DNA Methylation Inhibitor 5-Azacytidine Promotes Leaf Senescence in Pak Choi (Brassica rapa subsp. chinensis) by Regulating Senescence-Related Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatment
2.2. Surface Color and Chl Analyses
2.3. Chl-Degrading Enzyme Activity
2.4. Chl-Degradation Derivative Content
2.5. RNA Isolation and Transcript Quantification
2.6. DNA Methylation-Sensitive Restriction Enzyme Test
2.7. Bisulfite Sequencing Detection
2.8. Statistical Analysis
3. Results
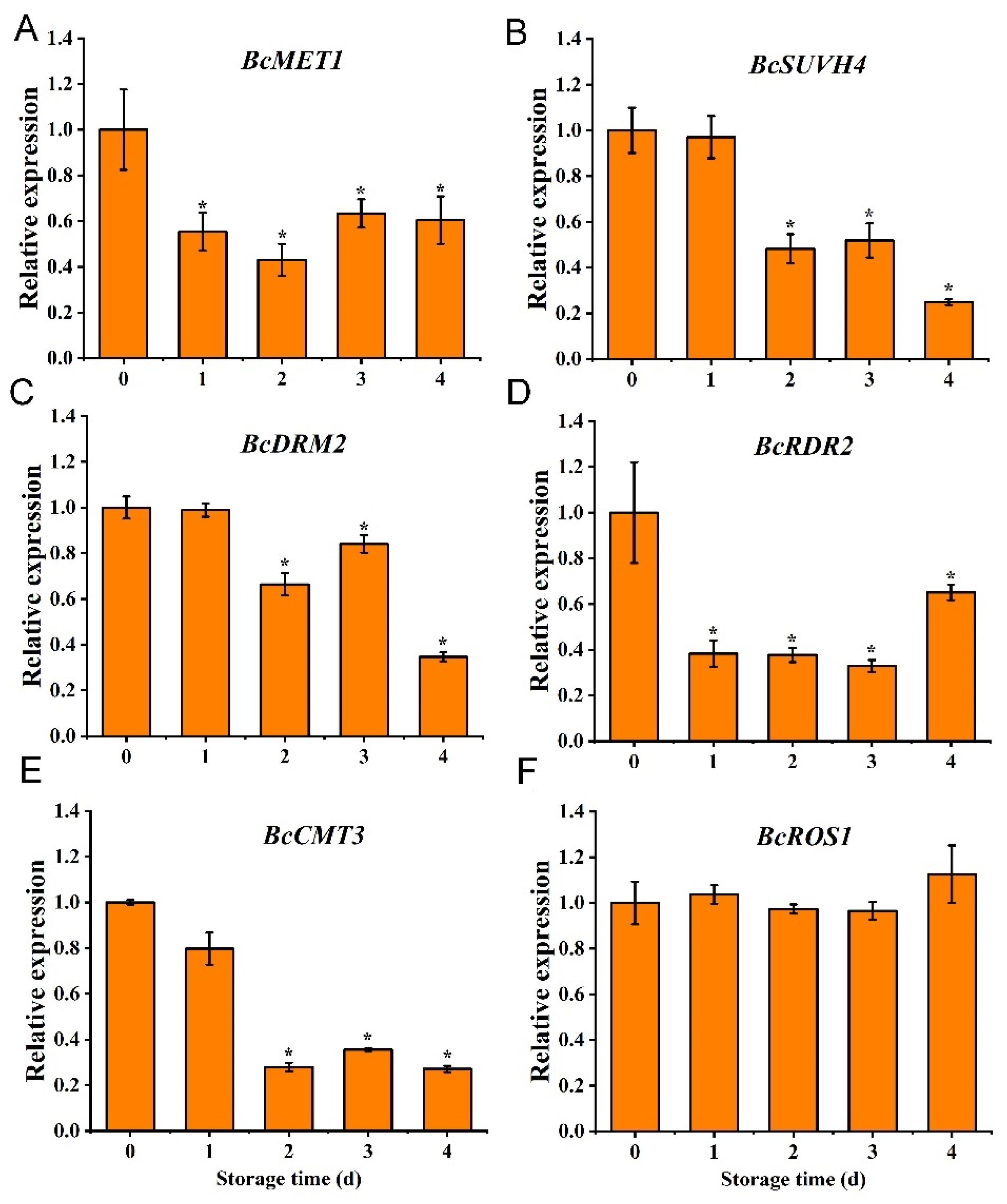
3.1. Expression of DNA Methyltransferase
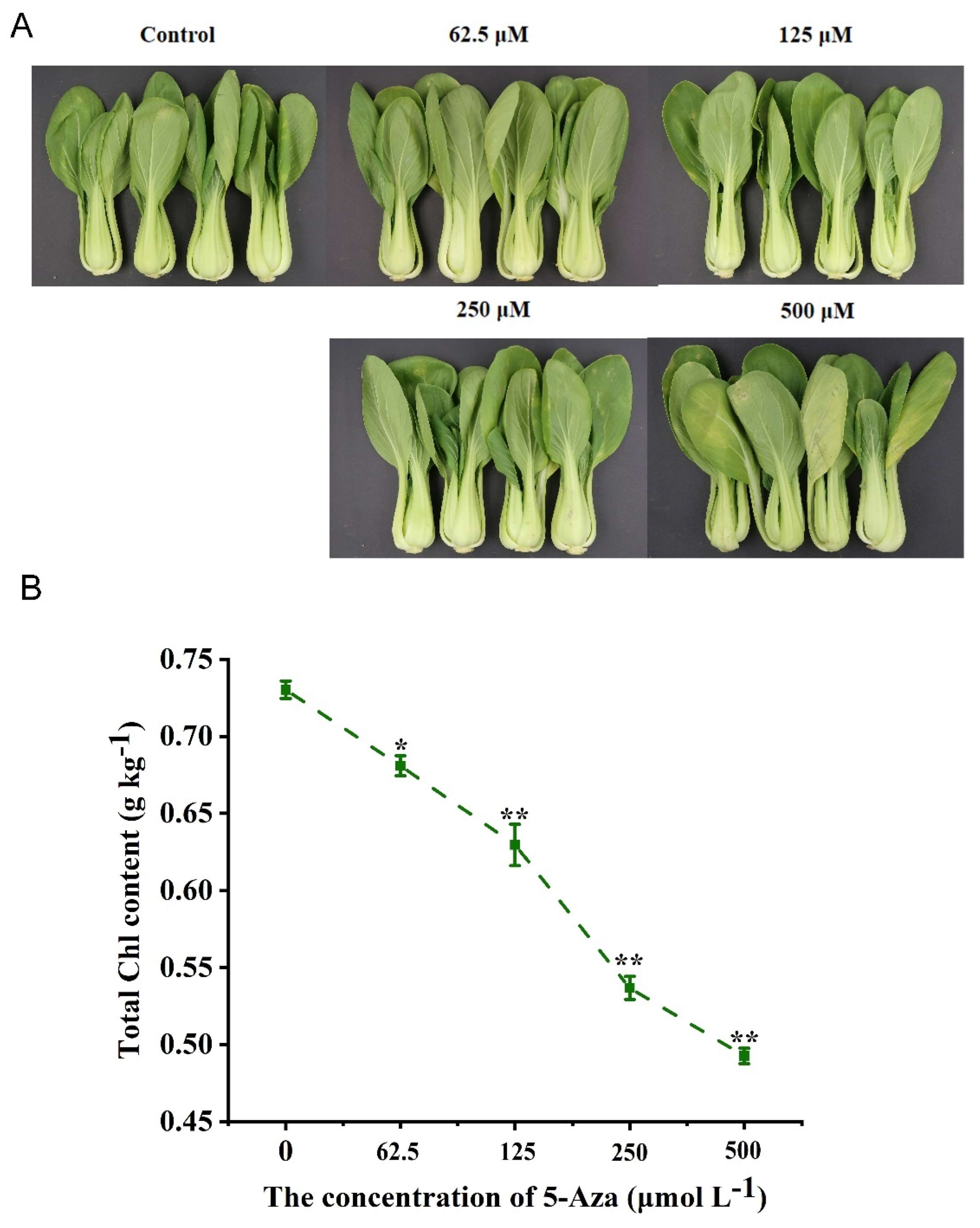
3.2. Treatment with Different Concentrations of 5-Aza on the Phenotypic Characteristics and Total Chl Content
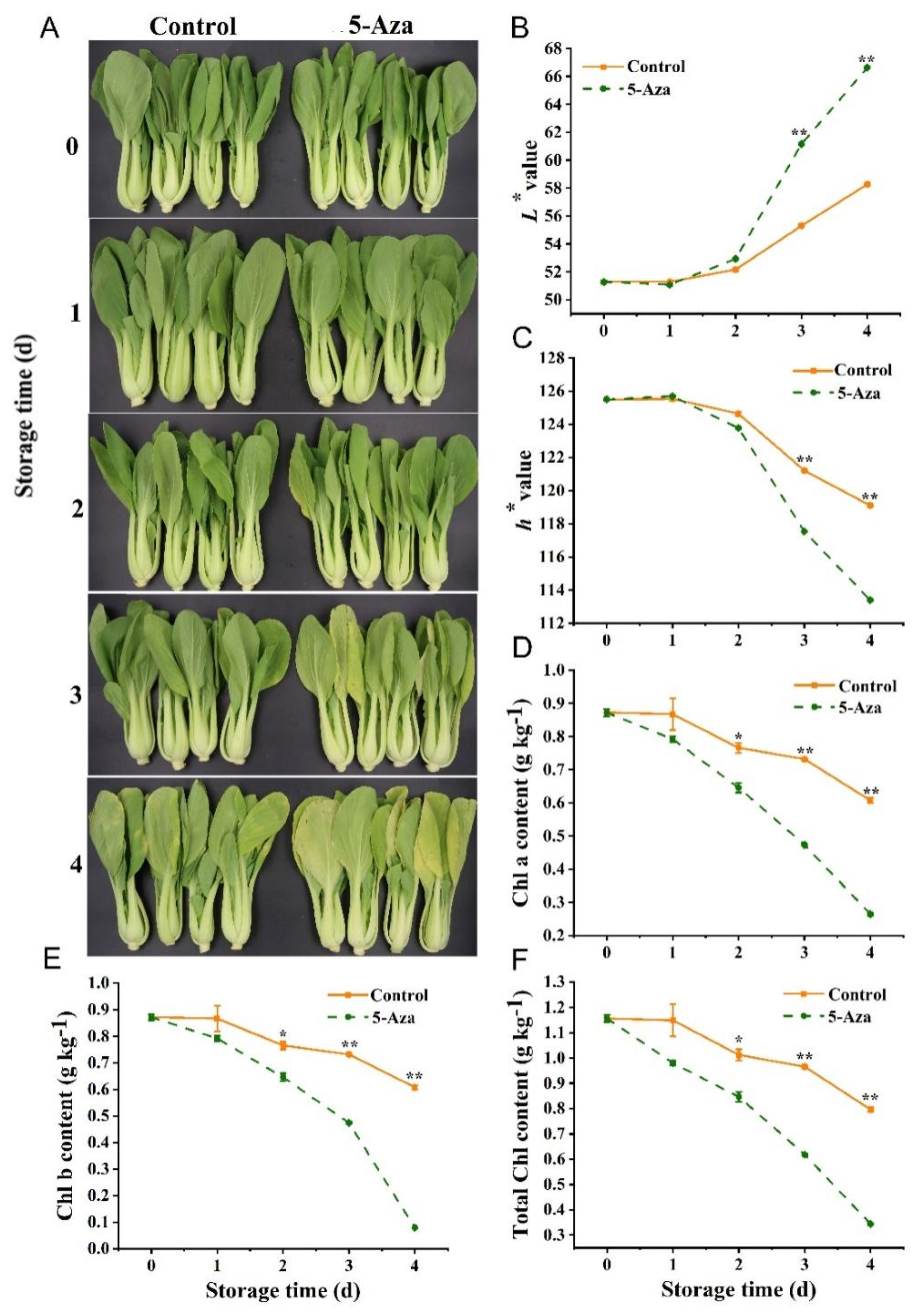
3.3. Surface Color and Total Chl Content
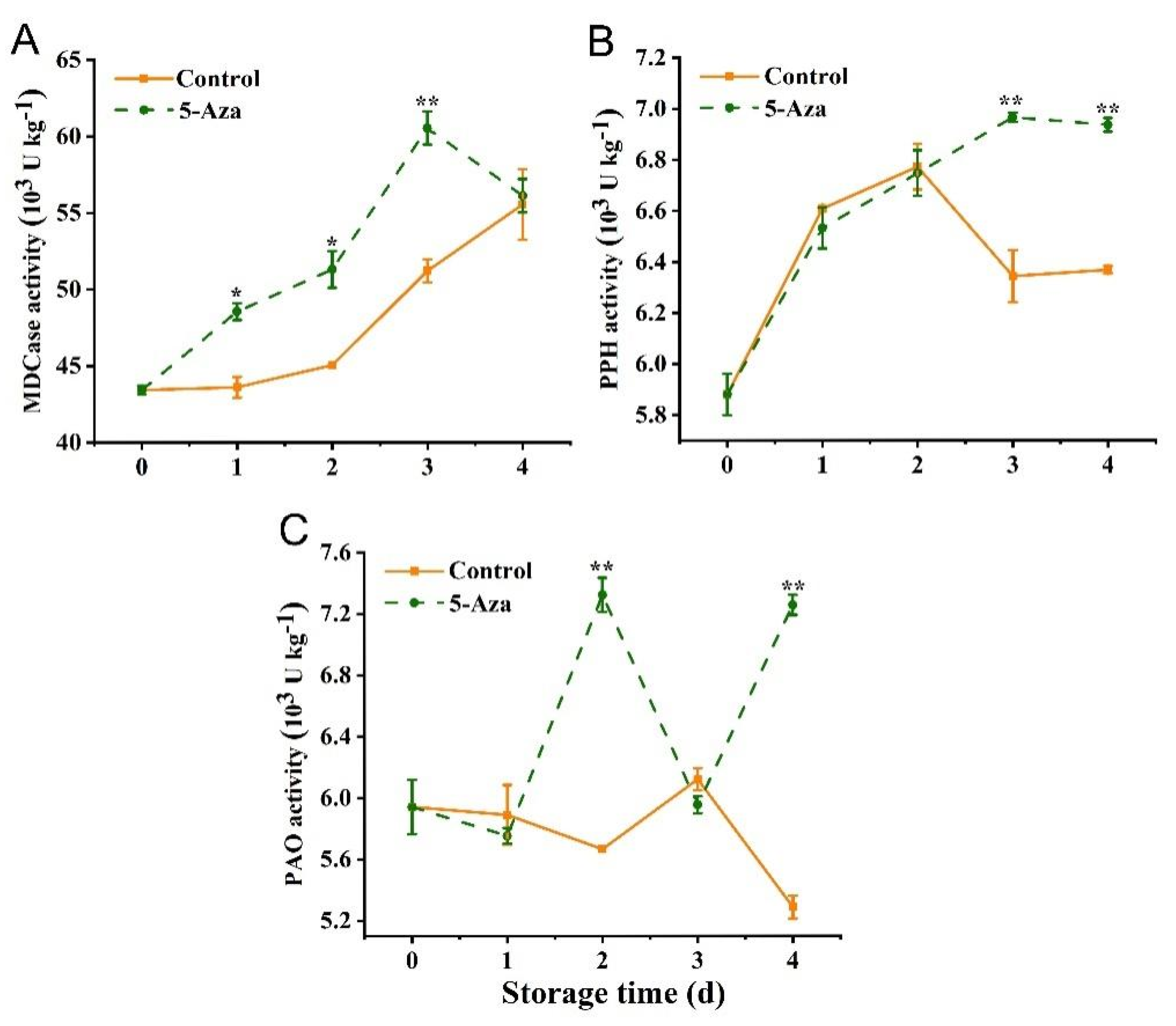
3.4. Activity of Chl-Degrading Enzymes
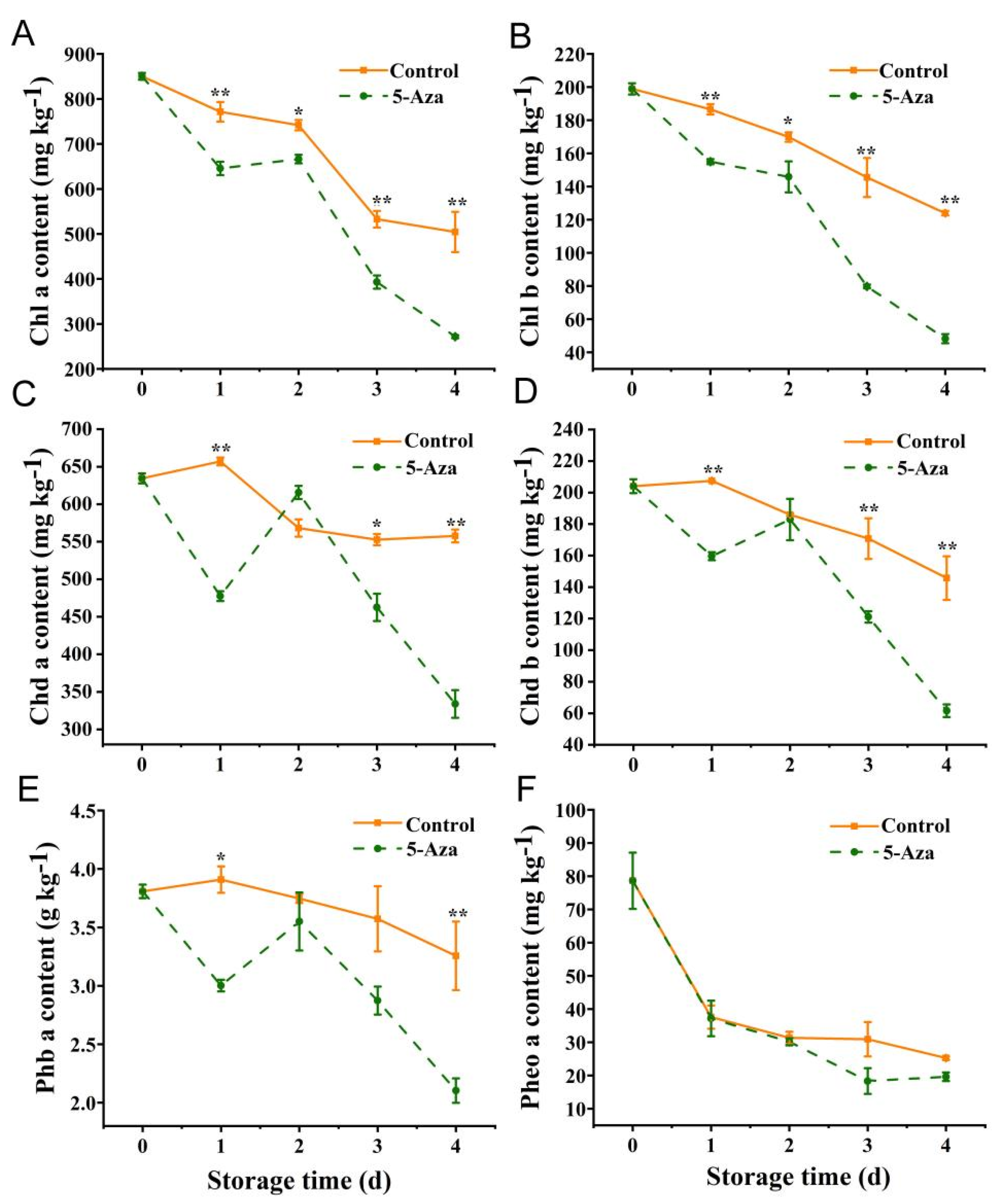
3.5. Chl-Degradation Derivative Content
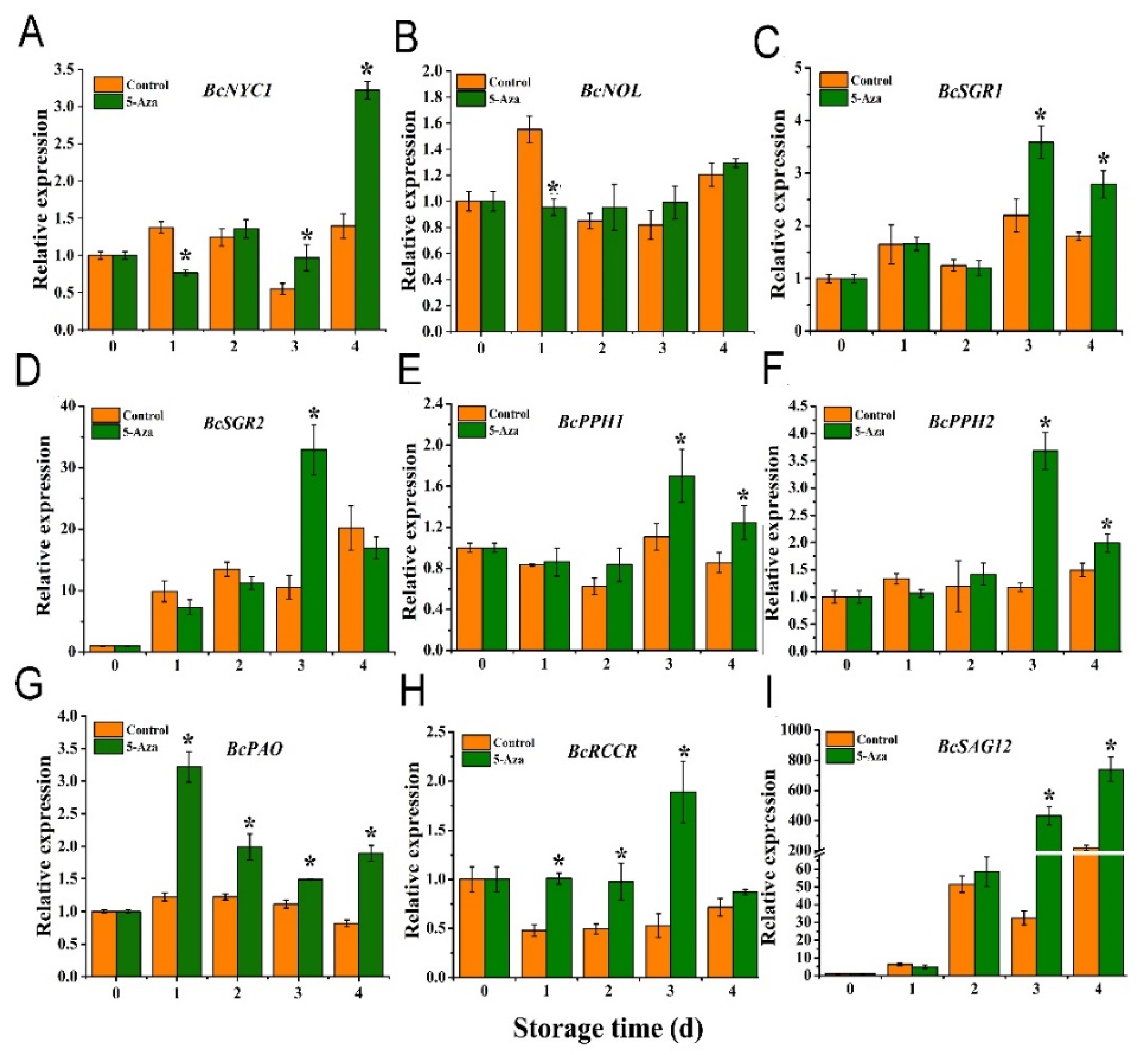
3.6. Expression of Senescence-Related Genes
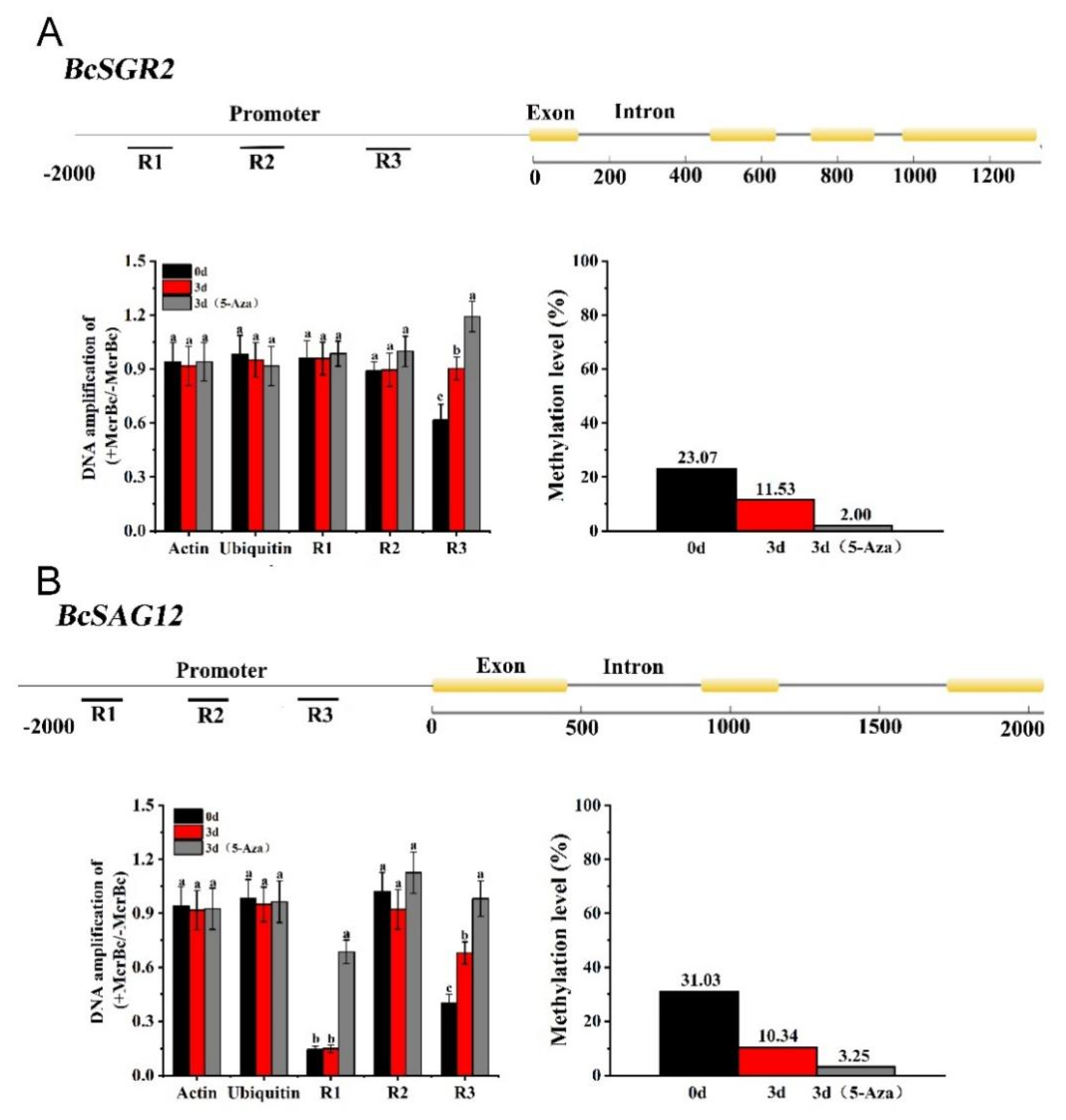
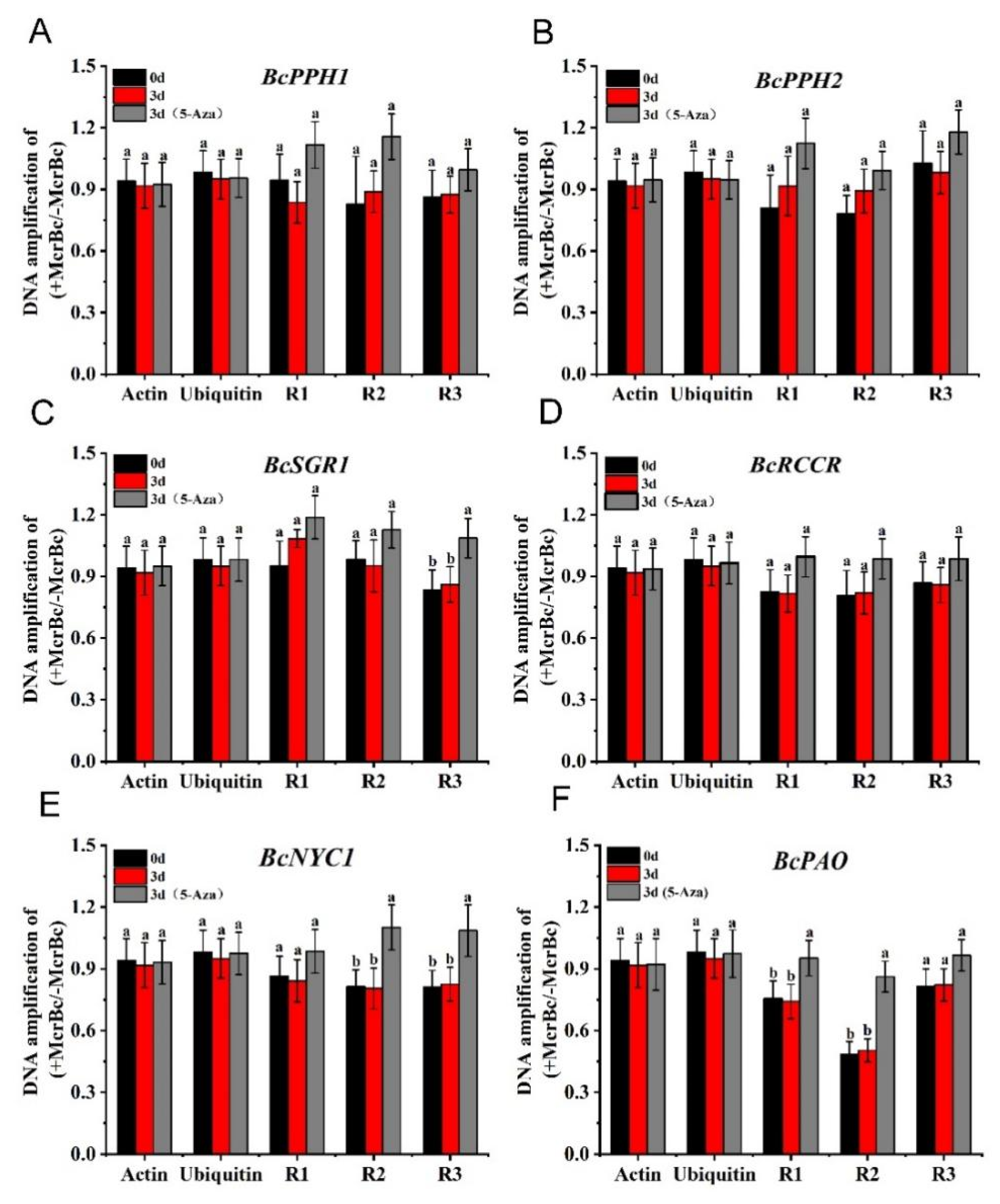
3.7. DNA Methylation of Senescence-Related Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- An, R.H.; Luo, S.F.; Zhou, H.S.; Zhang, Y.T.; Zhang, L.G.; Hu, H.L.; Li, P.X. Effects of hydrogen-rich water combined with vacuum precooling on the senescence and antioxidant capacity of pakchoi (Brassica rapa subsp. Chinensis). Sci. Hortic. 2021, 289, 110469. [Google Scholar] [CrossRef]
- Ubeed, H.M.S.; Wills, R.B.H.; Bowyer, M.C.; Vuong, Q.V.; Golding, J.B. Interaction of exogenous hydrogen sulphide and ethylene on senescence of green leafy vegetables. Postharvest Biol. Technol. 2017, 133, 81–87. [Google Scholar] [CrossRef]
