1. Introduction
Accurate and timely geographical knowledge of crop types on local, regional, and global scales provides a valuable source of information for various agricultural, environmental, social, and economic applications [
1,
2]. The new generation of satellite Earth observation systems (e.g., PlanetScope, RapidEye, and Sentinel 2) provide massive amounts of data with very high spatial and temporal resolutions. These data and a robust classification framework can be used for accurate crop mapping applications at regional and national scales [
3,
4].
Crop types experience high levels of spectral, spatial, and temporal dynamics during their growth period from farming activities, local climate, soil fertility, pest and disease, and the phenological stage [
5,
6,
7]. Consequently, high intra-class and low inter-class variability problems arise when classifying crop types [
8]. As a result, pixels of a class with high spectral heterogeneity may be classified as different classes, which shows the salt-and-pepper effect in the resulting classification maps. Using multitemporal and multispectral data and powerful classification frameworks to exploit spatial information appropriately can mitigate these problems. Traditional machine learning methods such as Linear Discriminant Analysis (LDA) [
9,
10], Maximum Likelihood (ML) [
11,
12,
13], Support Vector Machine (SVM) [
6,
13,
14,
15], and Random Forest (RF) [
16,
17,
18,
19] have been primarily used for crop type mapping. Pena et al. assessed the potential of Spectro-temporal indices derived from satellite image time series in improving the fruit-tree crop classification accuracy [
9]. They used LDA on a Landsat-8 time series of nine images from the 2014–2015 growing season. Arvor et al. presented two successive classifications based on Moderate Resolution Imaging System (MODIS)/Enhance Vegetation Index (EVI) time series first to determine agricultural areas and then classify five different crop types [
11]. They used ML as the classifier after a feature selection/extraction process. Ashourloo et al. provided an automatic canola mapping framework using sentinel-2 image time series. They presented a new spectral index to discriminate canola from other crop types during its flowering date [
20]. Wang et al. proposed an automatic large-scale cotton mapping technique based on LDA using the white bolls index (WBI) extracted from the cotton canopy at the bolls opening stage [
10]. They demonstrated the potential of the proposed approach using Sentinel-2 image time series. Niazmardi et al. presented three multiple kernel representations of satellite image time series to extract the most classification-related information from these data [
6]. The validity of the proposed representations was evaluated on 10 RapidEye imageries of an agricultural area using an SVM classifier. Song et al. presented an in-season crop-type mapping technique using object-based spectral/textural features and vegetation indices [
19]. The mapping strategy was evaluated on multi-temporal GaoFen satellite data with a spatial resolution of 16-m. The results showed that an object-based image analysis integrated with RF has considerable potential for in-season crop mapping. All traditional machine learning methods use pre-defined features, such as spectral and vegetation indices, which are extracted manually from satellite imagery. Therefore, the success of these methods is highly dependent on the choice of input features.
In the last decade, Deep Learning (DL) has been successfully applied in a wide range of applications in the remote sensing community. DL methods are trained in an end-to-end manner, which means they are trained to extract features and perform classification simultaneously. This eliminates the need for manual feature extraction and selection for the traditional machine learning methods that require the user’s knowledge and experience and a robust method. The other advantage is extracting features that are optimized for the task. In recent years, DL techniques have also been noticed in crop mapping. Kussul et al. proposed multi-level Convolutional Neural Networks (CNN) architecture to map crop types from multi-temporal Landsat-8 and Sentinel-1A images [
21]. Cué La Rosa et al. applied three types of DL methods, including auto-encoder (AE), CNN, and fully-CNN (FCNN), for crop-type mapping in multi-temporal Sentinel-1 images of tropical regions [
22]. They used prior knowledge about crop dynamics in the post-processing step, which improved classification accuracy. Qu et al. implemented a depth-wise separable convolution recurrent neural network (DSCRNN) for cop mapping from polarimetric Sentinel-1 time-series data [
23]. The results were compared to the classical random forest method and proved the superiority of DL algorithms. Adrian et al. fused multi-temporal Sentinel-1 Radar data with multi-temporal optical Sentinel-2 data using a DL approach (3D U-Net) for crop mapping application using the Google Earth Engine framework [
24]. Ge et al. proposed a DL-based crop mapping model, which can be transferred through location based on the phenological matching principle [
25]. They exploited Landsat data in the experiments. Ofori-Ampofo et al. performed crop-type mapping from Sentinel-2 optical and Sentinel-1 Radar time series using attention-based DL strategies [
26]. Turkoglu et al. developed a crop classification framework based on a convolutional Recurrent Neural Network (convRNN), which can produce multi-scale label hierarchies [
27]. The model was validated on multi-temporal Sentinel-2 image patches, and significant improvements were observed in mapping rare crop types. Chen et al. proposed stacking of two-dimensional (2D) gridded spectral features as input to 2D CNNs for crop mapping from Hyper-Spectral Imagery (HSI) [
28]. Chen et al. developed a deep learning model based on Bidirectional Long Short-term Memory network (BiLSTM) to jointly impute missing data in time series and crop classification [
29]. They have tested their model on the Sentinel-2 image time series. In this study, we will consider Sparse Auto-Encoders (SAE) as a successful type of DL methods and will focus on improving its performance in crop mapping applications.
As we mentioned earlier, considering spatial information in the classification framework is of great importance to alleviate high intra-class and low inter-class variability problems when the classification of crop types is the issue. Spatial data can be extracted in two stages: feature extraction and post-processing. Classification methods that exploit spatial information in the feature extraction phase can be divided into object-based and pixel-based categories. Object-based methods use multi-resolution segmentation to find homogenous image segments or parts having the same class label. Various spatial features are then extracted manually from each segment and then fed to a classifier to provide class labels, which are then associated with all pixels of that segment [
30]. Despite its excellent results and popularity, there is a conceptual problem in the definition of this method. It tries to find image segments of pixels having the same class labels while no information about the classes is available. Accordingly, the classification results depend highly on the segmentation’s hyper-parameters [
12]. Window-based algorithms use spatial information in pixel-based methods to use neighborhood information around pixels. Spatial features are extracted from each window and then fed to a classifier to find the class labels allocated to the window’s center pixel. This commonly used method in remote sensing applications can improve performance to some degree. However, according to its smoothing nature it will produce classification maps which are over-smoothed in object boundary areas.
The post-processing of classification maps is another commonly used method for spatial information included in the classification process. Conditional Random Fields (CRFs) are widely used for correcting classification maps (especially in object border areas) using spatial information when very high spatial resolution data are available [
31,
32,
33,
34]. Wei et al. proposed a crop classification model based on CRF for Unmanned Aerial Vehicle (UAV)-borne hyperspectral remote sensing imagery [
32]. They tested their method on two datasets with 10- and 40-cm spatial resolutions and 270 spectral channels. Pan et al. proposed a CRF-based model to correct the resulting map of high-resolution remote sensing image semantic segmentation in urban areas where the images had a 9 cm spatial resolution [
31]. However, CRF models have difficulties when applied to remote sensing images, and overcorrection phenomena may occur [
31,
32].
The main objective of this study is to improve the classification performance of crop mapping using deep neural networks and field boundary information extracted from multitemporal and multispectral imagery. The proposed framework can use spatial information to improve classification results in a post-processing step. More precisely, we use the edge-aware filtering process [
35], which can filter the classification probability maps, transferring the guidance image’s structures to filtered maps. We generate a guidance image with the structure of all images in time series as much as possible. Hence, the filtered classification maps are aware of structures latent in image data and produce more accurate maps in field boundary areas. Moreover, the filtering process is very fast in computations.
