De Novo Transcriptome Assembly and SNP Discovery for the Development of dCAPS Markers in Oat
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Total RNA Extraction and Transcriptome Sequencing
2.3. De Novo Assembly and Generation of the Unigene Set
2.4. Gene Functional Annotations
2.5. SNP Identification
2.6. SNP Primer Design and SNP Validation
2.7. Analysis of Genetic Diversity
3. Results
3.1. Sequencing and Assembly of the Transcriptome
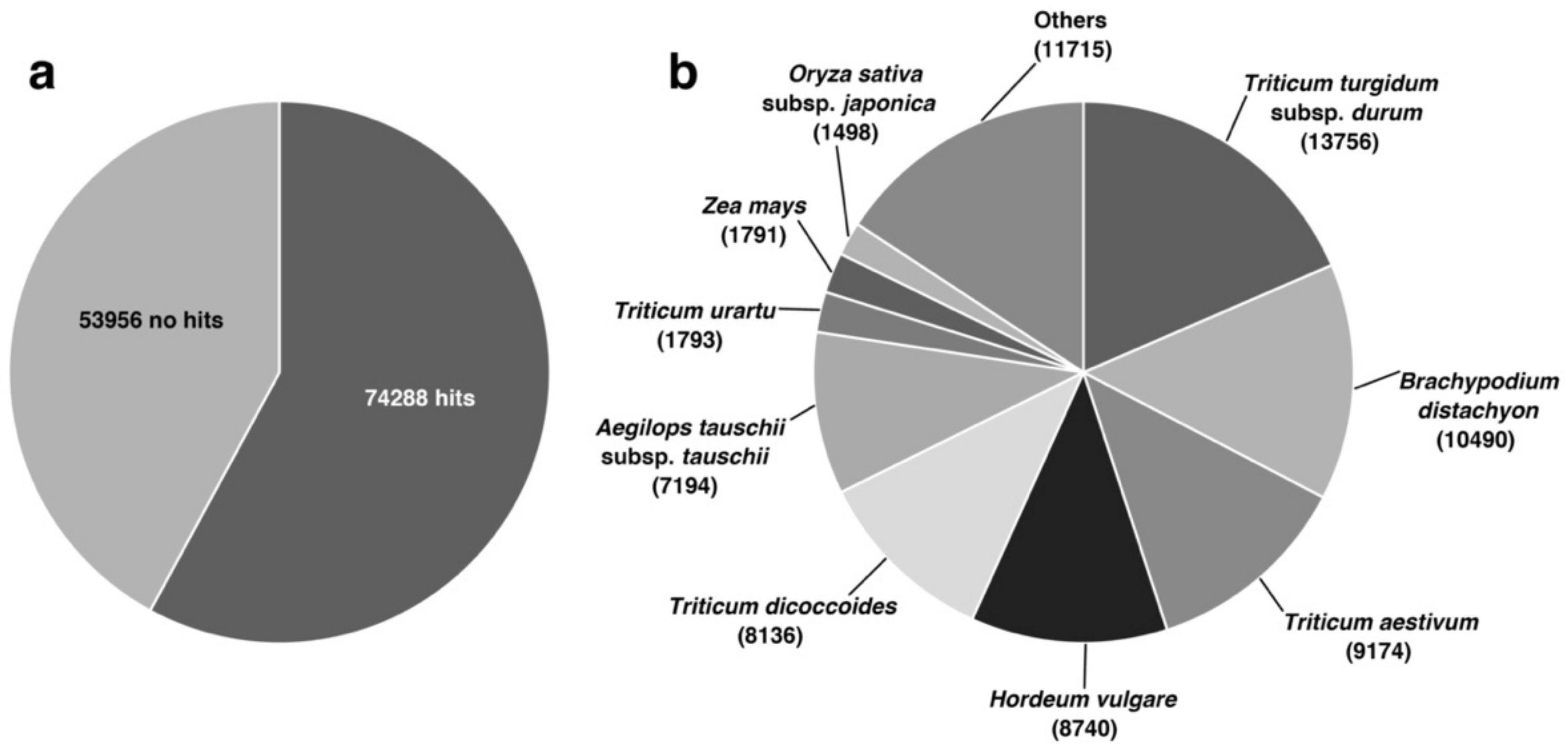
3.2. Functional Annotation
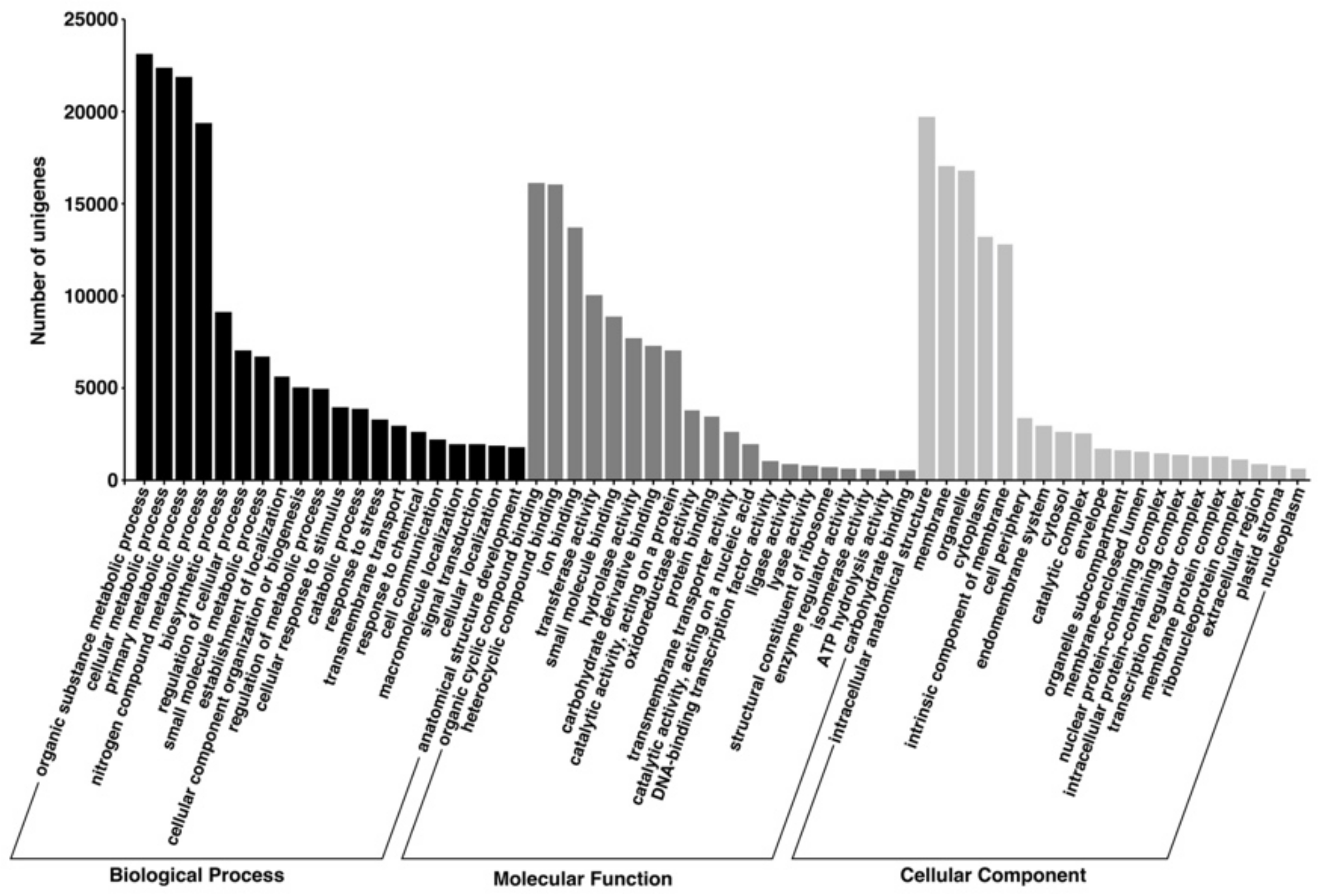
3.3. Gene Ontology (GO)
3.4. Cluster of Orthologous Groups (COG)
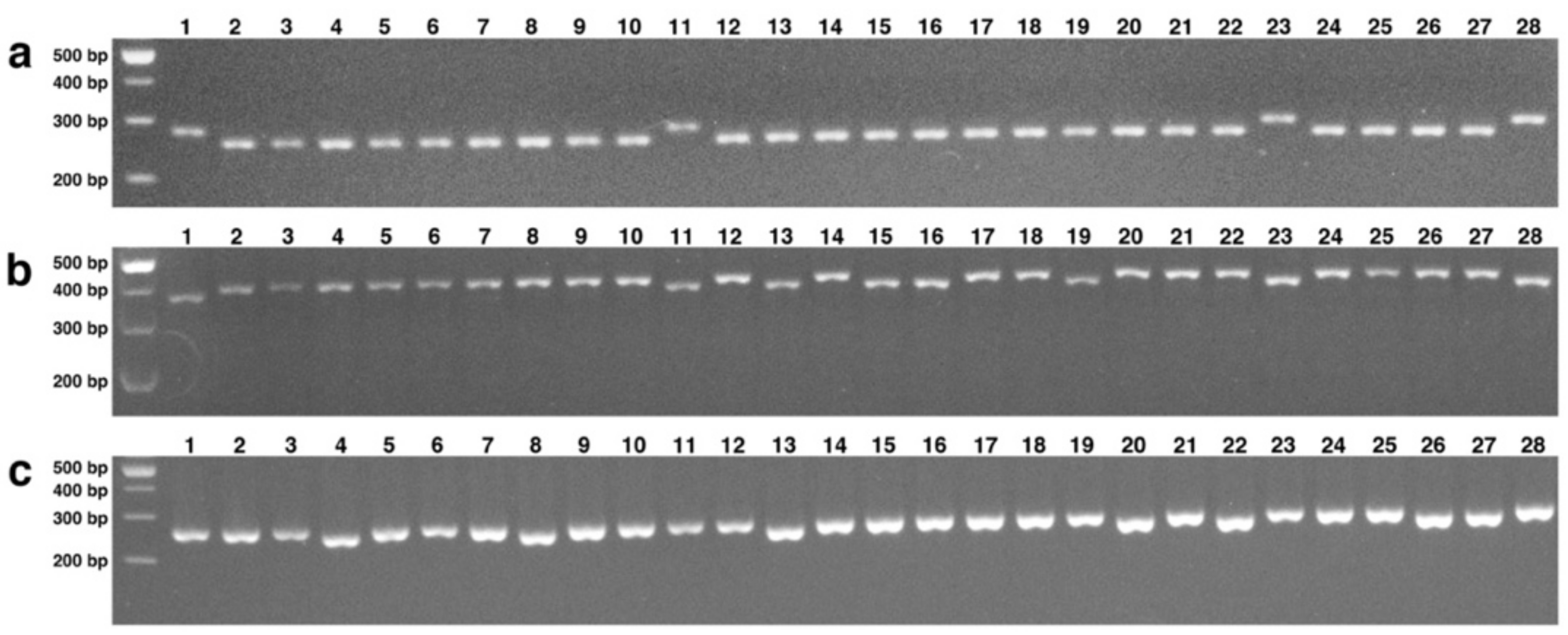
3.5. SNP Discovery and Validation
3.6. Analysis of Genetic Diversity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Strychar, R.; Webster, F.; Wood, P. World oat production, trade, and usage. In Oats: Chemistry and Technology; AACC International, Inc.: St. Paul, MN, USA, 2011; pp. 1–10. [Google Scholar]
- Hoffman, L.A. World production and use of oats. In The Oat Crop; Springer: Berlin/Heidelberg, Germany, 1995; pp. 34–61. [Google Scholar]
- Bekele, W.A.; Wight, C.P.; Chao, S.; Howarth, C.J.; Tinker, N.A. Haplotype-based genotyping-by-sequencing in oat genome research. Plant Biotechnol. J. 2018, 16, 1452–1463. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Hu, Y.; Huo, P.; Zhang, Q.; Chen, X.; Zhang, Z. Transcriptome analysis of hexaploid hulless oat in response to salinity stress. PLoS ONE 2017, 12, e0171451. [Google Scholar] [CrossRef]
- Ames, N.; Rhymer, C.; Storsley, J. Food oat quality throughout the value chain. In Oats Nutrition and Technology; Wiley: Hoboken, NJ, USA, 2013; pp. 33–70. [Google Scholar]
- Maki, K.; Galant, R.; Samuel, P.; Tesser, J.; Witchger, M.; Ribaya-Mercado, J.; Blumberg, J.; Geohas, J. Effects of consuming foods containing oat β-glucan on blood pressure, carbohydrate metabolism and biomarkers of oxidative stress in men and women with elevated blood pressure. Eur. J. Clin. Nutr. 2007, 61, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Pomeroy, S.; Tupper, R.; Cehun-Aders, M.; Nestel, P. Oat b-glucan lowers total and LDL-cholesterol. Aust. J. Nutr. Diet. 2001, 58, 51–55. [Google Scholar]
- Ripsin, C.M.; Keenan, J.M.; Jacobs, D.R.; Elmer, P.J.; Welch, R.R.; Van Horn, L.; Liu, K.; Turnbull, W.H.; Thye, F.W.; Kestin, M. Oat products and lipid lowering: A meta-analysis. JAMA 1992, 267, 3317–3325. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, V.S.; Samidurai, M.; Park, H.J.; Wang, M.; Park, R.Y.; Yu, S.Y.; Kang, H.K.; Hong, S.; Choi, W.-S.; Lee, Y.Y. Avenanthramide-C restores impaired plasticity and cognition in Alzheimer’s disease model mice. Mol. Neurobiol. 2020, 57, 315–330. [Google Scholar] [CrossRef] [PubMed]