- Zhou, F.H.; Zuo, J.H.; Xu, D.Y.; Gao, L.P.; Wang, Q.; Jiang, A.L. Low intensity white light-emitting diodes (LED) application to delay senescence and maintain quality of postharvest pakchoi (Brassica campestris L. ssp. chinensis (L.) Makino var. communis Tsen et Lee). Sci. Hortic. 2020, 262, 109060. [Google Scholar] [CrossRef]
- Kuai, B.K.; Chen, J.Y.; Hortensteiner, S. The biochemistry and molecular biology of chlorophyll breakdown. J. Exp. Bot. 2018, 69, 751–767. [Google Scholar] [CrossRef] [PubMed]
- Hortensteiner, S. Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci. 2009, 14, 155–162. [Google Scholar] [CrossRef]
- Shimoda, Y.; Ito, H.; Tanaka, A. Arabidopsis STAY-GREEN, Mendel’s Green Cotyledon Gene, Encodes Magnesium-Dechelatase. Plant Cell 2016, 28, 2147–2160. [Google Scholar] [CrossRef]
- Zhang, K.W.; Xia, X.Y.; Zhang, Y.Y.; Gan, S.S. An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J. 2012, 69, 667–678. [Google Scholar] [CrossRef]
- Shi, J.Y.; Gao, L.P.; Zuo, J.H.; Wang, Q.; Wang, Q.; Fan, L.S. Exogenous sodium nitroprusside treatment of broccoli florets extends shelf life, enhances antioxidant enzyme activity, and inhibits chlorophyll-degradation. Postharvest Biol. Technol. 2016, 116, 98–104. [Google Scholar] [CrossRef]
- Sato, T.; Shimoda, Y.; Matsuda, K.; Tanaka, A.; Ito, H. Mg-dechelation of chlorophyll a by Stay-Green activates chlorophyll b degradation through expressing Non-Yellow Coloring 1 in Arabidopsis thaliana. J. Plant Physiol. 2018, 222, 94–102. [Google Scholar] [CrossRef]
- Vanyushin, B.F.; Ashapkin, V.V. DNA methylation in higher plants: Past, present and future. Biochim. Biophys. Acta. 2011, 1809, 360–368. [Google Scholar] [CrossRef]
- Yolcu, S.; Li, X.J.; Li, S.B.; Kim, Y.J. Beyond the genetic code in leaf senescence. J. Exp. Bot. 2018, 69, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.W.; Henderson, I.R.; Jacobsen, S.E. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 2005, 6, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Penterman, J.; Zilberman, D.; Huh, J.H.; Ballinger, T.; Henikoff, S.; Fischer, R.L. DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. USA 2007, 104, 6752–6757. [Google Scholar] [CrossRef]
