoS ONE 2013, 8, e67467. [Google Scholar] [CrossRef]
- Appiano, M.; Pavan, S.; Catalano, D.; Zheng, Z.; Bai, Y. Identification of candidate MLO powdery mildew susceptibility genes in cultivated Solanaceae and functional characterization of tobacco NtMLO1. Transgenic Res. 2015, 24, 847–858. [Google Scholar] [CrossRef]
- Lian, Q.; Meng, Y.; Zhao, X.; Xu, Y.; Wang, Y.; Day, B.; Ma, Q. ShNPSN11, a vesicle-transport-related gene, confers disease resistance in tomato to Oidium neolycopersici. Biochem. J. 2020, 477, 3851–3866. [Google Scholar] [CrossRef]
- Tu, C.; Xu, P.; Han, R.; Luo, J.; Xu, L. Defining Suitable Reference Genes for qRT-PCR in Plagiodera versicolora (Coleoptera: Chrysomelidae) under Different Biotic or Abiotic Conditions. Agronomy 2022, 12, 1192. [Google Scholar] [CrossRef]
- Lin, Y.; Lai, Z. Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci. 2010, 178, 359–365. [Google Scholar] [CrossRef]

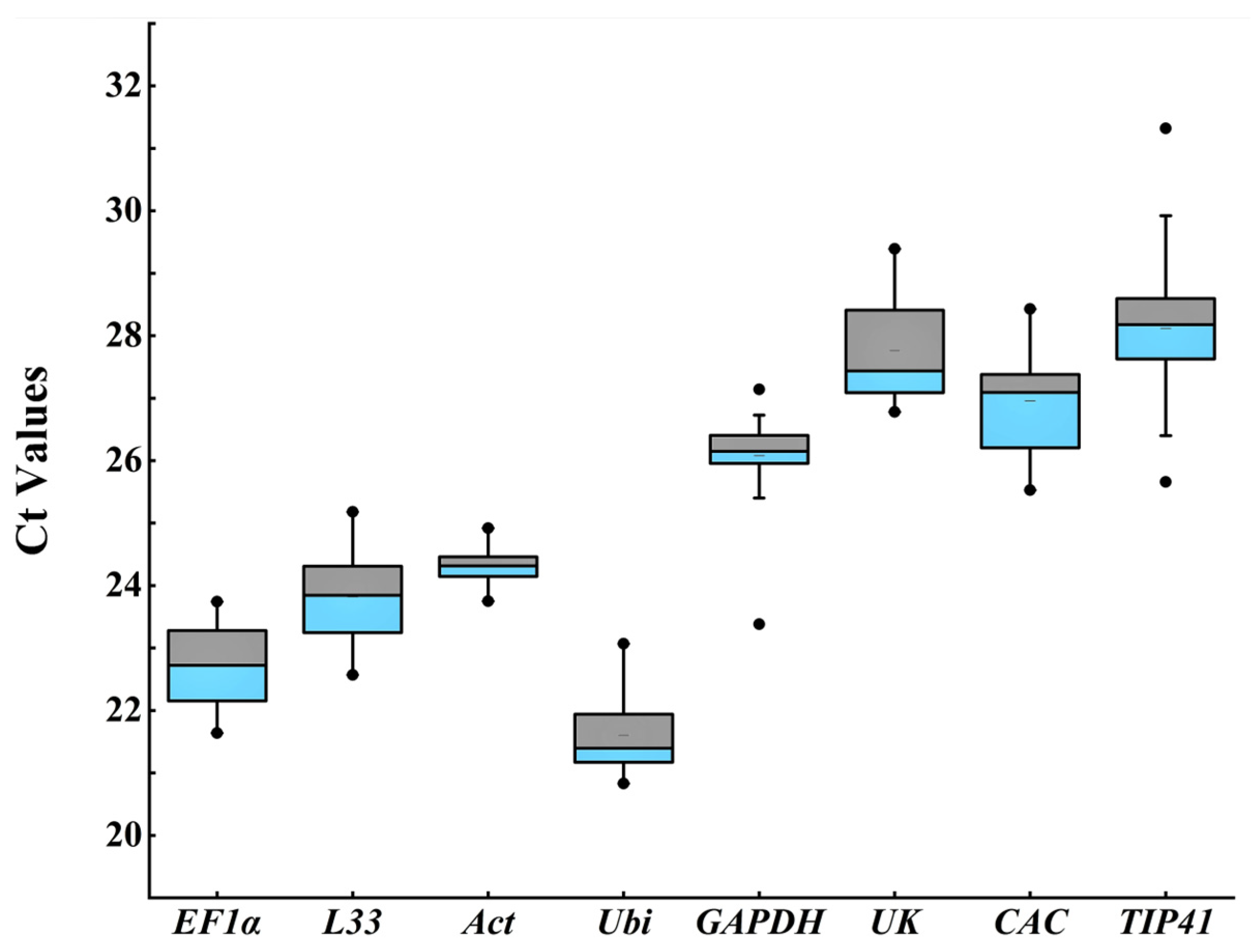

| Ordinal | Gene Name | Accession Number | Primer Sequence (5′-3′) | Amplicon (bp) | R2 | E (%) | Reference |
|---|---|---|---|---|---|---|---|
| 1 | EF1α | X14449 | F: ACAGGCGTTCAGGTAAGGAA | 120 | 0.998 | 99.6 | [26] |
| R: GAGGGTATTCAGCAAAGGTCTC | |||||||
| 2 | L33 | Q2MI79 | F: GGGAAGAGGCTGGGATACATC | 138 | 0.998 | 98.4 | [26] |
| R: AGGAGGCAAATTGGACTTGAAC | |||||||
| 3 | Act | NP_001317048.1 | F: GCTCCACCAGAGAGGAAATACAGT | 107 | 0.998 | 101.5 | [17] |
| R: CATACTCTGCCTTTGCAATCCA | |||||||
| 4 | Ubi | XP_004248311 | F: GGACGGACGTACTCTAGCTGAT | 134 | 0.996 | 103.2 | [26] |
| R: AGCTTTCGACCTCAAGGGTA | |||||||
| 5 | GAPDH | U93208 | F: ACCACAAATTGCCTTGCTCCCTTG | 110 | 0.998 | 97.6 | [27] |
| R: ATCAACGGTCTTCTGAGTGGCTGT | |||||||
| 6 | UK | LOC101267587 | F: TGGTAAGGGCACCCAATGTGCTAA | 114 | 0.990 | 106.7 | [28] |
| R: ATCATCGTCCCATTCTCGGAACCA | |||||||
| 7 | CAC | SGN-U314153 | F: CCTCCGTTGTGATGTAACTGG | 173 | 0.993 | 107.1 | [29] |
| R: ATTGGTGGAAAGTAACATCATCG | |||||||
