Determination and Metabolite Profiling of Mixtures of Triterpenoid Saponins from Seeds of Chilean Quinoa (Chenopodium quinoa) Germplasm
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. C. quinoa Diversity Panel
2.3. Extraction and Hydrolysis of Saponins from C. quinoa Germplasm
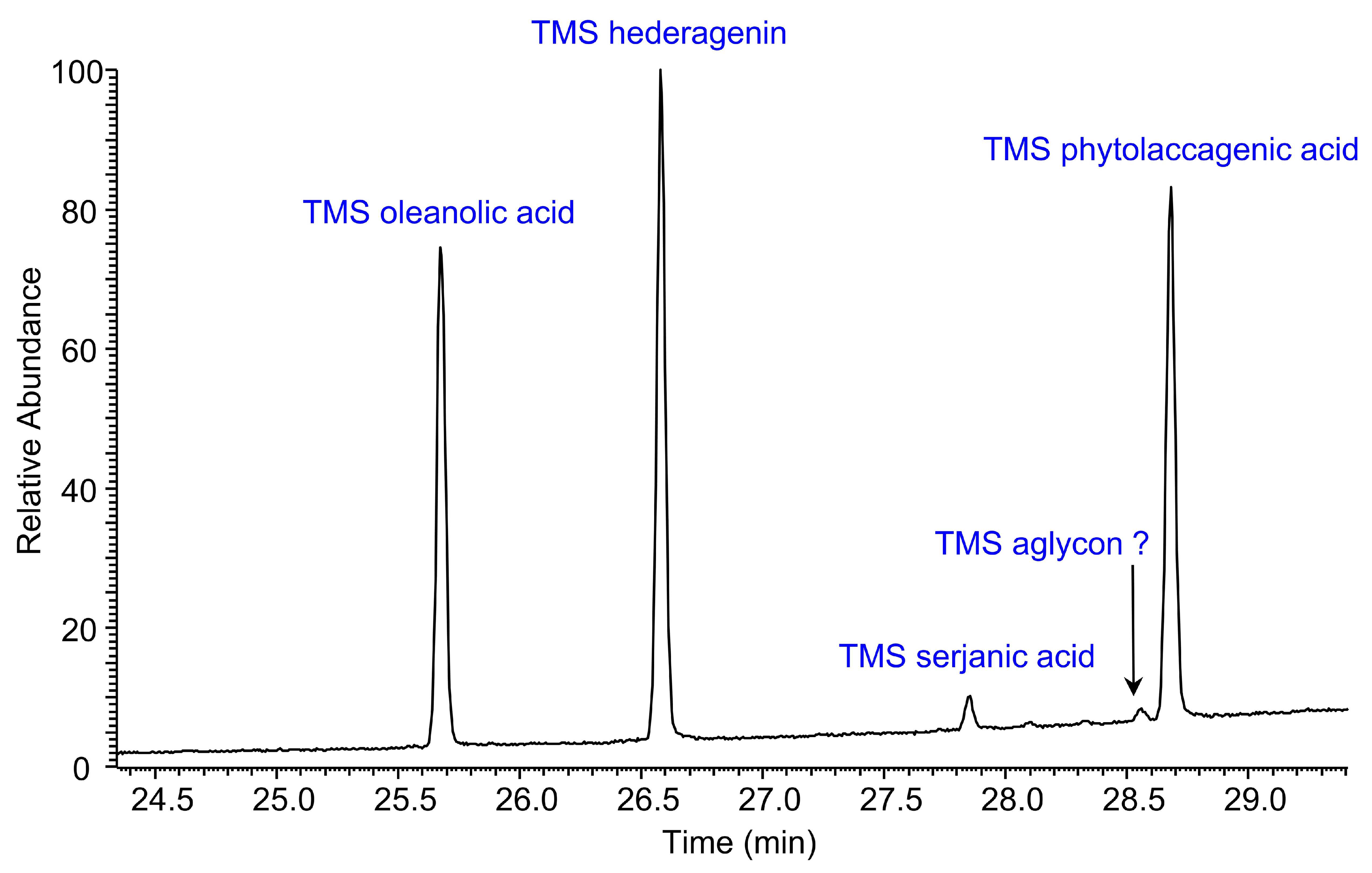
2.4. Quantification of Sapogenins by Gas Chromatography-Mass Spectrometry (GC-MS) Analysis
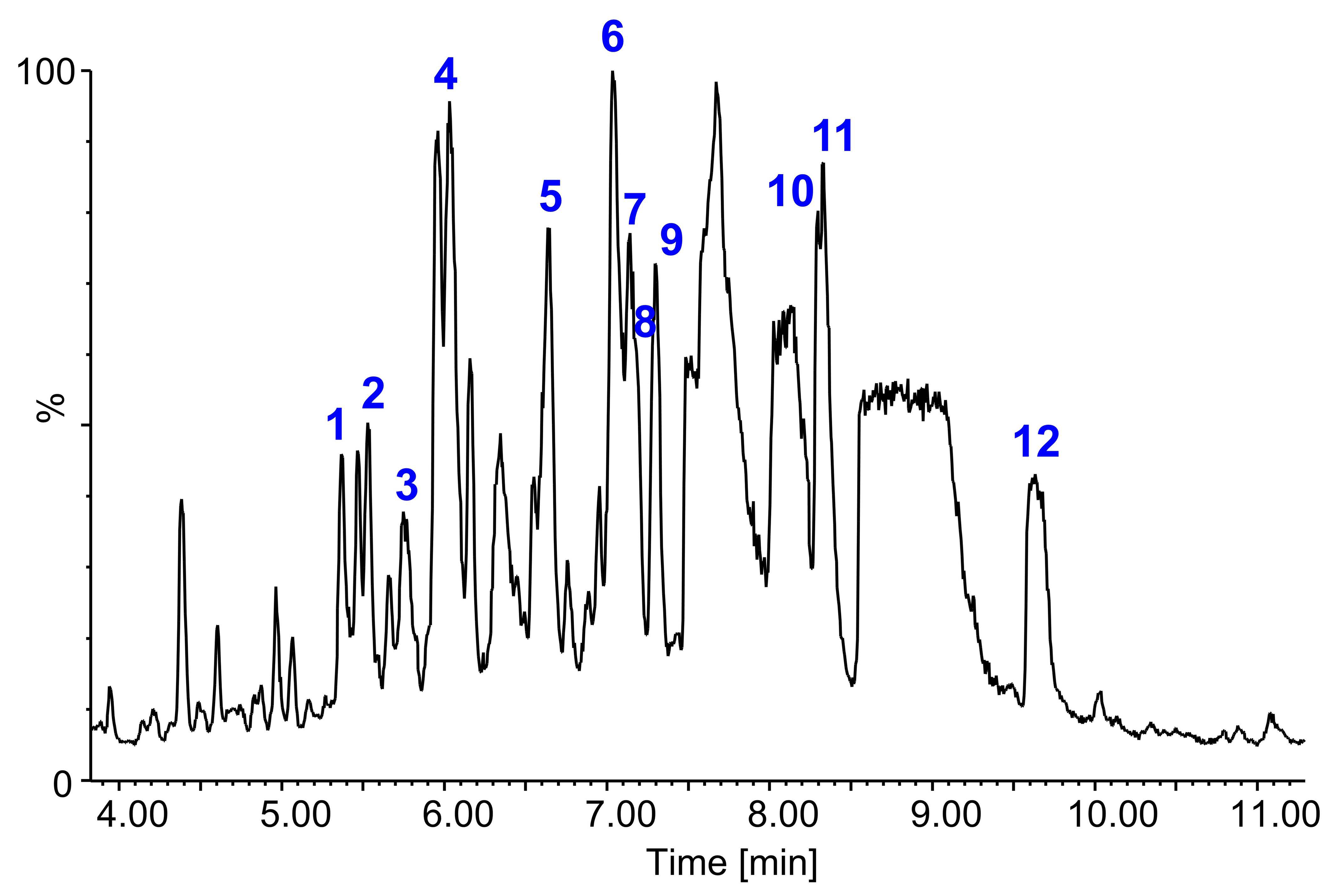
2.5. Identification of Saponins by LC-FTICR-MS and LC-MS/MS
2.6. Statistical Analysis
3. Results and Discussion
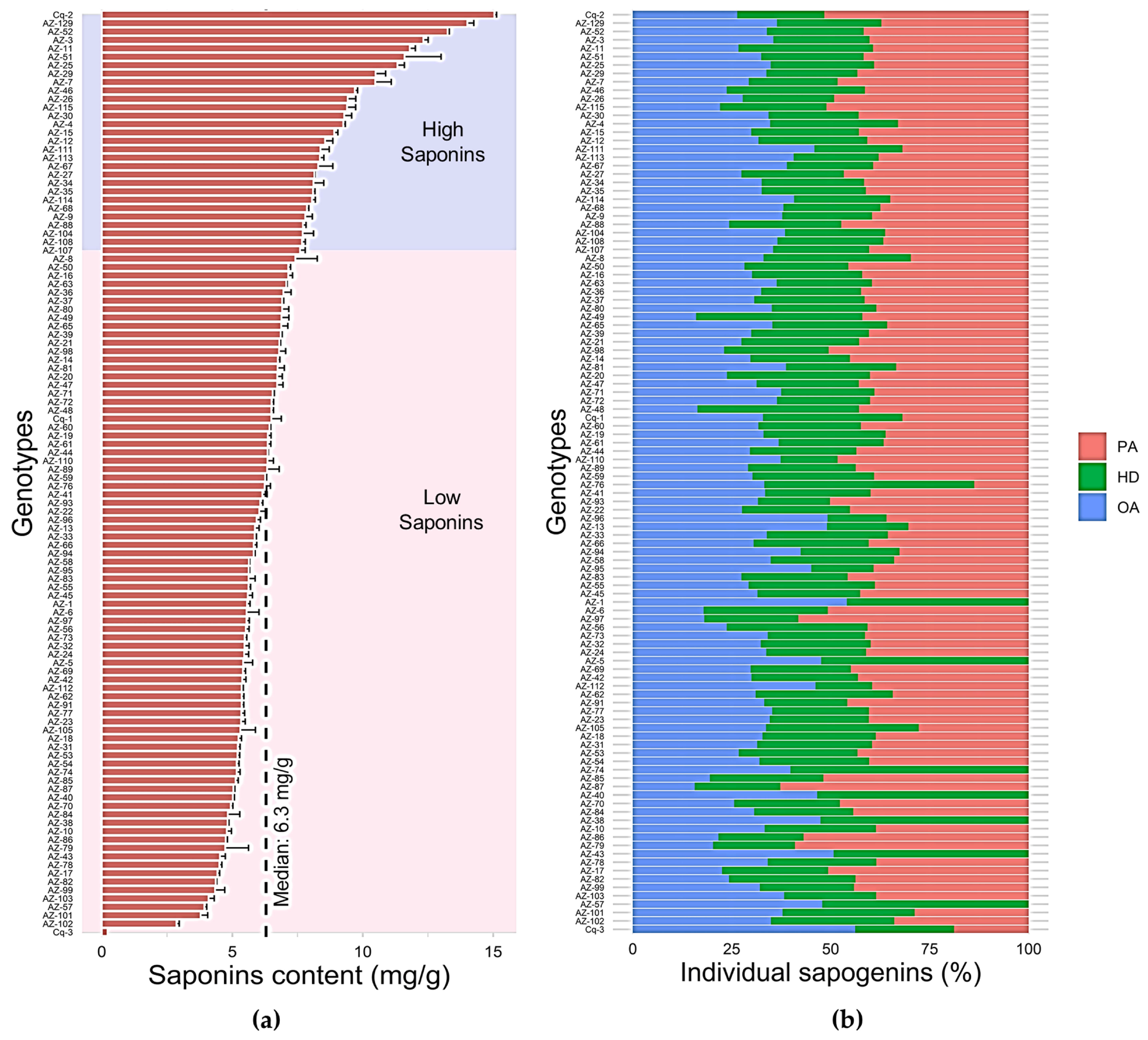
3.1. Extraction and Quantification of Triterpenoid Saponins in C. quinoa Germplasm
3.2. Evaluation of Triterpenoid Saponins of C. quinoa Seeds
3.3. Principal Component Analysis and Clustering
3.4. Variance by the Genotypic Effect
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vincken, J.P.; Heng, L.; de Groot, A.; Gruppen, H. Saponins, classification and occurrence in the plant kingdom. Phytochem 2007, 68, 275–297. [Google Scholar] [CrossRef]
- Cheok, C.Y.; Salman, H.A.K.; Sulaiman, R. Extraction and quantification of saponins: A review. Food Res. Int. 2014, 59, 16–40. [Google Scholar] [CrossRef]
- Stuardo, M.; Martin, R.S. Antifungal properties of quinoa (Chenopodium quinoa Willd.) alkali treated saponnins against Botrytis cinerea. Ind. Crop. Prod. 2008, 27, 296–302. [Google Scholar] [CrossRef]