- Chandler, V.L.; Stam, M. Chromatin conversations: Mechanisms and implications of paramutation. Nat. Rev. Genet. 2004, 5, 532–544. [Google Scholar] [CrossRef]
- Du, J.; Johnson, L.M.; Jacobsen, S.E.; Patel, D.J. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 2015, 16, 519–532. [Google Scholar] [CrossRef]
- Kim, M.; Hyonhwa, O.; Jee Woong, L.; Youbong, H.; Robert, L.F.; Yeonhee, C. Temporal and Spatial Downregulation of Arabidopsis MET1 Activity Results in Global DNA Hypomethylation and Developmental Defects. Mol. Cell 2008, 26, 611–615. [Google Scholar]
- Ay, N.; Janack, B.; Humbeck, K. Epigenetic control of plant senescence and linked processes. J. Exp. Bot. 2014, 65, 3875–3887. [Google Scholar] [CrossRef]
- Ogneva, Z.V.; Dubrovina, A.S.; Kiselev, K.V. Age-associated alterations in DNA methylation and expression of methyltransferase and demethylase genes in Arabidopsis thaliana. Biol. Plant. 2016, 60, 628–634. [Google Scholar] [CrossRef]
- Yuan, L.; Wang, D.; Cao, L.W.; Yu, N.N.; Liu, K.; Guo, Y.F.; Gan, S.S.; Chen, L.P. Regulation of Leaf Longevity by DML3-Mediated DNA Demethylation. Mol. Plant 2020, 13, 1149–1161. [Google Scholar] [CrossRef]
- Gao, S.; Gao, J.; Zhu, X.Y.; Song, Y.; Li, Z.P.; Ren, G.D.; Zhou, X.; Kuai, B.K. ABF2, ABF3, and ABF4 Promote ABA-Mediated Chlorophyll Degradation and Leaf Senescence by Transcriptional Activation of Chlorophyll Catabolic Genes and Senescence-Associated Genes in Arabidopsis. Mol. Plant 2016, 9, 1272–1285. [Google Scholar] [CrossRef] [PubMed]
- Dissanayake, P.K.; Yamauchi, N.; Shigyo, M. Chlorophyll degradation and resulting catabolite formation in stored Japanese bunching onion (Allium fistulosum L.). J. Sci. Food Agric. 2008, 88, 1981–1986. [Google Scholar] [CrossRef]