| 8 | TIP41 | SGN-U584254 | F: ATGGAGTTTTTGAGTCTTCTGC | 235 | 0.994 | 101.8 | [30] |
| R: GCTGCGTTTCTGGCTTAGG | |||||||
| 9 | SlMLO1 | KU759512 | F: CTTTGGGCAGGCTAAAGATG | 128 | 0.998 | 102.1 | [26] |
| R: AATGCCTACGTCCAAACGAG |
| Ranking Order | ‘MM’ and ‘62579′ | ‘MM’ | ‘62579′ | |||
|---|---|---|---|---|---|---|
| Gene Name | Stability Value (S) | Gene Name | Stability Value (S) | Gene Name | Stability Value (S) | |
| 1 | Act | 0.279 | GAPDH | 0.133 | Act | 0.223 |
| 2 | EF1α | 0.395 | Act | 0.154 | EF1α | 0.356 |
| 3 | L33 | 0.397 | L33 | 0.275 | L33 | 0.426 |
| 4 | Ubi | 0.466 | CAC | 0.280 | Ubi | 0.528 |
| 5 | CAC | 0.520 | EF1α | 0.371 | UK | 0.625 |
| 6 | GAPDH | 0.540 | TIP41 | 0.377 | CAC | 0.670 |
| 7 | UK | 0.770 | Ubi | 0.385 | GAPDH | 0.774 |
| 8 | TIP41 | 1.118 | UK | 0.918 | TIP41 | 1.567 |
| Dataset | Ranking Order | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|---|
| ‘MM’ and ‘62579′ | Gene name | Act | GAPDH | Ubi | EF1α | CAC | L33 | UK | TIP41 |
| geo Mean [CP] | 24.32 | 26.07 | 21.60 | 22.73 | 26.95 | 23.81 | 27.75 | 28.09 | |
| min [CP] | 23.75 | 23.38 | 20.83 | 21.64 | 25.53 | 22.57 | 26.78 | 25.66 | |
| max [CP] | 24.92 | 27.14 | 23.07 | 23.74 | 28.43 | 25.18 | 29.39 | 31.32 | |
| std dev [±CP] | 0.23 | 0.41 | 0.48 | 0.52 | 0.58 | 0.62 | 0.72 | 0.83 | |
| CV [% CP] | 0.95 | 1.56 | 2.22 | 2.29 | 2.16 | 2.61 | 2.60 | 2.96 | |
| coeff. of corr. [r] | 0.768 | 0.673 | 0.707 | 0.794 | 0.732 | 0.866 | 0.527 | 0.560 | |
| p-value | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.008 | 0.004 | |
| ‘MM’ | Gene name | Act | CAC | GAPDH | Ubi | L33 | EF1α | TIP41 | UK |
| geo Mean [CP] | 24.30 | 27.24 | 26.24 | 21.66 | 24.14 | 23.05 | 28.19 | 27.84 | |
| min [CP] | 23.75 | 26.82 | 25.69 | 21.27 | 23.47 | 22.53 | 27.21 | 26.78 | |
| max [CP] | 24.92 | 28.01 | 26.73 | 22.35 | 24.94 | 23.64 | 29.33 | 29.39 | |
| std dev [±CP] | 0.23 | 0.23 | 0.23 | 0.30 | 0.32 | 0.35 | 0.39 | 0.77 | |