- FAO & CIRAD. State of the Art Report of Quinoa in the World in 2013; Bazile, D., Bertero, D., Nieto, C., Eds.; FAO & CIRAD: Rome, Italy, 2015. [Google Scholar]
- Güclü-Üstündag, Ö.; Mazza, G. Saponins: Properties, applications and processing. Crit. Rev. Food Sci. Nutr. 2007, 47, 231–258. [Google Scholar] [CrossRef] [PubMed]
- Vega-Galvez, A.; Miranda, M.; Vergara, J.; Uribe, E.; Puente, L.; Martinez, E.A. Nutrition facts and functional potential of quinoa (Chenopodium quinoa Willd.), an ancient Andean grain: A review. J. Sci. Food Agric. 2010, 90, 2541–2547. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez Gómez, M.J.; Matías Prieto, J.; Cruz Sobrado, V.; Calvo Magro, P. Nutritional characterization of six quinoa (Chenopodium quinoa Willd) varieties cultivated in Southern Europe. J. Food Compos. Anal. 2021, 99, 103876. [Google Scholar] [CrossRef]
- Mhada, M.; Metougui, M.L.; El Hazzam, K.; El Kacimi, K.; Yasri, A. Variations of Saponins, Minerals and Total Phenolic Compounds Due to Processing and Cooking of Quinoa (Chenopodium quinoa Willd.) Seeds. Foods 2020, 9, 660. [Google Scholar] [CrossRef]
- Adolf, V.I.; Shabala, S.; Andersen, M.N.; Razzaghi, F.; Jacobsen, S.E. Varietal differences of quinoa’s tolerance to saline conditions. Plant Soil 2012, 357, 117–129. [Google Scholar] [CrossRef]
- Hariadi, Y.; Marandon, K.; Tian, Y.; Jacobsen, S.E.; Shabala, S. Ionic and osmotic relations in quinoa (Chenopodium quinoa Willd.) plants grown at various salinity levels. J. Exp. Bot. 2011, 62, 185–193. [Google Scholar] [CrossRef] [Green Version]