- Zhong, S.; Fei, Z.; Chen, Y.R.; Zheng, Y.; Huang, M.; Vrebalov, J.; McQuinn, R.; Gapper, N.; Liu, B.; Xiang, J.; et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 2013, 31, 154–159. [Google Scholar] [CrossRef]
- Popova, O.V.; Dinh, H.Q.; Aufsatz, W.; Jonak, C. The RdDM pathway is required for basal heat tolerance in Arabidopsis. Mol. Plant 2013, 6, 396–410. [Google Scholar] [CrossRef] [PubMed]
- Eichten, S.R.; Springer, N.M. Minimal evidence for consistent changes in maize DNA methylation patterns following environmental stress. Front. Plant Sci. 2015, 6, 308. [Google Scholar] [CrossRef]
- Miryeganeh, M. Synchronization of senescence and desynchronization of flowering in Arabidopsis thaliana. AoB Plants 2020, 12, 3–14. [Google Scholar] [CrossRef]
- Tariq, M.; Paszkowski, J. DNA and histone methylation in plants. Trends Genet. 2014, 20, 244–251. [Google Scholar] [CrossRef]
- Saze, H.; Tsugane, K.; Kanno, T.; Nishimura, T. DNA methylation in plants: Relationship to small RNAs and histone modifications, and functions in transposon inactivation. Plant Cell Physiol. 2012, 53, 766–784. [Google Scholar] [CrossRef]
- Miryeganeh, M.; Saze, H. Epigenetic inheritance and plant evolution. Popul. Ecol. 2019, 62, 17–27. [Google Scholar] [CrossRef]
- Zhang, H.M.; Deng, X.Y.; Miki, D.; Cutler, S.; La, H.G.; Hou, Y.J.; Oh, J.; Zhu, J.K. Sulfamethazine suppresses epigenetic silencing in Arabidopsis by impairing folate synthesis. Plant Cell 2012, 24, 1230–1241. [Google Scholar] [CrossRef]
- Duan, H.Y.; Liu, W.X.; Li, J.Y.; Ding, W.K.; Zhou, Y.Q.; Wang, H.N.; Jiang, L.N.; Zhou, Y.Q. Growth and DNA methylation level of Triticum aestivum seedlings treated with 5-azacytidine. Pak. J. Bot. 2016, 48, 1585–1591. [Google Scholar]
