Comparative Analysis of Pedigree-Based BLUP and Phenotypic Mass Selection for Developing Elite Inbred Lines, Based on Field and Simulated Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Populations and Trials
2.2. Efficacy of Pedigree-Based BLUP and Statistical Analysis
3. Results
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Henderson, C.R. Sire evaluation and genetic trends. J. Anim. Sci. 1973, 1973, 10–41. [Google Scholar] [CrossRef]
- Henderson, C.R. General flexibility of linear model techniques for sire evaluation. J. Dairy Sci. 1974, 57, 963–972. [Google Scholar] [CrossRef]
- Blasco, A. The Bayesian controversy in animal breeding. J. Anim. Sci. 2001, 79, 2023–2046. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bernardo, R. Prediction of Maize Single-Cross Performance Using RFLPs and Information from Related Hybrids. Crop Sci. 1994, 34, 20–25. [Google Scholar] [CrossRef]
- Gianola, D.; Rosa, G.J. One hundred years of statistical developments in animal breeding. Annu. Rev. Anim. Biosci. 2015, 3, 19–56. [Google Scholar] [CrossRef] [PubMed]
- Viana, J.M.S.; Faria, V.R.; Fonseca e Silva, F.; Vilela de Resende, M.D. Best Linear Unbiased Prediction and Family Selection in Crop Species. Crop Sci. 2011, 51, 2371–2381. [Google Scholar] [CrossRef]
- Viana, J.M.S.; de Almeida, I.F.; Vilela de Resende, M.D.; Faria, V.R.; Fonseca e Silva, F. BLUP for genetic evaluation of plants in non-inbred families of annual crops. Euphytica 2010, 174, 31–39. [Google Scholar] [CrossRef]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Van Eenennaam, A.L.; Weigel, K.A.; Young, A.E.; Cleveland, M.A.; Dekkers, J.C.M. Applied Animal Genomics: Results from the Field. Annu. Rev. Anim. Biosci. 2014, 2, 105–139. [Google Scholar] [CrossRef]
- Meuwissen, T.; Hayes, B.; Goddard, M. Accelerating Improvement of Livestock with Genomic Selection. Annu. Rev. Anim. Biosci. 2013, 1, 221–237. [Google Scholar] [CrossRef]
- Gianola, D.; Cecchinato, A.; Naya, H.; Schon, C.C. Prediction of Complex Traits: Robust Alternatives to Best Linear Unbiased Prediction. Front. Genet. 2018, 9, 195. [Google Scholar] [CrossRef] [PubMed]