- US. Food and Drug Administration. Food labeling: Health claims; oats and coronary heart disease: Final rule. Fed. Regist. 1997, 62, 3584–3601. [Google Scholar]
- Nazco, R.; Villegas, D.; Ammar, K.; Pena, R.J.; Moragues, M.; Royo, C. Can Mediterranean durum wheat landraces contribute to improved grain quality attributes in modern cultivars? Euphytica 2012, 185, 1–17. [Google Scholar] [CrossRef]
- Fu, Y.B.; Peterson, G.W.; Scoles, G.; Rossnagel, B.; Schoen, D.J.; Richards, K.W. Allelic diversity changes in 96 Canadian oat cultivars released from 1886 to 2001. Crop Sci. 2003, 43, 1989–1995. [Google Scholar] [CrossRef]
- International Wheat Genome Sequencing Consortium. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Khurana, P.; Gaikwad, K. The map-based sequence of the rice genome. Nature 2005, 436, 793–800. [Google Scholar]
- The International Barley Genome Sequencing Consortium. A physical, genetic and functional sequence assembly of the barley genome. Nature 2012, 491, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef]
- Bentley, D.R. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006, 16, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Gonzalez, J.J.; Tu, Z.J.; Garvin, D.F. Analysis and annotation of the hexaploid oat seed transcriptome. BMC Genom. 2013, 14, 471. [Google Scholar] [CrossRef]
- O’Donoughue, L.; Souza, E.; Tanksley, S.; Sorrells, M. Relationships among North American oat cultivars based on restriction fragment length polymorphisms. Crop Sci. 1994, 34, 1251–1258. [Google Scholar] [CrossRef]
- Baohong, G.; Zhou, X.; Murphy, J. Genetic variation within Chinese and Western cultivated oat accessions. Cereal Res. Commun. 2003, 31, 339–346. [Google Scholar] [CrossRef]
- Achleitner, A.; Tinker, N.A.; Zechner, E.; Buerstmayr, H. Genetic diversity among oat varieties of worldwide origin and associations of AFLP markers with quantitative traits. Theor. Appl. Genet. 2008, 117, 1041–1053. [Google Scholar] [CrossRef]
- Subudhi, P.K.; Parami, N.P.; Harrison, S.A.; Materne, M.D.; Murphy, J.P.; Nash, D. An AFLP-based survey of genetic diversity among accessions of sea oats (Uniola paniculata, Poaceae) from the southeastern Atlantic and Gulf coast states of the United States. Theor. Appl. Genet. 2005, 111, 1632–1641. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Rossnagel, B.; Scoles, G. The development of oat microsatellite markers and their use in identifying relationships among Avena species and oat cultivars. Theor. Appl. Genet. 2000, 101, 1259–1268. [Google Scholar] [CrossRef]