- Zhong, L.; Xu, Y.H.; Wang, J.B. The effect of 5-azacytidine on wheat seedlings responses to NaCl stress. Biol. Plant. 2010, 54, 753–756. [Google Scholar] [CrossRef]
- Matthew, S.; McCabe, M.; Garratt, L.; Schepers, F.; Jordi, W.; Stoopen, G.; Davelaar, E.; van Rhijn, J.; Power, J.; Davey, M. Effects of P(SAG12)-IPT gene expression on development and senescence in transgenic lettuce. Plant Physiol. 2001, 127, 505–516. [Google Scholar]
- Nakajima, S.; Ito, H.; Tanaka, R.; Tanaka, A. Chlorophyll b reductase plays an essential role in maturation and storability of Arabidopsis seeds. Plant Physiol. 2012, 160, 261–273. [Google Scholar] [CrossRef]
- Xu, Y.M.; Xiao, X.M.; Zeng, Z.X.; Tan, X.L.; Liu, Z.L.; Chen, J.W.; Su, X.G.; Chen, J.Y. BrTCP7 Transcription Factor Is Associated with MeJA-Promoted Leaf Senescence by Activating the Expression of BrOPR3 and BrRCCR. Int. J. Mol. Sci. 2019, 20, 3963. [Google Scholar] [CrossRef]
- Tan, X.L.; Fan, Z.Q.; Shan, W.; Yin, X.R.; Kuang, J.F.; Lu, W.J.; Chen, J.Y. Association of BrERF72 with methyl jasmonate-induced leaf senescence of Chinese flowering cabbage through activating JA biosynthesis-related genes. Hortic. Res. 2018, 5, 2–11. [Google Scholar] [CrossRef]
- Hortensteiner, S. Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 2006, 57, 55–77. [Google Scholar] [CrossRef]
- Yaish, M.W. DNA Methylation-Associated Epigenetic Changes in Stress Tolerance of Plants. Physiol. Mol. Biol. Plants 2013, 2013, 427–440. [Google Scholar]
- Zhang, H.M.; Lang, Z.B.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Alleman, M.; Sidorenko, L.; McGinnis, K.; Seshadri, V.; Dorweiler, J.E.; White, J.; Sikkink, K.; Chandler, V.L. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 2006, 442, 295–298. [Google Scholar] [CrossRef]