- Vela-Avitua, S.; Meuwissen, T.H.E.; Luan, T.; Odegard, J. Accuracy of genomic selection for a sib-evaluated trait using identity-by-state and identity-by-descent relationships. Genet. Sel. Evol. 2015, 47, 9. [Google Scholar] [CrossRef] [PubMed]
- Scholtens, M.; Lopez-Villalobos, N.; Lehnert, K.; Snell, R.; Garrick, D.; Blair, H.T. Advantage of including Genomic Information to Predict Breeding Values for Lactation Yields of Milk, Fat, and Protein or Somatic Cell Score in a New Zealand Dairy Goat Herd. Animals 2021, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Velazco, J.G.; Malosetti, M.; Hunt, C.H.; Mace, E.S.; Jordan, D.R.; van Eeuwijk, F.A. Combining pedigree and genomic information to improve prediction quality: An example in sorghum. Theor. Appl. Genet. 2019, 132, 2055–2067. [Google Scholar] [CrossRef]
- Suontama, M.; Klapste, J.; Telfer, E.; Graham, N.; Stovold, T.; Low, C.; McKinley, R.; Dungey, H. Efficiency of genomic prediction across two Eucalyptus nitens seed orchards with different selection histories. Heredity 2019, 122, 370–379. [Google Scholar] [CrossRef]
- Kainer, D.; Stone, E.A.; Padovan, A.; Foley, W.J.; Kulheim, C. Accuracy of Genomic Prediction for Foliar Terpene Traits in Eucalyptus polybractea. G3-Genes Genomes Genet. 2018, 8, 2573–2583. [Google Scholar] [CrossRef]
- Seno, L.D.; Guidolin, D.G.F.; Aspilcueta-Borquis, R.R.; do Nascimento, G.B.; da Silva, T.B.R.; de Oliveira, H.N.; Munari, D.P. Genomic selection in dairy cattle simulated populations. J. Dairy Res. 2018, 85, 125–132. [Google Scholar] [CrossRef]
- Viana, J.M.S.; Garcia, A.A.F. Significance of linkage disequilibrium and epistasis on genetic variances in noninbred and inbred populations. BMC Genom. 2022, 23, 286. [Google Scholar] [CrossRef]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics, 4th ed.; Longman: London, UK, 1996. [Google Scholar]
- Viana, J.M.S.; Faria, V.R.; Fonseca e Silva, F.; Vilela de Resende, M.D. Combined selection of progeny in crop breeding using best linear unbiased prediction. Can. J. Plant Sci. 2012, 92, 553–562. [Google Scholar] [CrossRef][Green Version]
- Cockerham, C.C. Covariances of relatives from self-fertilization. Crop Sci. 1983, 23, 1177–1180. [Google Scholar] [CrossRef]