- Boczkowska, M.; Nowosielski, J.; Nowosielska, D.; Podyma, W. Assessing genetic diversity in 23 early Polish oat cultivars based on molecular and morphological studies. Genet. Resour. Crop. Evol. 2014, 61, 927–941. [Google Scholar] [CrossRef][Green Version]
- Nersting, L.G.; Andersen, S.B.; von Bothmer, R.; Gullord, M.; Jørgensen, R.B. Morphological and molecular diversity of Nordic oat through one hundred years of breeding. Euphytica 2006, 150, 327–337. [Google Scholar] [CrossRef]
- Tinker, N.A.; Kilian, A.; Wight, C.P.; Heller-Uszynska, K.; Wenzl, P.; Rines, H.W.; Bjørnstad, Å.; Howarth, C.J.; Jannink, J.-L.; Anderson, J.M. New DArT markers for oat provide enhanced map coverage and global germplasm characterization. BMC Genom. 2009, 10, 39. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhou, P.; Peng, Y.; Bekele, W.A.; Ren, C.; Tinker, N.A.; Peng, Y. Genetic diversity and genome-wide association analysis in Chinese hulless oat germplasm. Theor. Appl. Genet. 2020, 133, 3365–3380. [Google Scholar] [CrossRef]
- Huang, Y.-F.; Poland, J.A.; Wight, C.P.; Jackson, E.W.; Tinker, N.A. Using genotyping-by-sequencing (GBS) for genomic discovery in cultivated oat. PLoS ONE 2014, 9, e102448. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Tian, L.; Bu, F.; Sun, Q.; Zhao, X.; Han, Y. RNA-seq-based identification of potential resistance genes against the soybean cyst nematode (Heterodera glycines) HG Type 1.2.3.5.7 in “Dongnong L-10”. Physiol. Mol. Plant Pathol. 2021, 114, 101627. [Google Scholar] [CrossRef]
- Arora, K.; Panda, K.K.; Mittal, S.; Mallikarjuna, M.G.; Rao, A.R.; Dash, P.K.; Thirunavukkarasu, N. RNAseq revealed the important gene pathways controlling adaptive mechanisms under waterlogged stress in maize. Sci. Rep. 2017, 7, 10950. [Google Scholar] [CrossRef]
- Yousefirad, S.; Soltanloo, H.; Ramezanpour, S.S.; Zaynali Nezhad, K.; Shariati, V. The RNA-seq transcriptomic analysis reveals genes mediating salt tolerance through rapid triggering of ion transporters in a mutant barley. PLoS ONE 2020, 15, e0229513. [Google Scholar] [CrossRef]
- Iquebal, M.A.; Sharma, P.; Jasrotia, R.S.; Jaiswal, S.; Kaur, A.; Saroha, M.; Angadi, U.; Sheoran, S.; Singh, R.; Singh, G. RNAseq analysis reveals drought-responsive molecular pathways with candidate genes and putative molecular markers in root tissue of wheat. Sci. Rep. 2019, 9, 13917. [Google Scholar] [CrossRef]