- Lang, Z.B.; Wang, Y.H.; Tang, K.; Tang, D.G.; Datsenka, T.; Cheng, J.F.; Zhang, Y.J.; Handa, A.K.; Zhu, J.K. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. USA 2017, 114, 4511–4519. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Yazaki, J.; Sundaresan, A.; Cokus, S.; Chan, S.W.; Chen, H.M.; Henderson, I.R.; Shinn, P.; Pellegrini, M.; Jacobsen, S.E.; et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 2006, 126, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.K. Active DNA demethylation in plants and animals. Cold Spring Harb. Symp. Quant. Biol. 2012, 77, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.G.; Zhang, H.M.; Julian, R.; Tang, K.; Xie, S.J.; Zhu, J.K. Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, 3553–3557. [Google Scholar] [CrossRef]
- Huang, H.; Liu, R.; Niu, Q.; Tang, K.; Zhang, B.; Zhang, H.; Chen, K.; Zhu, J.-K.; Lang, Z. Global increase in DNA methylation during orange fruit development and ripening. Proc. Natl. Acad. Sci. USA 2019, 116, 1430–1436. [Google Scholar] [CrossRef]
- Sicilia, A.; Scialo, E.; Puglisi, I.; Lo Piero, A.R. Anthocyanin Biosynthesis and DNA Methylation Dynamics in Sweet Orange Fruit [Citrus sinensis L. (Osbeck)] under Cold Stress. J. Agric. Food Chem. 2020, 68, 7024–7031. [Google Scholar] [CrossRef]
- Mao, H.D.; Wang, H.W.; Liu, S.X.; Li, Z.G.; Yang, X.H.; Yan, J.B.; Li, J.S.; Tran, L.S.P.; Qin, F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 2015, 6, 8326. [Google Scholar] [CrossRef]
| Primer Name | Forward (5′–3′) | Reverse (5′–3′) | |
|---|---|---|---|
| qRT-PCR | BcNYC1 | GGCTTGGTGGAGTTATCATTGG | ACGGATTCAGAACTGCGAGAT |
| BcNOL | ACACAATCTATCGCCTGGAATG | AGATACTCAGCAACCACTTCAG | |
| BcSGR1 | AGCTTATTCAGACAAACGGGGT | TGGTGGATGCTTCTTGTCATCA | |
| BcSGR2 | ACAGTGACATAACCGCTAAGC | CTCCGCTAATGTGGCAATGAA | |
| BcPPH1 | GTGGTCGGTGAGAATGAGGA | CGCAGTGAGAAGTAGTGATTCG | |
| BcPPH2 | TGTGGTTGGTGAGAATGATGAC | TCGCAGTGAGAAGGAGTGATT | |
| BcPAO | TCTCTGAAGGAAGGTTGGATGA | TGAAGTAGCAGCCTGTGGAA | |
| BcRCCR | TCATCGTCAGTCACTCCTCAA | AACCTCAAGAACTTGCGTAGC | |