2. Materials and Methods
2.1. Study Area and Data
We examined the performance of the proposed method over two agricultural areas in Canada.
Dataset 1: The first study area was located at the Central Experimental Farm (CEF) in the central part of the City of Ottawa, Ontario, Canada. It covered about 330 ha of agricultural fields (
Figure 1a). The image dataset was a 6-date time series of multispectral PlanetScope ortho-scene analytic products (Level 3-B) acquired over three months during the 2018 growing season (2 June, 15 June, 3 July, 12 July, 29 July, and 16 August) in an approximately two-week time intervals. The true color composite image of the July 3th date is displayed in
Figure 1a. PlanetScope ortho-scene products are corrected for radiometric, sensor-related, spacecraft-related, and geometric effects and projected to a cartographic map projection system [
36]. The images used here have the horizontal datum of WGS 84 with the map projection of UTM-Zone 18N. They are orthorectified with a pixel size of 3 m and have four spectral bands (i.e., blue, green, red, and near-infrared).
The ground reference map was obtained in two steps. In the first step, field observations were performed to identify the cultivated crop types in each field, and the results were manually inscribed on a freehand sketch. In the second step, the multi-temporal high-resolution georeferenced image datasets were manually digitized. This step was performed with the previously obtained freehand drawing utilizing the ArcMap 10.4.1 software. As a result, accurate field boundaries were obtained with the specific crop type attended to each field. This area covers nine major crop types, including corn, soybean, oat, barley, spring wheat, winter wheat, sunflower, hay/pasture, and grass, in the 2018 period.
Dataset 2: The second study area was an agricultural region in the City of Winnipeg, southwest Manitoba state in Canada, covering about 5500 ha (
Figure 1b). A time series of RapidEye sensors with a 5 m spatial resolution and five spectral bands (blue, green, red, red-edge, and near-infrared) were used as image data. The images were atmospherically corrected, orthorectified, and acquired during the 2012 growing season (14 May, 5 July, 14 July, and 19 August). The 14 July image is displayed in
Figure 1b.
Seven different crop classes, including corn, pea, canola, soybean, oat, wheat, and broadleaf, were considered in the classification scheme. These data were collected to support the Soil Moisture Active-Passive Mission Validation Experiment (SMAPVEX) 2012 campaign of the National Aeronautics and Space Administration (NASA) [
37].
2.2. Problem Statement
Let us suppose we have q high spatial resolution multispectral images () acquired on q imaging dates during the growing season from an agricultural area. Where m, n, and s are the number of rows, columns, and spectral bands of the image, respectively. Consider be the train set, where is the ith train sample with the first two rows corresponding to its position in the image (its row and column indices as and ) and the other f rows corresponding to its extracted features (), and is its corresponding class label. Validation set is defined similarly with its corresponding features ().
Moreover, consider as the test set, where is the ith test sample associated with the label . Again, the first two rows of corresponds to its position in the image ( and ) and the other f rows corresponding to its extracted features (). We want to train a classifier using the train and validation sets so that the estimated class labels for the test set () be equal to their true lables () as much as possible. The classification accuracy could be obtained by comparing and .
2.3. Proposed Method
This study proposes a two-step DL-based framework (Guided Filtered SAE; GFSAE) that can adequately exploit spectral, spatial, and temporal information of high-resolution image time series for accurate crop mapping. Firstly, we use a window-based strategy in the feature extraction phase. This process generates spatial-spectral features, which are then fed to an SAE for generating class labels. Secondly, we filter the obtained classification maps utilizing the edge information of the input image data in a post-processing step. Accordingly, they are related to the boundaries of crop fields and, therefore, can be very helpful in the classification. The framework of the proposed method is displayed in
Figure 2.
2.3.1. Preprocessing
In the first step, all q images () are stacked consecutively to obtain an image data cube . This data cube is then normalized using the maximum and minimum values of each band to obtain a data cube with values between 0 and 1.
In the next step, the training, validation, and test samples are selected from the ground reference map, and their coordinates and also class labels are saved, i.e., [], for the train set, [], for the validation set, and [], for the test samples. The sample selection is based on a patch-wise random selection method which generates data patches with a maximum size of wp × wp pixels (where wp is patch size and was set to 15 pixels for both datasets).
2.3.2. Feature Extraction
Corresponding features for all samples are extracted using the normalized image data cube (
) to generate
and
sets. The feature extraction process is represented graphically in
Figure 3 for
w = 5 pixels. This process extracts spectral-spatial-temporal features for all pixels. For each pixel
in the train set with the image coordinates of
and
, we consider a neighbourhood window of odd size
w centered on it. Then, features associated with that pixel are obtained from the normalized data cube (
) considering all the band values for all of its neighbour pixels reshaped to a
feature vector.
vector is then generated, attaching its positional indices and the extracted features as
. This process is repeated for all pixels in the validation and test sets.
The next step is training the classifier using the training and validation data. Validation data prevents model overfitting to the training data by breaking the training process when the validation error grows. Unlike the training set error, which decreases steadily over time, the validation set error increases after some iterations when the model overfits the training data. The model parameters are saved and returned as the final trained parameters at this point if, after some successive iterations, for example, 20 or 30, no improvements will be observed in the validation set error. Otherwise, the new point is considered a new potential breaking point of the training process, and the steps are repeated. This process is known as early-stopping and is a well-known generalization strategy in machine learning concepts. The objective is to increase the model performance on new unseen data, even with the price of increasing the training data error. Early stopping can also be considered an efficient hyper-parameter selection technique as it can determine the number of training epochs. After the training, the test data are used to calculate the quality indices and generate the probability maps for all classes. According to the high potential of SAE in classifying complex areas, it is considered the classifier.
2.3.3. Classification with SAEs
Sparse Auto-Encoders (SAEs) are a specific type of deep network which uses Auto-Encoders (AEs) as their building blocks [
38]. AE is a neural network that is trained so that it can encode an input
x to a latent underlying representation (the encoder part) in a way such that the input can be reconstructed from that representation (the decoder part). The encoder and decoder formulas are represented in Equations (1) and (2), respectively:
where,
x is the vector of input values, W
1, b
1, W
2, and b
2 are the weight and bias parameters for the encoder and decoder parts, respectively,
f is the activation function (usually the ReLU function), h is the latent representation (activation values of the hidden units), and
is the reconstructed vector.
Generally, some constraints are applied to the AE networks to exploit useful structures from the input data [
39], and most of the data variabilities can be modeled [
38]. According to the used constraints, specific types of AEs have been developed. One of the successful types is sparse AE which enforces a sparsity constraint on the activation values of the hidden units averaged over the training samples:
where,
is the activation of the
jth hidden unit on the
ith training sample, and
m is the total number of training samples. The sparsity constraint can be modelled by adding a penalty term to the optimization function, which penalizes significant differences between the averaged activation values (
) and the sparsity parameter (
):
where,
is the cost function (e.g., squared-error or cross-entropy function [
39]),
KL is the Kullback–Leibler divergence function [
39],
s is the number of hidden units in the hidden layer, and
is a hyperparameter which controls the level of sparsity.
Training SAEs as deep multi-level networks is a two-step procedure. Each constituent AE is trained separately in the first step, layer-wise pre-training. In the second fine-tuning step, the entire network is trained and initialized by the parameter values found in the pre-training step.