- Jacobsen, S.E.; Mujica, A.; Jensen, C.R. The resistance of quinoa (Chenopodium quinoa Willd.) to adverse abiotic factors. Food Rev. Int. 2003, 19, 99–109. [Google Scholar] [CrossRef]
- Burnoufradosevich, M.; Delfel, N.E.; England, R. Gas chromatography-mass spectrometry of olealane-type and ursane-type triterpenes—Application to chenopodium-quinoa triterpenes. Phytochem 1985, 24, 2063–2066. [Google Scholar] [CrossRef]
- Cuadrado, C.; Ayet, G.; Burbano, C.; Muzquiz, M.; Camacho, L.; Cavieres, E.; Lovon, M.; Osagie, A.; Price, K.R. Occurence of saponins and sapogenols in andean crops. J. Sci. Food Agric. 1995, 67, 169–172. [Google Scholar] [CrossRef]
- Mastebroek, H.D.; Limburg, H.; Gilles, T.; Marvin, H.J.P. Occurrence of sapogenins in leaves and seeds of quinoa (Chenopodium quinoa Willd). J. Sci. Food Agric. 2000, 80, 152–156. [Google Scholar] [CrossRef]
- Ridout, C.L.; Price, K.R.; Dupont, M.S.; Parker, M.L.; Fenwick, G.R. Quinoa saponins—Analysis and preliminary investigations into the effects of reduction by processing. J. Sci. Food Agric. 1991, 54, 165–176. [Google Scholar] [CrossRef]
- Madl, T.; Sterk, H.; Mittelbach, M. Tandem mass spectrometric analysis of a complex triterpene saponin mixture of Chenopodium quinoa. J. Am. Soc. Mass Spectrom. 2006, 17, 795–806. [Google Scholar] [CrossRef] [Green Version]
- Dini, I.; Schettino, O.; Simioli, T.; Dini, A. Studies on the constituents of Chenopodium quinoa seeds: Isolation and characterization of new triterpene saponins. J. Agric. Food Chem. 2001, 49, 741–746. [Google Scholar] [CrossRef]