| BcSAG12 | CACTGGCGGCTTAACCACTGAA | GAAGATTGGCTGTATCCTACGGC | |
| BcMET1 | TGGTTTGGTTCTCGACGGAG | CGAGTTGTACAGTGCCCAGT | |
| BcSUVH4 | ATGATTGGTGACCTGCCAGG | ACACCAAGCCCTGAATCTCG | |
| BcDRM2 | TTCCAAACGAGCCAGGACTC | GACGCTCGGTCCTACTCATG | |
| BcRDR2 | AGAGCCTATGTTACGCCTTCA | TAGCCTTCCTTGGAGTTCACA | |
| BcCMT3 | GCAACTTGTTCGCTCAATCTC | GCCACTTCCGCTGTTACTC | |
| BcROS1 | TTTGCTGCAGGACTAGCTCC | GGTACTGGATACTCCGCAGC | |
| BcActin | TCTCTTCCACACGCCATCC | GTCTCCATCTCCTGCTCATAGT | |
| BcUbiquitin | GAGGTGGAGAGCAGTGACAC | GCTGTTTTCCGGCGAAGATC | |
| Primers used for qPCR of enzyme digestion | BcSGR2-R1 | CTCCTTTACCCGAACCAACAAT | AGAGTACCCAATCTCCCTAACG |
| BcSGR2-R2 | ATAGATAAGTTCCGACCGAAGC | GTAGCGTTGACGAGTTCTCTT | |
| BcSGR2-R3 | CACCTCGTCAGAGCGGATT | TTGTTGTTTGCGTGTTGGAGT | |
| BcSAG12-R1 | TACACCCATACATCAGCATTGT | TCAGATTCCAGTAGGCAAAGAT | |
| BcSAG12-R2 | GAAGAAGACTGACCAGCGATG | GAACAGACGAGCCGATCCT | |
| BcSAG12-R3 | TGAACCGAATAAACCGAATTGG | AAGCCCGAAGCACAAACTG | |
| BcRCCR-R1 | GGTCGGTGGAGAACATGGT | TTCATCTGCTCGGTCAAGAAC | |
| BcRCCR-R2 | AAGCACGGGATTAGATTTGGT | ACGGAAACTACCTACTAATTGC | |
| BcRCCR-R3 | CCAATTAAGTCGCTCTTGAGTC | TACACTAAACCGAACCCGTTAA | |
| BcNOL-R1 | AGACAGCAACCAAGAGGAACA | CTACCAACCTGGCAGATCAATG | |
| BcNOL-R2 | AGAGATGGCTGAGGCAAGG | GCTTCTTCCACACGCTTCC | |
| BcNOL-R3 | GTGAAGAGAAGAAGTTGATGGT | GAGCAGAAGATGAGGAACAGA | |
| BcNYC1-R1 | TTACTTCTCAGTGGTGCCTTCA | ATGGTTGCCTGCTGCTCTC | |
| BcNYC1-R2 | ACAGCTCTTGCGACCGTAG | CGTGGTGGTGTCTCTTGAATC | |
| BcNYC1-R3 | CGAGATGAGGTTGCCGTAAC | TCGGAGAAGGAAAGAGATGAGG | |
| BcPAO-R1 | CAGCGGGACTTAGGTTACAGA | GACCAGTTAAGCATCCAACAGT | |
| BcPAO-R2 | GGGTTTGGTTCTGATCGGTTT | CGCAAAGATCCAAATCGAACTC | |
| BcPAO-R3 | TGGATCGGTATCGGTTATGTTC | TGGCACTTGGCATAATAAGAAC | |
| BcPPH1-R1 | TGTAAGCAGCGTCCATAGAGA | TCCGTTCCTGAGCCTAAGC | |
| BcPPH1-R2 | TGTCATCGACCTGCTGAAGAA | CGGTGAGGATGCGATTGTTAT | |