In classification applications, successive latent representations are obtained by stacking encoder parts of constituent AEs, and a softmax layer is added at the end. The softmax layer has an n-unit equivalent to the total number of classes. It uses an exponential function to generate class probabilities based on features obtained from the last hidden layer:
where,
is the probability of the input
being in class
k,
e is the exponential function,
is the activation of the input
i on the unit
j of the softmax layer, and
n is the total number of the units in this layer. The final class index of the input
is the class which obtained the maximum probability (i.e.,
).
The probability maps for the image cube are obtained as the output of the network (Equation (5)), where each map P(i) corresponds to the class i representing the probability of classifying belonging pixels into that class. Each probability map i filled with values of 0 and 1, 1 when the specified pixel labelled as the class i, and 0 otherwise.
Although SAE performs well in discrimination between crop types, it does not consider field boundary information. Consequently, we use the boundary information of the input data to improve classification accuracy. For this purpose, we filter the probability maps obtained in the previous step, guided by a guidance image that includes the edge information content of the input imagery as much as possible.
2.3.4. Guided Filtering
The guided filter, proposed in [
35], is one of the fastest and most efficient edge-preserving algorithms. It is a local linear model that involves the content of a guidance image in the filtering process in a way such that the filtered image inherits edges from the guidance. Using a guidance image that contains most of the edge information from the input image data cube, we can filter the probability maps obtained from the classification process, preserving the edge information of the input imagery. We briefly introduce the guided filter for our edge-aware filtering application in the following. Interested reader refers to the original paper [
35] for a complete explanation and other applications.
The guided filter assumes that the filtering output
is a linear transform of the guidance image G in a window
. This local linearity is defined as Equation (6) at a pixel
i assuming constant linear coefficients
in
:
This local linearity ensures identical edges in
and G because of
[
35]. Assuming the probability map, P, as the input image, the linear coefficients can be determined to minimize the following energy function in the window
:
where
is a regularization parameter to penalize largely
. This energy function will minimize the difference between the input (
) and the output (
) of the filter, and at the same time, the linear transformation assumption will be assured. The solution is obtained from linear functions of the input and the guidance images as:
where,
and
are the mean and variance of G in
,
is the number of pixels in
, and
is the mean of P in
.
Considering all windows overlapping pixel
i, different values of
will be obtained. The final filtered image could be obtained using the averaged coefficients
:
where,
and
. Equations (8)–(10) define the guided filter. As we see, the guided filter has two hyperparameters, which are needed to be optimized, including the neighbourhood size
, and the regularization parameter
.
The guided filter has been used in this study utilizing the PCA transformation of the input image data cube for generating the guidance image. We can use the grayscale PC1 image or the RGB-coloured image from PC1 to PC3 as the guidance image (G). We tested both choices and obtained slightly better results when using RGB-colored PCs image as (G). In the final step, the filtered probability maps are used for class allocation for each pixel using the arg-max function, and the final quality assessment indices are calculated.
3. Results and Discussion
3.1. Experimental Setup
The validity and superiority of the proposed GFSAE are analysed compared to the original SAE and two traditional commonly used classifiers, i.e., SVM and RF. All experiments were implemented using MATLAB v2017.b on a Desktop Server with Intel(R) Xeon(R) CPU E5-2697 v4 @ 2.30 GHz, 2295 MHz, 18 Core(s), 36 Logical Processor(s), 48-GB RAM.
For the CEF area, we selected 20, 5, and 5 percent of pixels of the ground reference map as the training, validation, and test data, respectively. For the Winnipeg dataset, these values were 4, 1, and 1 percent, respectively. The data selection was performed using a stratified random patch-based sampling method in which a patch size of 15 pixels is considered for both datasets.
Figure 1a shows the geographical extent of training, validation, and test data imposed on image data. The number of training, test, and validation samples in each class is displayed in
Table 1 and
Table 2 for the CEF and Winnipeg datasets.
In the experiments, the classifier’s hyper-parameters were set as follows. After testing different structures, we selected three network layers, i.e., input, hidden, and softmax. The number of units in the first layer equals the number of input features (e.g., 24 and 20 for the CEF and Winnipeg datasets, respectively, in the case of the
w = 1 setting). The number of units in the last layer equals the number of classes (i.e., 9 and 7 for the CEF and Winnipeg datasets, respectively). We have used the Bayesian optimization algorithm for the other parameters to find optimum parameters. The results of Bayesian optimization are displayed in
Table 3 for each dataset.
Two hyperparameters of the guided filter were optimized as follows. A grid search method was used for selecting the neighborhood size (z) considering window sizes (i.e., ) in the range of [2, 35] pixels and evaluating the results on the validation data. Finally, 31 pixels were selected as the best window size, and hence z = . The regularization parameter () were set as ; where G is the guidance image, range (X) is a function that gives the range of values in X, and diff (X) is a function that calculates differences between adjacent elements of X.
For accuracy analysis, we consider statistical indices, namely the Overall Accuracy (OA), the Kappa coefficient, class F-scores, and macro-averaged F-score over the classes. We also compared Omission and Commission errors of different crop types as Producer and User Accuracies (PA and UA) functions. PA and UA for field boundary areas are also calculated. Visualizations of crop classification maps and boundary maps are provided as well.
3.2. Results on the CEF Dataset
Accuracy analysis metrics of SAE and GFSAE are displayed in
Figure 4 for the CEF dataset. It shows that using spatial information (either with increasing neighborhood window size, i.e.,
w, or guided filtering) improves classification accuracy. Considering SAE, in the case
w = 1, the worst accuracy is achieved, while by increasing neighborhood size (to 5 pixels), classification accuracy increases more than 1%. However, classification accuracy decreases when the window size is more significant than 5 pixels because the over-smoothing effect of huge windows occurs, i.e., including spectral information of non-related far pixels in the classification of the target pixel.
Comparing SAE metrics with ones of the GFSAE proves that the proposed method causes considerable performance improvement in terms of all accuracy metrics. It increases classification accuracy by up to 5%. The accuracy metrics and training time of the proposed GFSAE with respect to the conventional SAE and traditional SVM and RF classifiers are represented in
Table 4 for the CEF dataset.
Table 4 shows that increasing window size (
w) does not guarantee per-class performance improvements. Considering the first part (i.e., SAE), we see that F-scores in some classes improve with increasing
w from 1 to 5 (such as the classes Corn, Soybean, and Oats), while degradations in class F-scores are observed for other classes (such as Summer Wheat and Winter Wheat). The second part of the table (i.e., GFSAE) shows the same dissimilar results for different classes. On the other hand, comparing the results of the GFSAE with SAE proves the superior performance of the proposed GFSAE method in terms of per-class F-scores. It has almost equal calculation time to SAE (see the last column) with much better accuracy. Comparing the accuracy metrics of the GFSAE with the traditional SVM and RF classifiers also proves the validity and superiority of the proposed method. Even though the traditional SVM and RF classifiers have produced slightly better results than the original SAE on the CEF dataset, the GFSAE performs much better than all the compared classifiers. It also has a comparable calculation time with respect to these traditional methods.