- Butler, D.G.; Cullis, B.R.; Gilmour, A.R.; Gogel, B.G.; Thompson, R. ASReml-R Reference Manual Version 4; VSN International Ltd.: Hemel Hempstead, UK, 2017. [Google Scholar]
- Patterson, H.D.; Thompson, R. Recovery of inter-block information when block sizes are unequal. Biometrika 1971, 58, 545–554. [Google Scholar] [CrossRef]
- Mrode, R.A. Linear Models for the Prediction of Animal Breeding Values, 2nd ed.; CABI Publishing: Wallingford, UK, 2005. [Google Scholar]
- Mehrban, H.; Naserkheil, M.; Lee, D.; Ibanez-Escriche, N. Multi-Trait Single-Step GBLUP Improves Accuracy of Genomic Prediction for Carcass Traits Using Yearling Weight and Ultrasound Traits in Hanwoo. Front. Genet. 2021, 12, 692356. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, J.; Li, Q.H.; Wang, Q.; Wen, J.; Zhao, G.P. Comparison of the Efficiency of BLUP and GBLUP in Genomic Prediction of Immune Traits in Chickens. Animals 2020, 10, 419. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, I.; Fernandez, E.N.; Blasco, A.; Ravagnolo, O.; Legarra, A. Effects of ignoring inbreeding in model-based accuracy for BLUP and SSGBLUP. J. Anim. Breed. Genet. 2020, 137, 356–364. [Google Scholar] [CrossRef]
- Dunne, F.L.; Kelleher, M.M.; Walsh, S.W.; Berry, D.P. Characterization of best linear unbiased estimates generated from national genetic evaluations of reproductive performance, survival, and milk yield in dairy cows. J. Dairy Sci. 2018, 101, 7625–7637. [Google Scholar] [CrossRef] [PubMed]
- Viana, J.M.S.; Pereira, H.D.; Mundim, G.B.; Piepho, H.P.; Silva, F.F.E. Efficiency of genomic prediction of non-assessed single crosses. Heredity 2018, 120, 283–295. [Google Scholar] [CrossRef]
- Viana, J.M.S.; Pereira, H.D.; Piepho, H.P.; Silva, F.F.E. Efficiency of Genomic Prediction of Nonassessed Testcrosses. Crop Sci. 2019, 59, 2020–2027. [Google Scholar] [CrossRef]
- Jibrila, I.; ten Napel, J.; Vandenplas, J.; Veerkamp, R.F.; Calus, M.P.L. Investigating the impact of preselection on subsequent single-step genomic BLUP evaluation of preselected animals. Genet. Sel. Evol. 2020, 52, 42. [Google Scholar] [CrossRef]