- Hsu, S.-K.; Tung, C.-W. RNA-Seq analysis of diverse rice genotypes to identify the genes controlling coleoptile growth during submerged germination. Front. Plant Sci. 2017, 8, 762. [Google Scholar] [CrossRef]
- Zhang, J.; Liang, S.; Duan, J.; Wang, J.; Chen, S.; Cheng, Z.; Zhang, Q.; Liang, X.; Li, Y. De novo assembly and characterisation of the transcriptome during seed development, and generation of genic-SSR markers in peanut (Arachis hypogaea L.). BMC Genom. 2012, 13, 90. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Wu, M.; Pei, W.; Wang, X.; Zhai, H.; Wang, W.; Li, X.; Zhang, J.; Yu, J.; Yu, S. RNA-seq-mediated transcriptome analysis of a fiberless mutant cotton and its possible origin based on SNP markers. PLoS ONE 2016, 11, e0151994. [Google Scholar] [CrossRef]
- Xu, S.; Wang, J.; Shang, H.; Huang, Y.; Yao, W.; Chen, B.; Zhang, M. Transcriptomic characterization and potential marker development of contrasting sugarcane cultivars. Sci. Rep. 2018, 8, 1683. [Google Scholar] [CrossRef]
- Salgado, L.R.; Koop, D.M.; Pinheiro, D.G.; Rivallan, R.; Le Guen, V.; Nicolás, M.F.; De Almeida, L.G.P.; Rocha, V.R.; Magalhães, M.; Gerber, A.L. De novo transcriptome analysis of Hevea brasiliensis tissues by RNA-seq and screening for molecular markers. BMC Genom. 2014, 15, 236. [Google Scholar] [CrossRef] [PubMed]
- Oliver, R.E.; Lazo, G.R.; Lutz, J.D.; Rubenfield, M.J.; Tinker, N.A.; Anderson, J.M.; Morehead, N.H.W.; Adhikary, D.; Jellen, E.N.; Maughan, P.J. Model SNP development for complex genomes based on hexaploid oat using high-throughput 454 sequencing technology. BMC Genom. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Zhang, X.; Zhang, C. mRIN for direct assessment of genome-wide and gene-specific mRNA integrity from large-scale RNA-sequencing data. Nat. Commun. 2015, 6, 7816. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and samtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Neff, M.M.; Turk, E.; Kalishman, M. Web-based primer design for single nucleotide polymorphism analysis. TRENDS Genet. 2002, 18, 613–615. [Google Scholar] [CrossRef]
- Korbie, D.J.; Mattick, J.S. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat. Protoc. 2008, 3, 1452–1456. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Sneath, P.H.A.; Sokal, R.R. Unweighted pair group method with arithmetic mean. In Numerical Taxonomy; Freeman: San Francisco, CA, USA, 1973; pp. 230–234. [Google Scholar]
- Mora-Ortiz, M.; Swain, M.T.; Vickers, M.J.; Hegarty, M.J.; Kelly, R.; Smith, L.M.; Skøt, L. De-novo transcriptome assembly for gene identification, analysis, annotation, and molecular marker discovery in Onobrychis viciifolia. BMC Genom. 2016, 17, 756. [Google Scholar] [CrossRef]