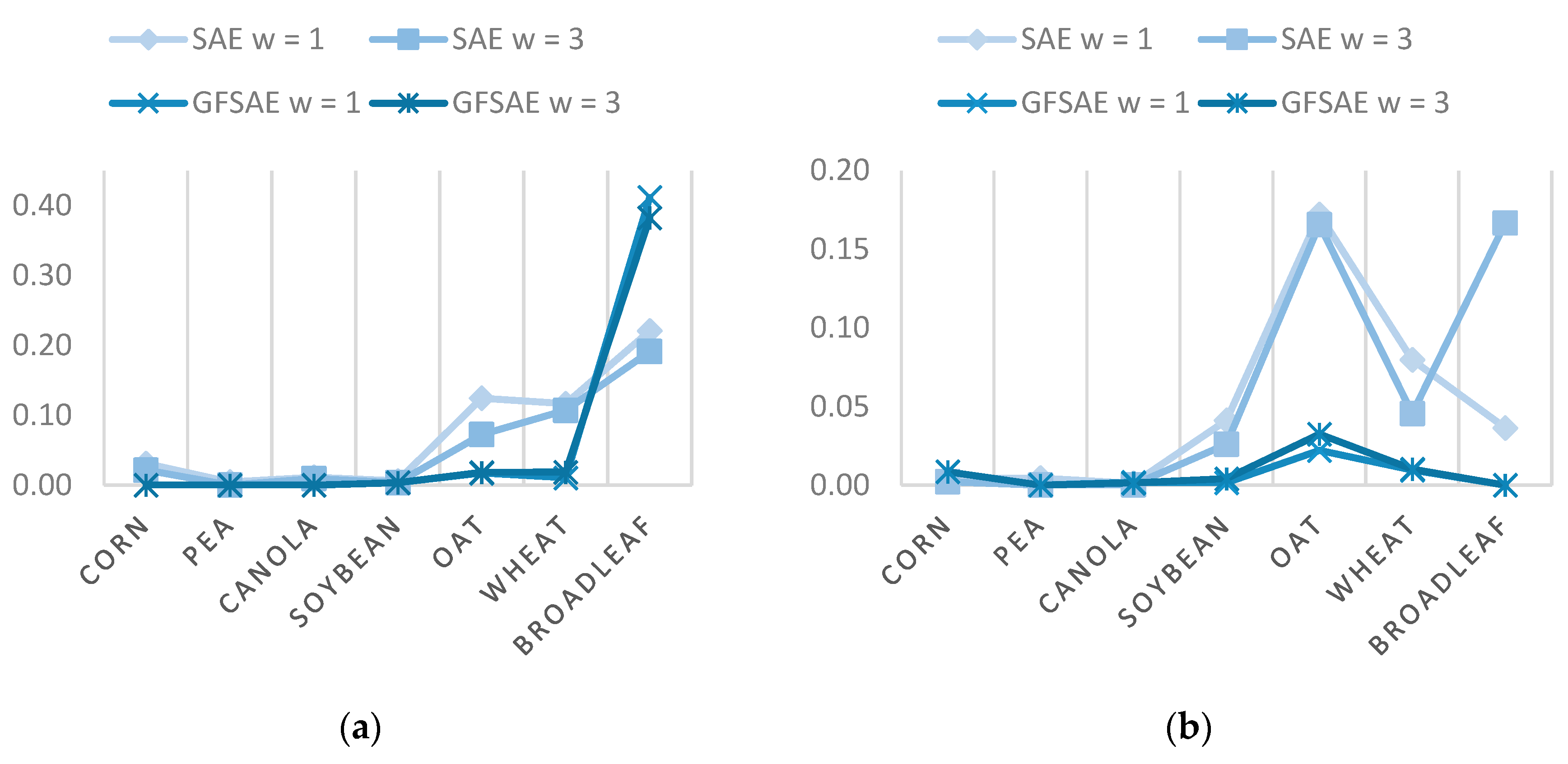
Figure 5 shows commission and omission errors of different classes obtained from SAE and the proposed GFSAE classifiers for the CEF dataset. These errors are calculated from the UA and PA as:
Omission error represents the number of pixels in a certain class that the algorithm wrongly classified as other crop classes. Conversely, commission error refers to the pixels classified in a certain class but do not truly belong to that class type. These errors arise from high intra-class and low inter-class variability problems in crop mapping from multispectral imagery. As we see, superior performance of the proposed GFSAE framework is observed in reducing both omission and commission errors of different crop types. In other words, the proposed GFSAE could better deal with the aforementioned high intra-class and low inter-class variability problems.
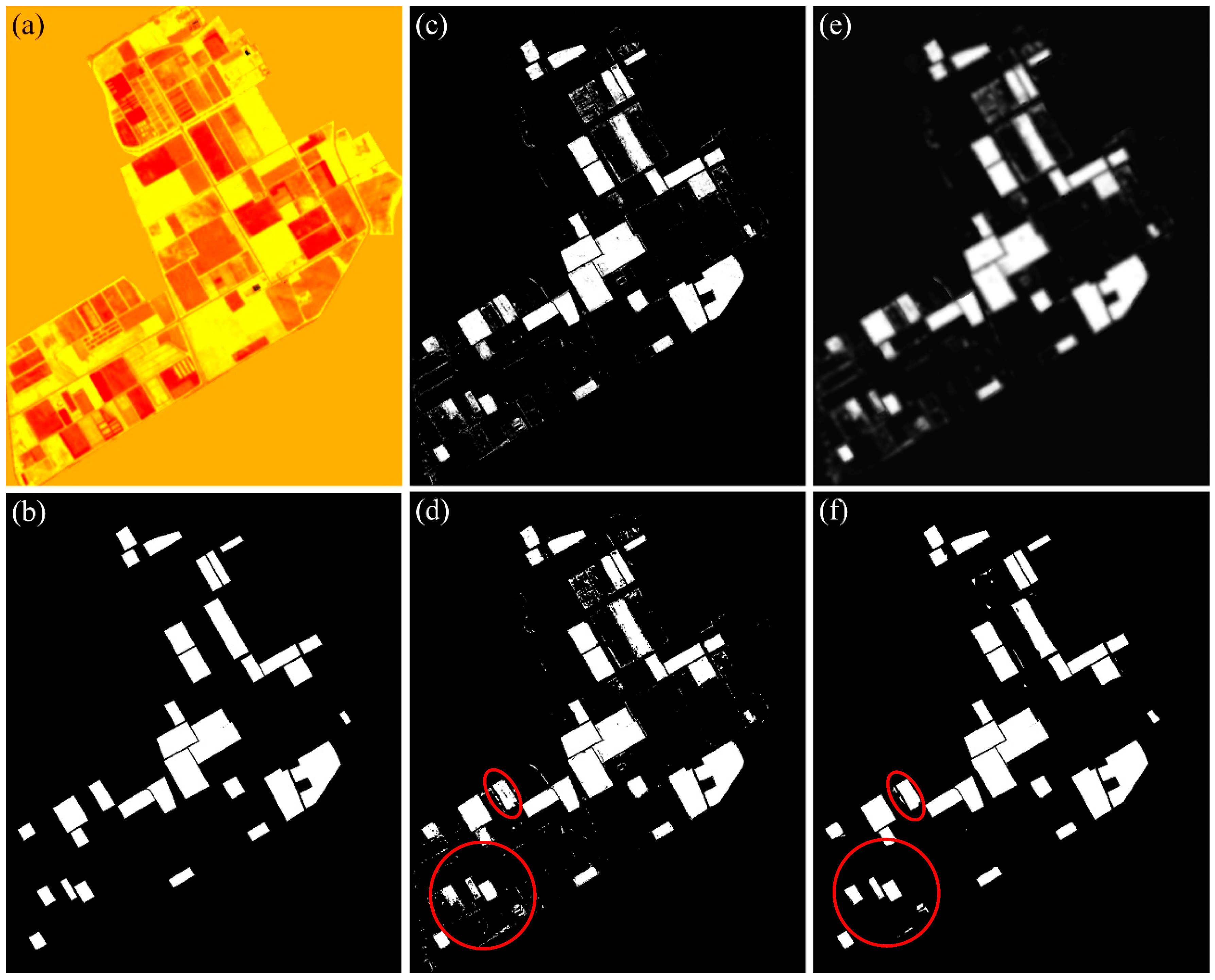
The edge-aware filtering process is represented graphically in
Figure 6. This figure shows the process for corn in the case of the
w = 1 setting for the CEF dataset.