- El-Attrouny, M.M.; Manaa, E.A.; Ramadan, S.I. Genetic evaluation and selection correlated response of growth traits in Japanese quail. S. Afr. J. Anim. Sci. 2020, 50, 325–333. [Google Scholar] [CrossRef]
- D’Ambrosio, J.; Morvezen, R.; Brard-Fudulea, S.; Bestin, A.; Perez, A.A.; Guemen, D.; Poncet, C.; Haffray, P.; Dupont-Nivet, M.; Phocas, F. Genetic architecture and genomic selection of female reproduction traits in rainbow trout. BMC Genom. 2020, 21, 558. [Google Scholar] [CrossRef]
- Cobo, E.; Raoul, J.; Bodin, L. Genetic parameters of litter weight, an alternative criterion to prolificacy and pre-weaning weight for selection of French meat sheep. Livest. Sci. 2021, 250, 104596. [Google Scholar] [CrossRef]
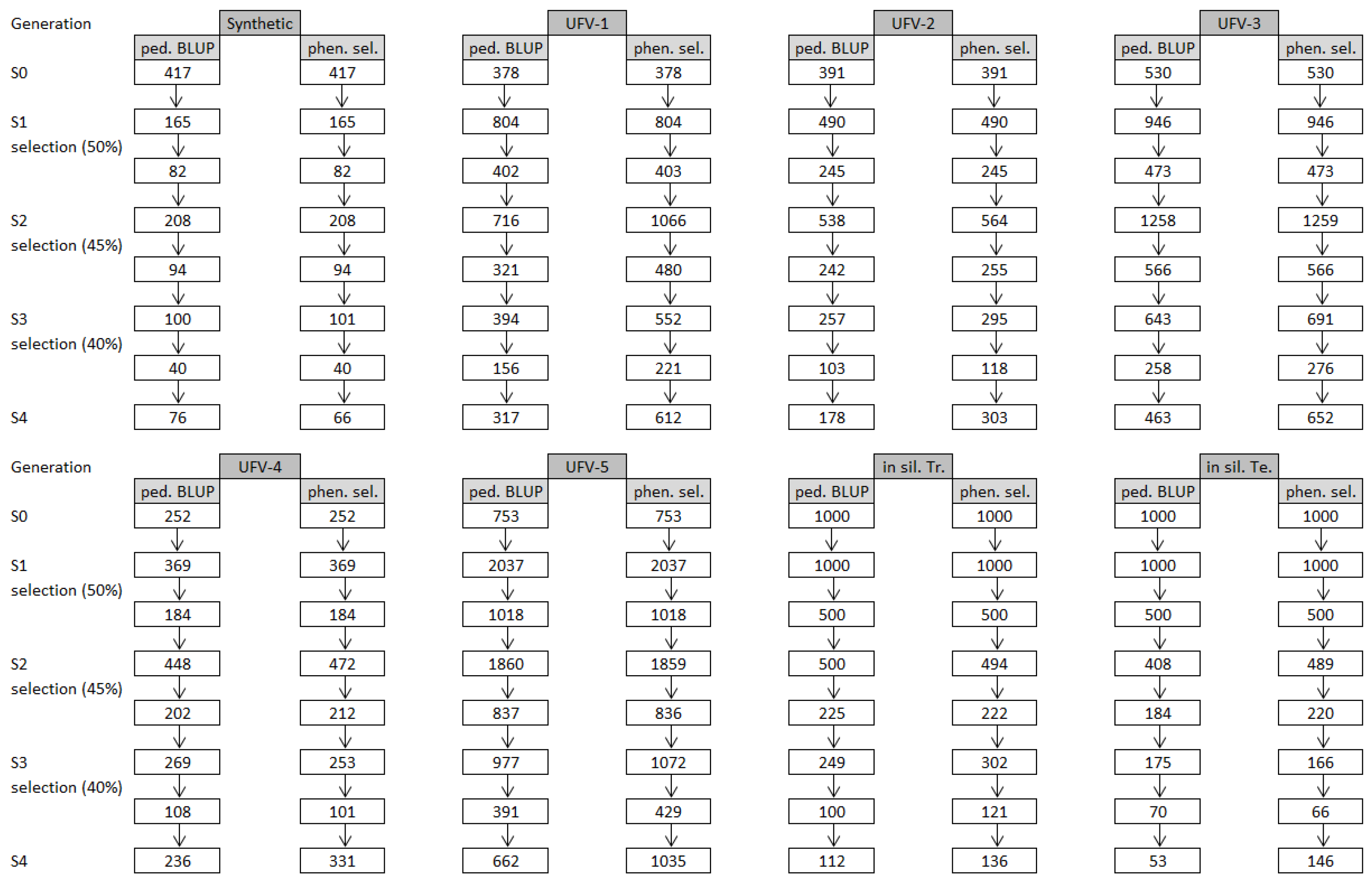
| Pop. | Gen. | Prog. | Size | EV | Grain Yield | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M/MF | M/MF | |||||||||||||||
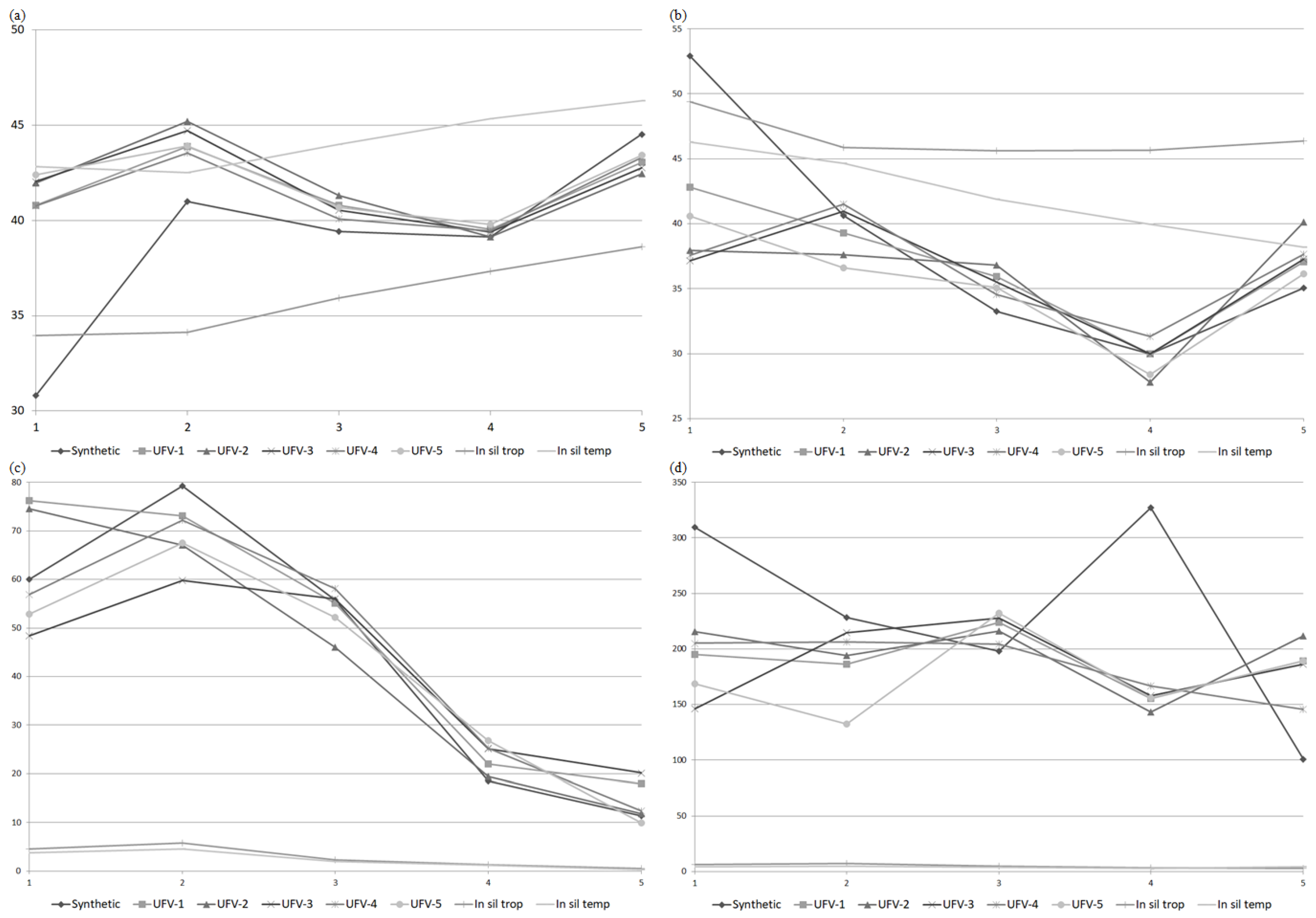
| Synthetic | S0 | - | 417 | 30.8 | 60.1 | 44.25 | - | - | 11.34 | 52.9 | 309.5 | 155.13 | - | - | 148.16 | −0.05 |
| S1 | 32 | 165 | 41.0 | 79.3 | 97.42 | - | - | 11.34 | 40.6 | 228.2 | 50.93 | - | - | 148.16 | −0.21 | |
| S2 | 59 | 290 | 39.3 | 51.0 | 43.91 | - | - | 11.34 | 32.3 | 209.8 | 36.10 | - | - | 148.16 | 0.57 | |
| S3 | 75 | 360 | 39.0 | 23.8 | 10.82 | - | - | 11.34 | 30.4 | 201.3 | 39.29 | - | - | 148.16 | −0.11 | |
| S4 | 76 | 367 | 43.3 | 11.8 | 0.50 | - | - | 11.34 | 36.9 | 168.2 | 11.86 | - | - | 148.16 | 0.11 | |
| UFV-1 | S0 | - | 378 | 40.8 | 76.2 | 32.11 | - | - | 17.90 | 42.8 | 195.2 | 52.92 | - | - | 141.26 | −0.25 |
| S1 | 215 | 804 | 43.9 | 73.0 | 69.59 | - | - | 17.90 | 39.3 | 186.0 | 30.61 | - | - | 141.26 | −0.40 | |
| S2 | 317 | 1130 | 40.7 | 54.0 | 37.82 | - | - | 17.90 | 35.9 | 225.7 | 46.58 | - | - | 141.26 | 0.02 | |
| S3 | 276 | 1134 | 39.6 | 25.4 | 3.85 | - | - | 17.90 | 29.8 | 166.6 | 15.68 | - | - | 141.26 | −0.58 | |
| S4 | 427 | 1981 | 43.1 | 17.3 | 0.26 | - | - | 17.90 | 36.2 | 177.0 | 17.62 | - | - | 141.26 | −1.00 | |
| UFV-2 | S0 | - | 391 | 42.0 | 74.5 | 34.96 | - | - | 20.21 | 37.9 | 215.8 | 44.68 | - | - | 142.84 | −0.17 |
| S1 | 168 | 490 | 45.2 | 67.0 | 47.00 | - | - | 20.21 | 37.6 | 194.0 | 29.96 | - | - | 142.84 | −0.13 | |