- Li, X.; Acharya, A.; Farmer, A.D.; Crow, J.A.; Bharti, A.K.; Kramer, R.S.; Wei, Y.; Han, Y.; Gou, J.; May, G.D. Prevalence of single nucleotide polymorphism among 27 diverse alfalfa genotypes as assessed by transcriptome sequencing. BMC Genom. 2012, 13, 568. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Senalik, D.A.; Grzebelus, D.; Bowman, M.; Cavagnaro, P.F.; Matvienko, M.; Ashrafi, H.; Van Deynze, A.; Simon, P.W. De novo assembly and characterization of the carrot transcriptome reveals novel genes, new markers, and genetic diversity. BMC Genom. 2011, 12, 389. [Google Scholar] [CrossRef]
- Wang, S.; Wang, X.; He, Q.; Liu, X.; Xu, W.; Li, L.; Gao, J.; Wang, F. Transcriptome analysis of the roots at early and late seedling stages using Illumina paired-end sequencing and development of EST-SSR markers in radish. Plant Cell Rep. 2012, 31, 1437–1447. [Google Scholar] [CrossRef]
- Wang, Z.; Fang, B.; Chen, J.; Zhang, X.; Luo, Z.; Huang, L.; Chen, X.; Li, Y. De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of cSSR markers in sweetpotato (Ipomoea batatas). BMC Genom. 2010, 11, 726. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Liu, Y.; Xie, J.; Bie, Z. Development of simple sequence repeat markers from de novo assembled transcriptomes of pumpkins. Plant Mol. Biol. Rep. 2020, 38, 130–136. [Google Scholar] [CrossRef]
- Liu, T.; Liu, X.; Zhou, R.; Chen, H.; Zhang, H.; Zhang, B. De novo Transcriptome Assembly and Comparative Analysis Highlight the Primary Mechanism Regulating the Response to Selenium Stimuli in Oats (Avena sativa L.). Front. Plant Sci. 2021, 12, 1220. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Cheng, Y.; Lu, Z.; Wang, J.; Ye, X.; Zhang, X.; Luo, X.; Wang, H.; Zhang, B. Global insights to drought stress perturbed genes in oat (Avena sativa L.) seedlings using RNA sequencing. Plant Signal. Behav. 2021, 16, 1845934. [Google Scholar] [CrossRef]
- Kingan, S.B.; Heaton, H.; Cudini, J.; Lambert, C.C.; Baybayan, P.; Galvin, B.D.; Durbin, R.; Korlach, J.; Lawniczak, M.K. A high-quality de novo genome assembly from a single mosquito using PacBio sequencing. Genes 2019, 10, 62. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, X.; Lu, X.; Zhao, B.; Yang, Y.; Liu, J. Integrative analysis of transcriptome and metabolome reveal mechanism of tolerance to salt stress in oat (Avena sativa L.). Plant Physiol. Biochem. 