- Dini, I.; Tenore, G.C.; Dini, A. Oleanane saponins in “kancolla”, a sweet variety of Chenopodium quinoa. J. Nat. Prod. 2002, 65, 1023–1026. [Google Scholar] [CrossRef]
- Dini, I.; Tenore, G.C.; Schettino, O.; Dini, A. New oleanane saponins in Chenopodium quinoa. J. Agric. Food Chem. 2001, 49, 3976–3981. [Google Scholar] [CrossRef]
- Kuljanabhagavad, T.; Thongphasuk, P.; Chamulitrat, W.; Wink, M. Triterpene saponins from Chenopodium quinoa Willd. Phytochem 2008, 69, 1919–1926. [Google Scholar] [CrossRef]
- Mizui, F.; Kasai, R.; Ohtani, K.; Tanaka, O. Saponins from bran of quinoa, Chenopodium quinoa WILLD. II. Chem. Pharm. Bull. 1990, 38, 375–377. [Google Scholar] [CrossRef] [Green Version]
- Zhu, N.Q.; Sheng, S.Q.; Sang, S.M.; Jhoo, J.W.; Bai, N.S.; Karwe, M.V.; Rosen, R.T.; Ho, C.T. Triterpene saponins from debittered quinoa (Chenopodium quinoa) seeds. J. Agric. Food Chem. 2002, 50, 865–867. [Google Scholar] [CrossRef]
- El Hazzam, K.; Hafsa, J.; Sobeh, M.; Mhada, M.; Taourirte, M.; El Kacimi, K.; Yasri, A. An Insight into Saponins from Quinoa (Chenopodium quinoa Willd): A Review. Molecules 2020, 25, 1059. [Google Scholar] [CrossRef] [Green Version]