Figure 6a shows the guidance image (G), (b) is the ground reference map for this class, (c) is the initial probability map for the class corn, i.e., P (1), (d) is the initial class map (i.e., the result of SAE), (e) is the probability map, P (1), after the guided filtering, and (f) is the final class map (i.e., the result of GFSAE). As we see, a more accurate map is obtained after the guided filtering process, removing isolated wrongly classified pixels from the class map. To describe precisely, non-corn pixels inside non-corn fields that were wrongly classified as corn class (commission error) and corn pixels inside corn fields that were wrongly classified as other classes (omission error) are corrected after the proposed filtering process. This could also be observed in
Figure 5 for corn class numerically. The filtering process is performed for all classes separately, and the final classification map is generated by merging the per-class classification maps.
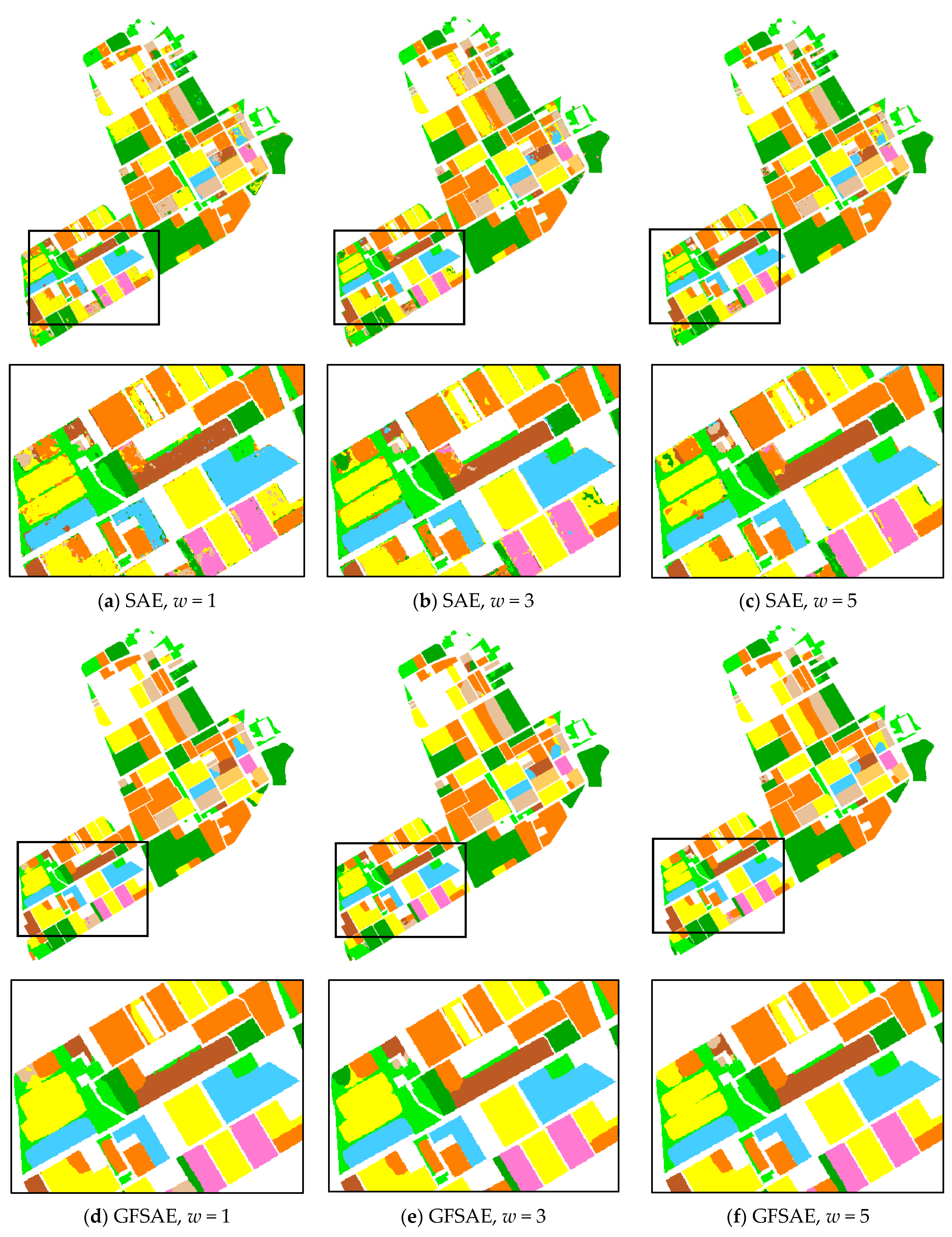
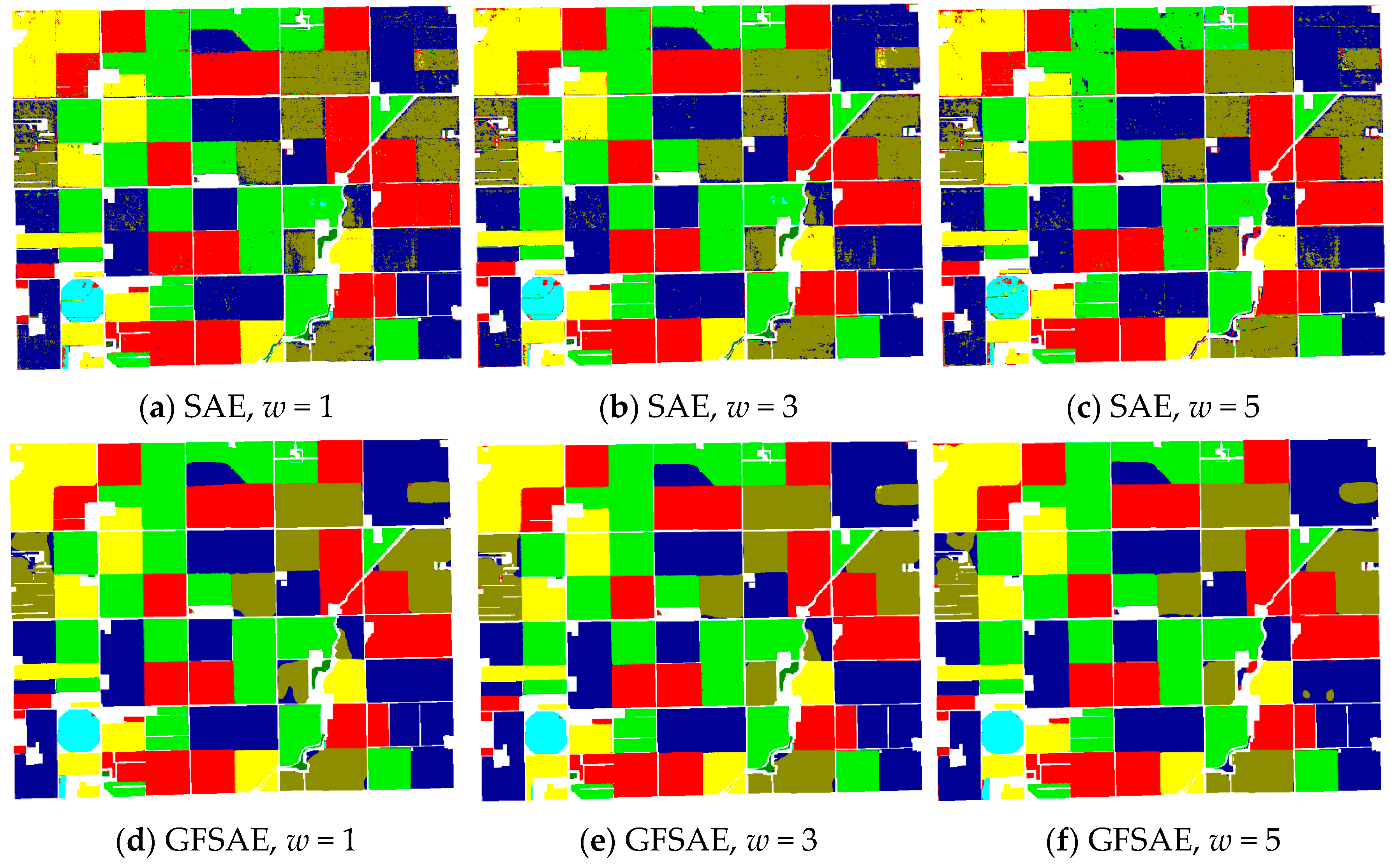
Classification maps obtained from SAE and GFSAE for all
w settings and SVM and RF classifiers are displayed in
Figure 7 for the CEF dataset, along with its ground reference map. As we see, in the case of SAE increasing spatial window size from 1 to 5 pixels has somewhat improved the final crop map. However, the classification maps obtained from GFSAE show more accurate field maps with less confusion with other classes compared to other classifiers. Specifically, GFSAE with
w = 1 and 3 settings have performed best among all classifiers.
3.3. Results on the Winnipeg Dataset
Accuracy metrics for the Winnipeg dataset are represented in
Figure 8. The results confirm that increasing spatial neighborhood window size (i.e.,