2021, 160, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Hou, R.; Bao, Z.; Wang, S.; Su, H.; Li, Y.; Du, H.; Hu, J.; Wang, S.; Hu, X. Transcriptome sequencing and de novo analysis for Yesso scallop (Patinopecten yessoensis) using 454 GS FLX. PLoS ONE 2011, 6, e21560. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, J.; Xiao, S.; Liu, X. De novo assembly and characterization of foot transcriptome and microsatellite marker development for Paphia textile. Gene 2016, 576, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.H.; Wei, D.D.; Shen, G.M.; Yuan, G.R.; Bai, P.P.; Wang, J.J. De novo characterization of the Dialeurodes citri transcriptome: Mining genes involved in stress resistance and simple sequence repeats (SSRs) discovery. Insect Mol. Biol. 2014, 23, 52–66. [Google Scholar] [CrossRef] [PubMed]
- Mardi, M.; Karimi Farsad, L.; Gharechahi, J.; Salekdeh, G.H. In-depth transcriptome sequencing of Mexican lime trees infected with Candidatus Phytoplasma aurantifolia. PLoS ONE 2015, 10, e0130425. [Google Scholar] [CrossRef] [PubMed]
- Oliver, R.E.; Tinker, N.A.; Lazo, G.R.; Chao, S.; Jellen, E.N.; Carson, M.L.; Rines, H.W.; Obert, D.E.; Lutz, J.D.; Shackelford, I. SNP discovery and chromosome anchoring provide the first physically-anchored hexaploid oat map and reveal synteny with model species. PLoS ONE 2013, 8, e58068. [Google Scholar] [CrossRef]
- Gutierrez-Gonzalez, J.J.; Garvin, D.F. Reference genome-directed resolution of homologous and homeologous relationships within and between different oat linkage maps. Plant Genome 2011, 4, 178–190. [Google Scholar] [CrossRef]
- Brookes, A.J. The essence of SNPs. Gene 1999, 234, 177–186. [Google Scholar] [CrossRef]
- Morin, P.A.; Luikart, G.; Wayne, R.K. SNPs in ecology, evolution and conservation. Trends Ecol. Evol. 2004, 19, 208–216. [Google Scholar] [CrossRef]
- Wang, K.; Wang, D.; Zheng, X.; Qin, A.; Zhou, J.; Guo, B.; Chen, Y.; Wen, X.; Ye, W.; Zhou, Y. Multi-strategic RNA-seq analysis reveals a high-resolution transcriptional landscape in cotton. Nat. Commun. 2019, 10, 4714. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Miao, H.; Li, C.; Duan, Y.; Niu, J.; Zhang, T.; Zhao, Q.; Zhang, H. Development of SNP and InDel markers via de novo transcriptome assembly in Sesamum indicum L. Mol. Breed. 2014, 34, 2205–2217. [Google Scholar] [CrossRef]
- Yates, S.A.; Swain, M.T.; Hegarty, M.J.; Chernukin, I.; Lowe, M.; Allison, G.G.; Ruttink, T.; Abberton, M.T.; Jenkins, G.; Skøt, L. De novo assembly of red clover transcriptome based on RNA-Seq data provides insight into drought response, gene discovery and marker identification. BMC Genom. 2014, 15, 453. [Google Scholar] [CrossRef]