| S2 | 169 | 617 | 41.2 | 48.2 | 24.72 | - | - | 20.21 | 36.6 | 219.6 | 30.72 | - | - | 142.84 | −0.16 | |
| S3 | 183 | 706 | 39.2 | 24.9 | 2.30 | - | - | 20.21 | 29.5 | 177.1 | 17.08 | - | - | 142.84 | −0.20 | |
| S4 | 315 | 1468 | 42.7 | 20.3 | 0.70 | - | - | 20.21 | 36.7 | 173.8 | 17.23 | - | - | 142.84 | 0.03 | |
| UFV-3 | S0 | - | 530 | 42.0 | 48.4 | 26.06 | - | - | 16.82 | 37.1 | 145.9 | 0.00 | - | - | 146.75 | - |
| S1 | 310 | 946 | 44.7 | 59.8 | 46.83 | - | - | 16.82 | 41.0 | 214.8 | 42.22 | - | - | 146.75 | −0.23 | |
| S2 | 357 | 1330 | 40.4 | 55.9 | 36.47 | - | - | 16.82 | 35.4 | 225.7 | 28.09 | - | - | 146.75 | 0.13 | |
| S3 | 337 | 1421 | 39.3 | 27.7 | 7.18 | - | - | 16.82 | 29.9 | 167.1 | 14.45 | - | - | 146.75 | −0.44 | |
| S4 | 502 | 2433 | 42.9 | 17.1 | 0.80 | - | - | 16.82 | 36.4 | 183.5 | 19.15 | - | - | 146.75 | −0.02 | |
| UFV-4 | S0 | - | 252 | 40.7 | 56.8 | 24.13 | - | - | 20.84 | 37.5 | 205.1 | 71.60 | - | - | 133.33 | −0.10 |
| S1 | 101 | 369 | 43.5 | 72.2 | 53.12 | - | - | 20.84 | 41.5 | 206.3 | 47.47 | - | - | 133.33 | −0.03 | |
| S2 | 116 | 517 | 40.6 | 61.6 | 38.44 | - | - | 20.84 | 35.5 | 218.1 | 32.94 | - | - | 133.33 | 0.12 | |
| S3 | 144 | 603 | 39.0 | 30.3 | 5.32 | - | - | 20.84 | 30.6 | 170.0 | 17.74 | - | - | 133.33 | −0.29 | |
| S4 | 218 | 1107 | 42.6 | 20.2 | 0.50 | - | - | 20.84 | 36.2 | 154.5 | 12.18 | - | - | 133.33 | −0.41 | |
| UFV-5 | S0 | - | 753 | 42.4 | 52.8 | 35.73 | - | - | 12.11 | 40.6 | 168.4 | 39.58 | - | - | 126.92 | −0.53 |
| S1 | 546 | 2037 | 43.9 | 67.5 | 78.68 | - | - | 12.11 | 36.6 | 132.6 | 9.63 | - | - | 126.92 | −1.00 | |
| S2 | 600 | 1913 | 40.7 | 51.7 | 41.12 | - | - | 12.11 | 35.1 | 231.3 | 50.00 | - | - | 126.92 | −0.26 | |
| S3 | 533 | 2163 | 39.8 | 29.8 | 13.21 | - | - | 12.11 | 28.8 | 151.6 | 15.92 | - | - | 126.92 | −0.63 | |
| S4 | 840 | 3914 | 43.2 | 12.6 | 0.59 | - | - | 12.11 | 37.0 | 191.3 | 46.52 | - | - | 126.92 | −1.00 | |
| In silico | S0 | 1000 | 42.9 | 14.0 | 4.04 | 0.17 | 0.00 | 9.84 | 48.2 | 26.5 | 4.40 | 0.30 | 0.00 | 18.82 | −0.65 | |
| temperate | 42.8 | 13.4 | 1.11 | - | - | 12.32 | 48.4 | 23.2 | 0.09 | - | - | 23.16 | −1.00 | |||