w from 1 to 3 pixels) increases the classification accuracy of SAE in all accuracy metrics (about 1%). Here, setting the window size larger than three pixels (about 15 m spatial extent), the accuracy decreases because of the over-smoothing effect. Specifically, for the broadleaf class, which has a minimal spatial extent, the algorithm could not identify any pixels correctly (See per-class F-score for the broadleaf class of
w = 5 settings in
Table 5). This is due to involving many non-broadleaf pixels in classifying target broadleaf pixels.
Confirming the CEF dataset results, the proposed GFSAE resulted in considerable performance improvement compared to the original SAE on the Winnipeg dataset. It increases classification accuracy by up to 5%. Accuracy metrics and training time are represented in
Table 5 for the Winnipeg dataset.
The first part of
Table 5 (i.e., SAE results) shows that all per-class F-scores increase by increasing
w to 3 pixels for the Winnipeg dataset except the broadleaf class, which has a minimal spatial extent. The second part proves the higher performance of the proposed GFSAE in terms of all per-class F-scores (except the narrow broadleaf class) and the mean F-score with comparable calculation time. The superiority of the proposed GFSAE is also proved by comparing accuracy metrics displayed in
Table 5. Both the original SAE and the proposed GFSAE have produced better results than the traditional classifiers (SVM and RF) on the Winnipeg dataset, and the GFSAE has much superior performance than all the compared methods. It also has a comparable calculation time with respect to these traditional classifiers.