- Jarvis, D.E.; Ho, Y.S.; Lightfoot, D.J.; Schmockel, S.M.; Li, B.; Borm, T.J.A.; Ohyanagi, H.; Mineta, K.; Michell, C.T.; Saber, N.; et al. The genome of Chenopodium quinoa. Nature 2017, 542, 307–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koziol, M.J. Afrosimetric estimation of threshold saponin concentration for bitterness in quinoa (Chenopodium quinoa Willd). J. Sci. Food Agric. 1991, 54, 211–219. [Google Scholar] [CrossRef]
- Medina-Meza, I.G.; Aluwi, N.A.; Saunders, S.R.; Ganjyal, G.M. GC-MS Profiling of Triterpenoid Saponins from 28 Quinoa Varieties (Chenopodium quinoa Willd.) Grown in Washington State. J. Agric. Food Chem. 2016, 64, 8583–8591. [Google Scholar] [CrossRef] [PubMed]
- Martinez, E.A.; Veas, E.; Jorquera, C.; San Martin, R.; Jara, P. Re-Introduction of Quinoa into Arid Chile: Cultivation of Two Lowland Races under Extremely Low Irrigation. J. Agro. Crop Sci. 2009, 195, 1–10. [Google Scholar] [CrossRef]
- Chauhan, G.S.; Eskin, N.A.M.; Tkachuk, R. Nutrients and antinutrients in quinoa seed. Cereal Chem. 1992, 69, 85–88. [Google Scholar]
- Gomez-Caravaca, M.A.; Iafelice, G.; Verardo, V.; Marconi, E.; Caboni, M.F. Influence of pearling process on phenolic and saponin content in quinoa (Chenopodium quinoa Willd). Food Chem. 2014, 157, 174–178. [Google Scholar] [CrossRef]
- Lundberg, L. Saponin Removal from Quinoa by Abrasion Processing. Ph.D. Thesis, California Polytechnic State University, San Luis Obispo, CA, USA, 2019. [Google Scholar]
- Zurita-Silva, A.; Fuentes, F.; Zamora, P.; Jacobsen, S.E.; Schwember, A.R. Breeding quinoa (Chenopodium quinoa Willd.): Potential and perspectives. Mol. Breeding 2014, 34, 13–30. [Google Scholar] [CrossRef]