| S1 | 247 | 1000 | 42.5 | 18.4 | 5.38 | 0.16 | −0.01 | 12.87 | 44.9 | 30.2 | 6.01 | 0.54 | −0.28 | 23.93 | −0.61 | |
| 42.5 | 16.3 | 3.13 | - | - | 12.32 | 44.5 | 16.74 | 3.44 | - | - | 23.16 | −0.74 | ||||
| S2 | 250 | 1000 | 42.3 | 20.0 | 5.91 | 0.12 | −0.02 | 13.96 | 43.2 | 31.8 | 6.69 | 0.44 | −0.43 | 25.09 | −0.65 | |
| 42.1 | 16.6 | 4.05 | - | - | 12.32 | 43.3 | 16.32 | 3.60 | - | - | 23.16 | −0.60 | ||||
| S3 | 220 | 1000 | 42.2 | 20.6 | 6.13 | 0.09 | −0.03 | 14.40 | 42.4 | 32.2 | 7.00 | 0.35 | −0.50 | 25.39 | −0.65 | |
| 42.0 | 16.5 | 3.36 | - | - | 12.32 | 42.1 | 17.37 | 5.16 | - | - | 23.16 | −0.59 | ||||
| S4 | 243 | 1000 | 42.1 | 20.9 | 6.23 | 0.08 | −0.03 | 14.58 | 42.0 | 32.4 | 7.14 | 0.30 | −0.53 | 25.49 | −0.52 | |
| 41.6 | 16.4 | 3.57 | - | - | 12.32 | 42.1 | 16.88 | 4.00 | - | - | 23.16 | −0.36 | ||||
| In silico | S0 | - | 1000 | 33.9 | 14.8 | 4.31 | 0.12 | 0.00 | 10.33 | 49.2 | 32.2 | 6.08 | 0.36 | 0.00 | 25.76 | 0.78 |
| tropical | 34.0 | 15.3 | 2.77 | - | - | 12.49 | 49.0 | 32.9 | 2.04 | - | - | 30.87 | 1.00 | |||
| S1 | 243 | 1000 | 33.8 | 19.3 | 5.71 | 0.11 | −0.01 | 13.51 | 45.5 | 41.6 | 8.19 | 0.64 | −0.28 | 33.06 | 0.75 | |
| 34.1 | 20.4 | 5.34 | - | - | 12.49 | 45.9 | 41.0 | 6.28 | - | - | 30.87 | 1.00 | ||||
| S2 | 227 | 1000 | 33.7 | 21.0 | 6.25 | 0.08 | −0.02 | 14.68 | 43.6 | 43.9 | 9.06 | 0.49 | −0.43 | 34.76 | 0.80 | |
| 33.9 | 21.0 | 4.87 | - | - | 12.49 | 43.9 | 44.9 | 8.29 | - | - | 30.87 | 0.75 | ||||
| S3 | 237 | 1000 | 33.9 | 21.7 | 6.48 | 0.06 | −0.02 | 15.15 | 42.7 | 44.5 | 9.43 | 0.37 | −0.50 | 35.20 | 0.81 | |
| 33.9 | 22.5 | 5.15 | - | - | 12.49 | 42.6 | 43.3 | 6.74 | - | - | 30.87 | 0.67 | ||||
| S4 | 217 | 1000 | 33.6 | 22.0 | 6.58 | 0.05 | −0.02 | 15.36 | 42.2 | 44.7 | 9.60 | 0.30 | −0.53 | 35.34 | 0.82 | |
| 34.0 | 23.7 | 5.45 | - | - | 12.49 | 42.9 | 42.4 | 6.11 | - | - | 30.87 | 0.62 | ||||
| Pop. | Gen. | Ac1 | Ac2 | Ac3 | Ac4 | Ac5 | %S | iDgd | iDgi |
|---|---|---|---|---|---|---|---|---|---|
| Synthetic | S0 | - | 0.89 | - | - | 0.88 | 0 | - | - |
| S1 | - | 0.95 | - | - | 0.92 | 50 | 0.00 | 0.00 | |
| S2 | - | 0.89 | - | - | 0.85 | 45 | −0.58 | 0.16 | |