Figure 9 shows commission and omission errors for the Winnipeg dataset. As we see again, the superior performance of the proposed GFSAE framework is proved in reducing both commission and omission errors of different crop types except the broadleaf class. The broadleaf class has a minimal spatial extent, and hence filtering process mixes its information with other classes. Its lower amount of training data compared to other classes (see
Table 2) aggravates this crop class’s challenging situation, leading to its higher omission error.
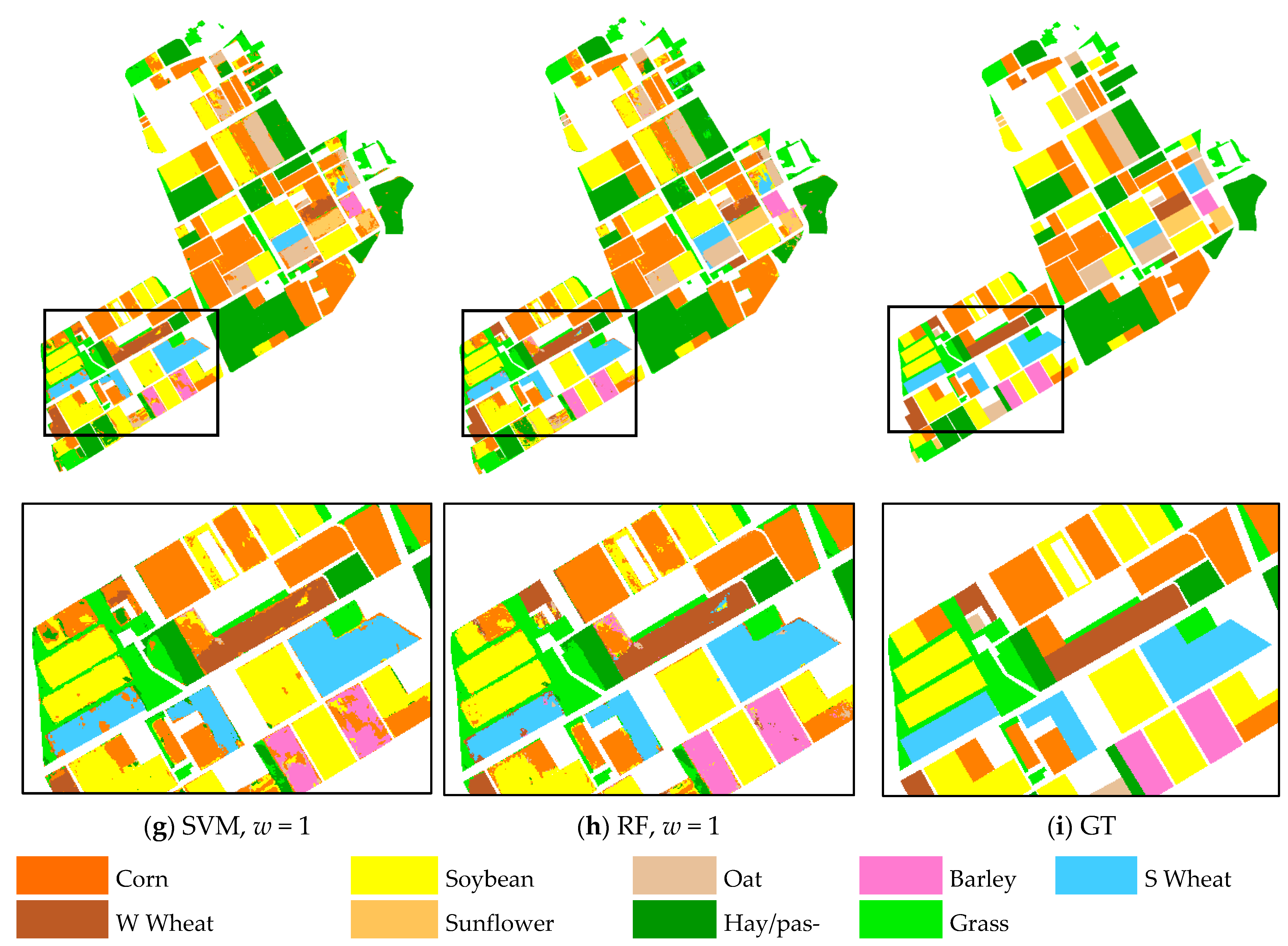
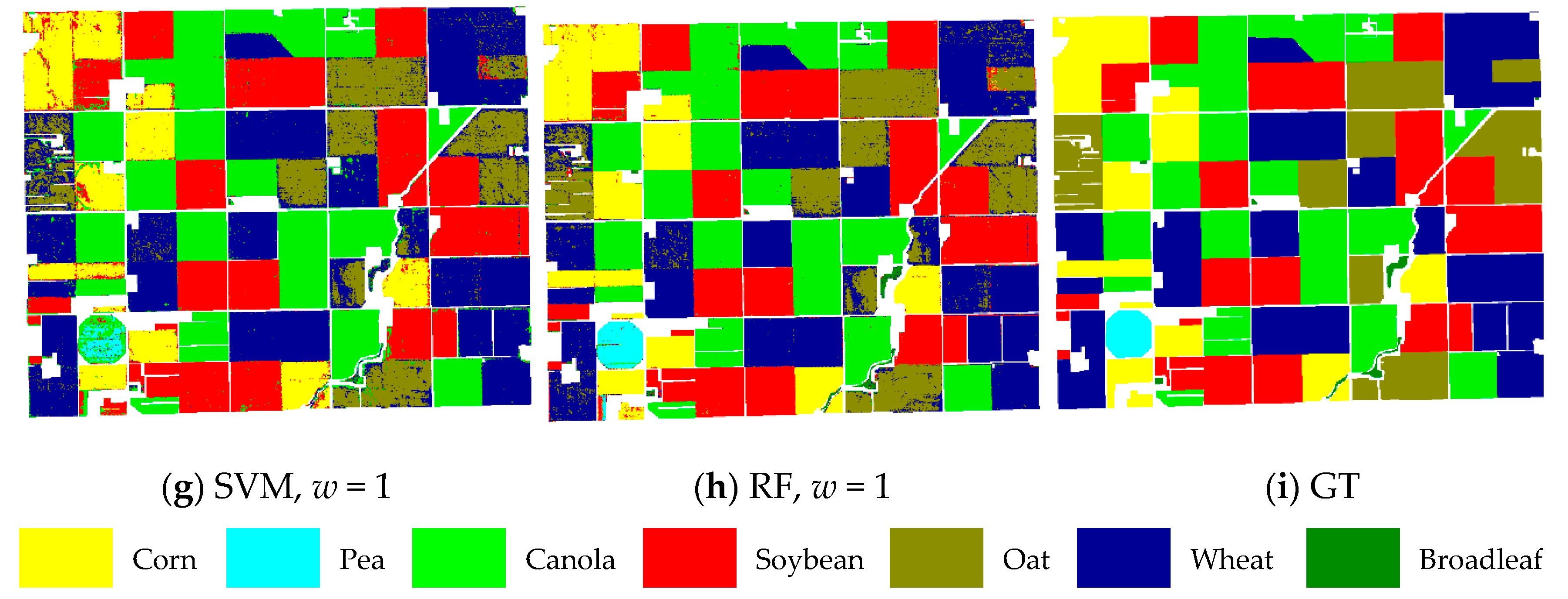
The ground reference map and final classification maps obtained from SAE and GFSAE for all
w settings and traditional SVM and RF classifiers are displayed in
Figure 10 for the Winnipeg dataset. As we can see, in the case of SAE increasing spatial window size did not have a considerable change in the result maps. However, the classification maps obtained from GFSAE significantly improve the original SAE method. They show more accurate field maps with less confusion with other classes compared to SAE results. Specifically, GFSAE with
w = 1 and 3 settings perform best. Comparing SAE- and GFSAE-obtained crop maps with ones of the traditional SVM and RF methods also proves the superiority of the proposed method. Specifically, more accurate and homogenous crop maps are obtained with GFSAE. This again proves the superior performance of the proposed GFSAE method in crop mapping compared to the original SAE and traditional methods.
3.4. Accuracy Analysis in Boundary Areas
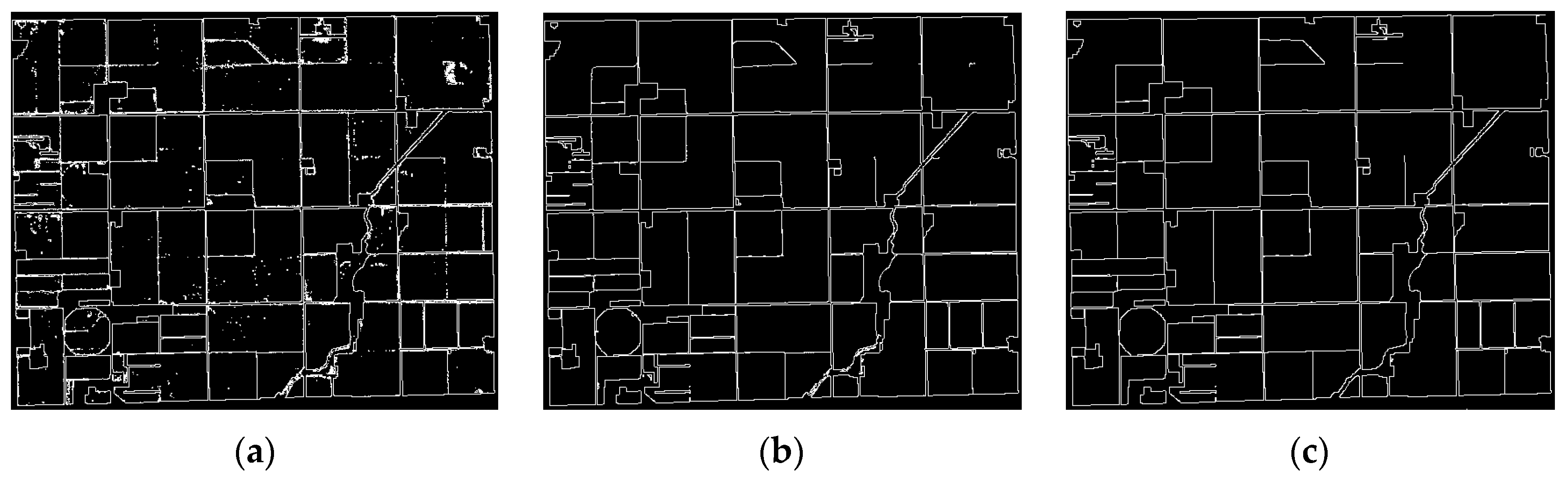
To evaluate the quality of obtained field boundaries, edge detection based on the Sobel operation was performed on classification maps from SAE and GFSAE (
w = 3) and the ground truth data (
Figure 11).