- Leonforte, A.; Sudheesh, S.; Cogan, N.O.; Salisbury, P.A.; Nicolas, M.E.; Materne, M.; Forster, J.W.; Kaur, S. SNP marker discovery, linkage map construction and identification of QTLs for enhanced salinity tolerance in field pea (Pisum sativum L.). BMC Plant Biol. 2013, 13, 161. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, G.; Jhanwar, S.; Priya, P.; Singh, V.K.; Saxena, M.S.; Parida, S.K.; Garg, R.; Tyagi, A.K.; Jain, M. Comparative analysis of kabuli chickpea transcriptome with desi and wild chickpea provides a rich resource for development of functional markers. PLoS ONE 2012, 7, e52443. [Google Scholar] [CrossRef] [PubMed]
- Izzah, N.K.; Lee, J.; Jayakodi, M.; Perumal, S.; Jin, M.; Park, B.-S.; Ahn, K.; Yang, T.-J. Transcriptome sequencing of two parental lines of cabbage (Brassica oleracea L. var. capitata L.) and construction of an EST-based genetic map. BMC Genom. 2014, 15, 149. [Google Scholar] [CrossRef]
- Allegre, M.; Argout, X.; Boccara, M.; Fouet, O.; Roguet, Y.; Bérard, A.; Thévenin, J.M.; Chauveau, A.; Rivallan, R.; Clement, D. Discovery and mapping of a new expressed sequence tag-single nucleotide polymorphism and simple sequence repeat panel for large-scale genetic studies and breeding of Theobroma cacao L. DNA Res. 2012, 19, 23–35. [Google Scholar] [CrossRef] [PubMed]



| Variety | Raw Data | Trimmed Data | Mapped Reads | Unique Hit PE Reads | Unmapped Reads | Read Mapping Rate (%) | Average Depth (x) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total Reads | Total Nucleotide Length (bp) | GC (%) | Q20 (%) | Average Length (bp) | Total Reads | Total Nucleotide Length (bp) | Average Length (bp) | ||||||
| Choyang | 34,091,906 | 5,147,877,806 | 54.82 | 98.27 | 151 | 31,937,598 | 4,679,320,810 | 146.5 | 27,965,238 | 10,594,206 | 1,681,828 | 87.6 | 15.11 |
| Daeyang | 35,130,420 | 5,304,693,420 | 53.79 | 98.26 | 151 | 32,872,618 | 4,802,284,508 | 146.1 | 28,806,502 | 11,247,262 | 1,873,278 | 87.6 | 15.07 |
| Darkhorse | 36,272,032 | 5,477,076,832 | 54.24 | 98.31 | 151 | 34,014,924 | 4,966,621,185 | 146.0 | 29,804,230 | 9,263,540 | 1,838,306 | 87.6 | 15.40 |
| Gehl | 43,776,694 | 6,610,280,794 | 54.40 | 98.46 | 151 | 41,304,176 | 5,998,734,784 | 145.2 | 36,786,406 | 10,428,980 | 1,723,984 | 89.1 | 18.81 |
| Gwanghan | 37,093,196 | 5,601,072,596 | 54.85 | 98.33 | 151 | 34,804,440 | 5,077,842,605 | 145.9 | 30,422,824 | 11,468,622 | 1,710,074 | 87.4 | 15.89 |
| Hispeed | 33,197,992 | 5,012,896,792 | 54.00 | 98.34 | 151 | 31,187,392 | 4,554,914,836 | 146.0 | 27,403,746 | 10,556,148 | 1,665,156 | 87.9 | 14.79 |