| S3 | - | 0.70 | - | - | 0.55 | 40 | 0.45 | −0.17 | |
| S4 | - | 0.20 | - | - | - 2 | - | - | - | |
| UFV-1 | S0 | - | 0.80 | - | - | - 2 | 0 | - | - |
| S1 | - | 0.89 | - | - | 0.62 | 50 | 0.13 | −0.08 | |
| S2 | - | 0.82 | - | - | - 2 | 45 | −0.20 | −0.38 | |
| S3 | - | 0.42 | - | - | - 2 | 40 | 0.06 | 0.10 | |
| S4 | - | 0.12 | - | - | - 2 | - | - | - | |
| UFV-2 | S0 | - | 0.80 | - | - | 0.75 | 0 | - | - |
| S1 | - | 0.84 | - | - | 0.85 | 50 | −0.01 | 0.03 | |
| S2 | - | 0.74 | - | - | 0.69 | 45 | −0.04 | −0.05 | |
| S3 | - | 0.32 | - | - | - 2 | 40 | 0.02 | 0.46 | |
| S4 | - | 0.18 | - | - | - 2 | - | - | - | |
| UFV-3 | S0 | - | 0.78 | - | - | 0.76 | 0 | - | - |
| S1 | - | 0.86 | - | - | 0.86 | 50 | 0.00 | −0.01 | |
| S2 | - | 0.83 | - | - | 0.83 | 45 | 0.05 | −0.01 | |
| S3 | - | 0.55 | - | - | - 2 | 40 | 0.00 | 0.18 | |
| S4 | - | 0.21 | - | - | - 2 | - | - | - | |
| UFV-4 | S0 | - | 0.73 | - | - | 0.68 | 0 | - | - |
| S1 | - | 0.85 | - | - | 0.86 | 50 | −0.25 | −0.10 | |
| S2 | - | 0.80 | - | - | 0.80 | 45 | 0.06 | −0.07 | |
| S3 | - | 0.45 | - | - | - 2 | 40 | 0.05 | −0.04 | |
| S4 | - | 0.15 | - | - | - 2 | - | - | - | |
| UFV-5 | S0 | - | 0.86 | - | - | 0.84 | 0 | - | - |
| S1 | - | 0.93 | - | - | 0.94 | 50 | −0.01 | 0.00 | |
| S2 | - | 0.88 | - | - | 0.87 | 45 | −0.04 | 0.00 | |
| S3 | - | 0.72 | - | - | 0.49 | 40 | 0.15 | −0.01 | |
| S4 | - | 0.21 | - | - | - 2 | - | - | - | |
| In silico | S0 | 0.54 | 0.29 | 0.52 | 0.52 | 0.28 | 0 | - | - |
| temperate | S1 | 0.54 | 0.45 | 0.52 | 0.67 | 0.42 | 50 | 0.07 | −0.04 |
| 0.46 3 | −0.31 3 | ||||||||
| 1.16 4 | −0.68 4 | ||||||||
| S2 | 0.54 | 0.50 | 0.49 | 0.72 | 0.45 | 45 | 0.14 | −0.04 | |
| 0.41 3 | −0.16 3 | ||||||||
| 0.32 4 | −0.27 4 | ||||||||
| S3 | 0.54 | 0.46 | 0.51 | 0.78 | 0.34 | 40 | 0.29 | −0.20 | |
| 0.83 3 | −0.96 3 | ||||||||
| 2.05 4 | −1.74 4 | ||||||||
| S4 | 0.55 | 0.47 | 0.48 | 0.78 | 0.34 | - | - | - | |
| In silico | S0 | 0.54 | 0.43 | 0.56 | 0.56 | 0.42 | 0 | - | - |
| tropical | S1 | 0.54 | 0.55 | 0.53 | 0.71 | 0.58 | 50 | 0.02 | 0.02 |
| 0.15 3 | 0.23 3 | ||||||||
| 0.75 4 | 0.48 4 | ||||||||
| S2 | 0.55 | 0.53 | 0.54 | 0.79 | 0.49 | 45 | 0.03 | −0.01 | |