| CV [% CP] | 0.93 | 0.84 | 0.89 | 1.41 | 1.33 | 1.52 | 1.39 | 2.76 | |
| coeff. of corr. [r] | 0.836 | 0.521 | 0.802 | 0.470 | 0.796 | 0.585 | 0.746 | 0.519 | |
| p-value | 0.001 | 0.082 | 0.002 | 0.123 | 0.002 | 0.046 | 0.005 | 0.083 | |
| ‘62579′ | Gene name | Act | EF1α | GAPDH | L33 | Ubi | UK | CAC | TIP41 |
| geo Mean [CP] | 24.34 | 22.41 | 25.90 | 23.48 | 21.54 | 27.65 | 26.65 | 28.00 | |
| min [CP] | 23.77 | 21.64 | 23.38 | 22.57 | 20.83 | 26.80 | 25.53 | 25.66 | |
| max [CP] | 24.83 | 23.74 | 27.14 | 25.18 | 23.07 | 29.08 | 28.43 | 31.32 | |
| std dev [±CP] | 0.24 | 0.53 | 0.59 | 0.62 | 0.64 | 0.66 | 0.77 | 1.29 | |
| CV [% CP] | 1.00 | 2.38 | 2.29 | 2.66 | 2.95 | 2.39 | 2.88 | 4.60 | |
| coeff. of corr. [r] | 0.926 | 0.824 | 0.631 | 0.862 | 0.781 | 0.610 | 0.723 | 0.555 | |
| p-value | 0.001 | 0.001 | 0.028 | 0.001 | 0.003 | 0.035 | 0.008 | 0.061 |
| Ranking Order | ‘MM’ and ‘62579′ | ‘MM’ | ‘62579′ | |||
|---|---|---|---|---|---|---|
| Gene Name | Average of STDEV | Gene Name | Average of STDEV | Gene Name | Average of STDEV | |
| 1 | Act | 0.673 | Act | 0.411 | Act | 0.763 |
| 2 | EF1α | 0.687 | GAPDH | 0.430 | EF1α | 0.768 |
| 3 | L33 | 0.690 | L33 | 0.457 | L33 | 0.789 |
| 4 | Ubi | 0.722 | EF1α | 0.490 | Ubi | 0.828 |
| 5 | CAC | 0.784 | Ubi | 0.509 | CAC | 0.950 |
| 6 | GAPDH | 0.796 | CAC | 0.517 | UK | 0.964 |
| 7 | UK | 0.960 | TIP41 | 0.562 | GAPDH | 1.031 |
| 8 | TIP41 | 1.223 | UK | 0.954 | TIP41 | 1.653 |
| Gene Name | geNorm | NormFinder | BestKeeper | ∆CT | Geomean of Ranking Values | RefFinder |
|---|---|---|---|---|---|---|
| Act | 4 | 1 | 1 | 1 | 1.41 | 1 |
| EF1α | 1 | 2 | 4 | 2 | 2.00 | 2 |
| L33 | 1 | 3 | 6 | 3 | 2.71 | 3 |
| Ubi | 3 | 4 | 3 | 4 | 3.46 | 4 |
| GAPDH | 5 | 6 | 2 | 6 | 4.36 | 5 |
| CAC | 6 | 5 | 5 | 5 | 5.23 | 6 |
| UK | 7 | 7 | 7 | 7 | 7.00 | 7 |
| TIP41 | 8 | 8 | 8 | 8 | 8.00 | 8 |
| Gene Name | geNorm | NormFinder | BestKeeper | ∆CT | Geomean of Ranking Values | RefFinder |
|---|---|---|---|---|---|---|