| Ilhan | 38,554,606 | 5,821,745,506 | 54.48 | 98.26 | 151 | 36,046,378 | 5,270,758,292 | 146.2 | 31,349,728 | 12,241,220 | 2,073,428 | 87.0 | 16.32 |
| Okhan | 33,311,918 | 5,030,099,618 | 55.15 | 98.28 | 151 | 31,244,814 | 4,557,668,933 | 145.9 | 27,511,954 | 10,208,568 | 1,410,194 | 88.1 | 14.51 |
| Samhan | 35,883,886 | 5,418,466,786 | 55.20 | 98.43 | 151 | 33,811,178 | 4,936,778,375 | 146.0 | 29,786,056 | 11,267,756 | 1,727,036 | 88.1 | 15.55 |
| Swan | 38,373,044 | 5,794,329,644 | 54.15 | 98.31 | 151 | 36,005,730 | 5,268,310,072 | 146.3 | 31,295,712 | 10,038,762 | 2,200,208 | 86.9 | 16.31 |
| Mean | 36,568,569 | 5,521,853,979 | 55.00 | 98.00 | 151 | 34,322,925 | 5,011,323,440 | 146.0 | 30,113,240 | 10,731,506 | 1,790,349 | 87.7 | 15.78 |
| Number of unigenes | 128,244 |
| Total read count | 301,132,396 |
| Total contig length | 137,438,033 bp |
| Mean contig length | 1071.7 bp |
| Max. contig length | 21,849 bp |
| Min. contig length | 187 bp |
| N50 | 1752 bp |
| Annotation of SNPs | Number of SNPs | Number of Associated Unigenes |
|---|---|---|
| Non-synonymous | 1228 | 777 |
| Synonymous | 1648 | 951 |
| Others a | 2372 | 1161 |
| Not determined b | 1386 | 648 |
| Total | 6634 | 3537 |
| Number of Unigenes | 128,244 |
| Total bases (bp) | 137,438,033 |
| Number of SNPs | 6634 |
| SNP frequency | 0.05 SNPs/kb |
| Transition | 3880 |
| A/G | 1941 |
| C/T | 1939 |
| Transversion | 2754 |
| A/C | 650 |
| A/T | 440 |
| C/G | 1018 |
| G/T | 646 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, T.-H.; Yoon, Y.-M.; Park, J.-C.; Park, J.-H.; Kim, K.-H.; Kim, Y.-K.; Son, J.-H.; Park, T.-I. De Novo Transcriptome Assembly and SNP Discovery for the Development of dCAPS Markers in Oat. Agronomy 2022, 12, 184. https://doi.org/10.3390/agronomy12010184
Kim T-H, Yoon Y-M, Park J-C, Park J-H, Kim K-H, Kim Y-K, Son J-H, Park T-I. De Novo Transcriptome Assembly and SNP Discovery for the Development of dCAPS Markers in Oat. Agronomy. 2022; 12(1):184. https://doi.org/10.3390/agronomy12010184
Chicago/Turabian StyleKim, Tae-Heon, Young-Mi Yoon, Jin-Cheon Park, Jong-Ho Park, Kyong-Ho Kim, Yang-Kil Kim, Jae-Han Son, and Tae-Il Park. 2022. "De Novo Transcriptome Assembly and SNP Discovery for the Development of dCAPS Markers in Oat" Agronomy 12, no. 1: 184. https://doi.org/10.3390/agronomy12010184
APA StyleKim, T.-H., Yoon, Y.-M., Park, J.-C., Park, J.-H., Kim, K.-H., Kim, Y.-K., Son, J.-H., & Park, T.-I. (2022). De Novo Transcriptome Assembly and SNP Discovery for the Development of dCAPS Markers in Oat. Agronomy, 12(1), 184. https://doi.org/10.3390/agronomy12010184