Visual interpretation of
Figure 11 proves the high ability of the proposed method in field boundary extraction. Although SAE applies spatial features in the classification process, there are still some issues in its boundary map. It loses some edges and creates extra edges. The obtained boundary map was compared to the real one (from GT) for accuracy assessment as a binary classification result. For this purpose, edges and non-edge pixels in a 30 m neighborhood of real boundaries were compared to the corresponding ones in SAE and GFSAE boundary maps.
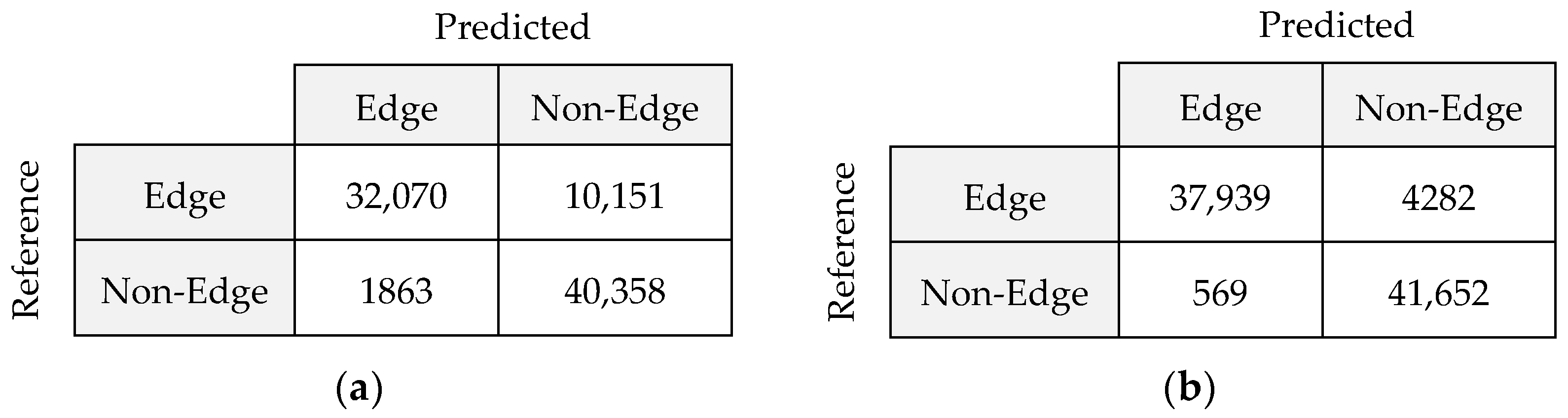
Figure 12 shows the confusion matrices for SAE and GFSAE.
Comparing
Figure 12a,b shows that using a guided filter in the post-process step can restore 42% of missing edges in SAE and also remove 62% of extra pixels.
Table 6 presents accuracy metrics obtained from confusion matrices.
Table 6 shows that 86.68% of pixels belonging to the field boundaries are assigned correctly in the proposed method. It improves the producer’s accuracy of edge class by more than 9% compared to the SAE method. Moreover, 98.58% of the assigned pixels to the edge class were truly in the edge class, up to 2% better than the SAE results. Generally, classification accuracy and Edge class F-score increase by approximately 7% and 6% when a guided filter is performed.
Figure 13 depicts the boundary maps of SAE and GFSAE (
w = 3) compared to the ground truth for the Winnipeg dataset.
Comparing the boundary maps of SAE and GFSAE with ground truth proves the high potential of the proposed method in field boundary extraction. Similar to the CEF dataset, applying a guided filter in the Winnipeg dataset eliminates redundant edges in the SAE map and adds missing edges.
Figure 14 shows the proposed method’s confusion matrix compared with SAE.
Type I error in the SAE method is 1863, meaning these pixels are incorrectly assigned to the edge class. By implementing a guided filter, 70% of those pixels are removed. Moreover, analyzing type II errors in both confusion matrices shows that the proposed method can restore 58% of edge pixels that are not detected in the SAE algorithm.
Table 7 presents accuracy metrics obtained from confusion matrices on the Winnipeg dataset.
Table 7 shows that the proposed method increases the producer’s accuracy of edge class to 89.86%, which is approximately 14% higher than the SAE algorithm. Moreover, 98.52% of pixels assigned to the edge class were indeed in the edge class, which increases the user’s accuracy by 4% compared to SAE results. According to F-score and overall accuracy metrics, the field boundary detection accuracy of the proposed method is 94%, approximately. It shows that using a guided filter improves boundary detection by 9% compared to the SAE method.
4. Conclusions
Field boundary information can be used as auxiliary information in the classification framework, which helps to separate pixels from different crop fields and improve crop mapping accuracy. However, these valuable data sources, which implicitly exist in image data, are usually ignored in most classification frameworks. This study presents a novel classification framework based on DL methods that can successfully involve field boundary information in accurate crop mapping applications. The proposed framework can exploit input data’s spectral, spatial, and temporal information and utilize field boundaries to improve mapping accuracy. We stacked multi-temporal image data to generate a data cube with spectro-temporal feature vectors. Spatial information is included in the classification framework via window-based feature extraction and a post-processing step that employs the guided filter as an edge-aware filtering method. It filters the classification maps using a guidance image extracted from the input data and maintains most of its edge information.
The proposed framework was evaluated using two time series of high-resolution PlanetLab’s PlanetScope and RapidEye images over two agricultural sites. The experiments were conducted to observe the performance of GFSAE in comparison to SAE and two traditional SVM and RF classifiers. Firstly, the efficiency of spatial information inclusion strategy considering different neighbourhood sizes and the effect of the post-processing step for filtering classification maps were evaluated.
The experimental results were conducted based on classification accuracy and field boundary accuracy. It showed that increasing window size in spatial feature extraction could increase the classification accuracy of SAE until a specific size and decrease. However, applying a guided filter improves classification accuracy by up to 5% compared to the SAE results. The experiments also demonstrated the superior performance of the GFSAE compared to the traditional SVM and RF methods. Moreover, field boundary extraction accuracy is evaluated. The obtained results prove the proposed method’s ability to reach 92.72% and 94.26% accuracy in field boundary extraction of two datasets, respectively.
Although the effect of the edge-aware filtering on the classification performance was higher than the window-based feature extraction method, using the two strategies together produced better results. The window-based strategy improved the overall accuracies by generating more homogeneous regions. In contrast, the filtering-based strategy reduced the commission and omission errors and improved the classification accuracy in boundary areas. The proposed GFSAE improved class scores from 0.44 to 17.01%. The proposed method showed its potential and efficiency in crop classification, specifically when available high-resolution satellite imagery.
The study results demonstrated the applicability and high performance of the proposed edge-aware crop mapping framework, GFSAE, in terms of accuracy and calculation time. It can improve final crop maps needing no additional data and with almost no increase in calculation time compared to the original SAE. Although we used SAE as the base classifier here, the guided filtering process could be used for any other classifier that can estimate class probabilities (DL-based or traditional classifiers). In addition, the proposed framework is extendable to all types of remote-sensing imagery. Hence, it introduces a general framework to improve crop mapping accuracy with no additional time or data (and therefore no additional cost). For future works, we will investigate using an edge-aware filtering process localized to consider field sizes in the filtering process. We will also experiment with applying this filter during the feature extraction phase.