| Act | 1 | 2 | 1 | 1 | 1.19 | 1 |
| GAPDH | 4 | 1 | 3 | 2 | 2.21 | 2 |
| EF1α | 1 | 5 | 6 | 4 | 3.31 | 3 |
| L33 | 3 | 3 | 5 | 3 | 3.41 | 4 |
| CAC | 6 | 4 | 2 | 6 | 4.12 | 5 |
| Ubi | 5 | 7 | 4 | 5 | 5.14 | 6 |
| TIP41 | 7 | 6 | 7 | 7 | 6.74 | 7 |
| UK | 8 | 8 | 8 | 8 | 8.00 | 8 |
| Gene Name | geNorm | NormFinder | BestKeeper | ∆CT | Geomean of Ranking Values | RefFinder |
|---|---|---|---|---|---|---|
| Act | 4 | 1 | 1 | 1 | 1.41 | 1 |
| EF1α | 1 | 2 | 2 | 2 | 1.68 | 2 |
| L33 | 1 | 3 | 4 | 3 | 2.45 | 3 |
| Ubi | 3 | 4 | 5 | 4 | 3.94 | 4 |
| GAPDH | 7 | 7 | 3 | 7 | 5.66 | 5 |
| CAC | 5 | 6 | 7 | 5 | 5.69 | 6 |
| UK | 6 | 5 | 6 | 6 | 5.73 | 7 |
| TIP41 | 8 | 8 | 8 | 8 | 8.00 | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, S.; Wang, X.; Guo, M.; Cheng, G.; Khan, A.; Yao, W.; Gao, Y.; Li, J. Selection and Evaluation of Reference Genes for Quantitative Real-Time PCR in Tomato (Solanum lycopersicum L.) Inoculated with Oidium neolycopersici. Agronomy 2022, 12, 3171. https://doi.org/10.3390/agronomy12123171
Bai S, Wang X, Guo M, Cheng G, Khan A, Yao W, Gao Y, Li J. Selection and Evaluation of Reference Genes for Quantitative Real-Time PCR in Tomato (Solanum lycopersicum L.) Inoculated with Oidium neolycopersici. Agronomy. 2022; 12(12):3171. https://doi.org/10.3390/agronomy12123171
Chicago/Turabian StyleBai, Shengyi, Xiaomin Wang, Meng Guo, Guoxin Cheng, Abid Khan, Wenkong Yao, Yanming Gao, and Jianshe Li. 2022. "Selection and Evaluation of Reference Genes for Quantitative Real-Time PCR in Tomato (Solanum lycopersicum L.) Inoculated with Oidium neolycopersici" Agronomy 12, no. 12: 3171. https://doi.org/10.3390/agronomy12123171
APA StyleBai, S., Wang, X., Guo, M., Cheng, G., Khan, A., Yao, W., Gao, Y., & Li, J. (2022). Selection and Evaluation of Reference Genes for Quantitative Real-Time PCR in Tomato (Solanum lycopersicum L.) Inoculated with Oidium neolycopersici. Agronomy, 12(12), 3171. https://doi.org/10.3390/agronomy12123171