| 0.15 3 | −0.06 3 | ||||||||
| 0.54 4 | 0.30 4 | ||||||||
| S3 | 0.55 | 0.54 | 0.56 | 0.82 | 0.48 | 40 | 0.26 | 0.31 | |
| 0.96 3 | 0.86 3 | ||||||||
| 1.25 4 | 0.81 4 | ||||||||
| S4 | 0.55 | 0.55 | 0.57 | 0.83 | 0.48 | - | - | - |
| Gen. | Ac | %S | iDgd | iDgi | ||
|---|---|---|---|---|---|---|
| S0 | 1.99 [0.83, 3.42] | 11.80 [10.71, 12.94] | 0.54 [0.52, 0.58] | - | - | - |
| S1 | 4.29 [3.06, 5.37] | 11.80 [10.71, 12.94] | 0.71 [0.63, 0.75] | 50 | −0.48 [−0.93, 0.59] 2 | 0.25 [−0.37, 0.74] |
| 0.08 [−0.28, 1.16] 3 | −0.03 [−0.68, 0.35] | |||||
| S2 | 4.36 [3.16, 5.35] | 11.80 [10.71, 12.94] | 0.75 [0.71, 0.81] | 45 | 0.32 [−0.04, 1.03] 2 | −0.23 [−0.77, 0.11] |
| 0.33 [0.06, 0.66] 3 | −0.28 [−0.71, 0.04] | |||||
| S3 | 4.57 [3.15, 6.76] | 11.80 [10.71, 12.94] | 0.78 [0.72, 0.82] | 40 | 0.50 [−0.40, 0.93] 2 | −0.33 [−1.12, 0.59] |
| 1.33 [0.80, 2.21] 3 | −0.76 [−1.74, 0.18] | |||||
| S4 | 4.32 [3.03, 5.87] | 11.80 [10.71, 12.94] | 0.80 [0.73, 0.83] | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Viana, J.M.S.; Dias, K.O.d.G.; Silva, J.P.A.d. Comparative Analysis of Pedigree-Based BLUP and Phenotypic Mass Selection for Developing Elite Inbred Lines, Based on Field and Simulated Data. Agronomy 2022, 12, 2560. https://doi.org/10.3390/agronomy12102560
Viana JMS, Dias KOdG, Silva JPAd. Comparative Analysis of Pedigree-Based BLUP and Phenotypic Mass Selection for Developing Elite Inbred Lines, Based on Field and Simulated Data. Agronomy. 2022; 12(10):2560. https://doi.org/10.3390/agronomy12102560
Chicago/Turabian StyleViana, José Marcelo Soriano, Kaio Olimpio das Graças Dias, and Jean Paulo Aparecido da Silva. 2022. "Comparative Analysis of Pedigree-Based BLUP and Phenotypic Mass Selection for Developing Elite Inbred Lines, Based on Field and Simulated Data" Agronomy 12, no. 10: 2560. https://doi.org/10.3390/agronomy12102560
APA StyleViana, J. M. S., Dias, K. O. d. G., & Silva, J. P. A. d. (2022). Comparative Analysis of Pedigree-Based BLUP and Phenotypic Mass Selection for Developing Elite Inbred Lines, Based on Field and Simulated Data. Agronomy, 12(10), 2560. https://doi.org/10.3390/agronomy12102560