| BcPPH1-R3 | TCTTTCCTCACCGTCCTTGTAA | TTCAGATTGCGGATGCTAGAAG | |
| BcPPH2-R1 | AATGGAAGGAGGAGGAGGATG | CACAGTTGACGGTTAGAGATTG | |
| BcPPH2-R2 | GCAACGGGTCTTTCAAATTGG | GCTGGCTTGGCTAACTTCTC | |
| BcPPH2-R3 | ACTTGGCTCTTACTGTCTGTGA | TTGTTAAGGTTGACGCACGAAT | |
| BcSGR1-R1 | TGATAACAGTGGACGGTCTTCT | GGTGGATGCGGTCATTGGA | |
| BcSGR1-R2 | TCTCTTCGAGTTTGCTCTGTTC | CAATCATACACCGTGACCTCAA | |
| BcSGR1-R3 | ACGCATCATCAGAAGAAGAACC | CTTTACCGAGGCTTGGAAACC | |
| BcActin | TCTCTTCCACACGCCATCC | GTCTCCATCTCCTGCTCATAGT | |
| BcUbiquitin | GAGGTGGAGAGCAGTGACA | CAAGGTACGACCGTCTTCAAG | |
| Primers for disulfite qPCR | BcSGR2 | ATGTAGTGAAGAAGTTGGATATTAA | ATCAAAAACTAAAAACCCTTAAAAA |
| BcSAG12 | AATTAATAGAGAAGAAGATTGATTAG | TAAACTAAATCAAATAAAAACAAAC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Zhu, J.; Xu, X.; Li, P.; Liu, X. DNA Methylation Inhibitor 5-Azacytidine Promotes Leaf Senescence in Pak Choi (Brassica rapa subsp. chinensis) by Regulating Senescence-Related Genes. Agronomy 2022, 12, 2568. https://doi.org/10.3390/agronomy12102568
Li Y, Zhu J, Xu X, Li P, Liu X. DNA Methylation Inhibitor 5-Azacytidine Promotes Leaf Senescence in Pak Choi (Brassica rapa subsp. chinensis) by Regulating Senescence-Related Genes. Agronomy. 2022; 12(10):2568. https://doi.org/10.3390/agronomy12102568
Chicago/Turabian StyleLi, Yuntong, Junzhen Zhu, Xiaoyang Xu, Pengxia Li, and Xuesong Liu. 2022. "DNA Methylation Inhibitor 5-Azacytidine Promotes Leaf Senescence in Pak Choi (Brassica rapa subsp. chinensis) by Regulating Senescence-Related Genes" Agronomy 12, no. 10: 2568. https://doi.org/10.3390/agronomy12102568
APA StyleLi, Y., Zhu, J., Xu, X., Li, P., & Liu, X. (2022). DNA Methylation Inhibitor 5-Azacytidine Promotes Leaf Senescence in Pak Choi (Brassica rapa subsp. chinensis) by Regulating Senescence-Related Genes. Agronomy, 12(10), 2568. https://doi.org/10.3390/agronomy12102568