- van Erp, W. Marker Development for Bitter-Tasting-Saponin Gene in Quinoa (Chenopodium quinoa): Final report; Wageningen UR: Wageningen, The Netherlands, 2016; Available online: http://edepot.wur.nl/394421 (accessed on 5 May 2021).
- Murphy, K.M.; Matanguihan, J.B.; Fuentes, F.F.; Gómez-Pando, L.R.; Jellen, E.N.; Maughan, P.J.; Jarvis, D.E. Quinoa Breeding and Genomics. In Plant Breeding Reviews; Goldman, I., Ed.; Wiley: New York, NY, USA, 2018; Volume 42. [Google Scholar]
- Reichert, R.D.; Tatarynovich, J.T.; Tyler, R.T. Abrasive dehulling of quinoa (Chenopodium-quinoa)—Effect on saponin content as determined by an adapted hemolytic assay. Cereal Chem. 1986, 63, 471–475. [Google Scholar]
- Ward, S.M. Response to selection for reduced grain saponin content in quinoa (Chenopodium quinoa Willd.). Field Crops Res. 2000, 68, 157–163. [Google Scholar] [CrossRef]
- Gómez-Caravaca, A.M.; Iafelice, G.; Lavini, A.; Pulvento, C.; Caboni, M.F.; Marconi, E. Phenolic Compounds and Saponins in Quinoa Samples (Chenopodium quinoa Willd.) Grown under Different Saline and Nonsaline Irrigation Regimens. J. Agric. Food Chem. 2012, 60, 4620–4627. [Google Scholar] [CrossRef] [PubMed]
- Ruales, J.; Nair, B.M. Saponins, phytic acid, tannins and protease inhibitors in quinoa (Chenopodium quinoa, Willd) seeds. Food Chem. 1993, 48, 137–143. [Google Scholar] [CrossRef]
- Woldemichael, G.M.; Wink, M. Identification and biological activities of triterpenoid saponins from Chenopodium quinoa. J. Agric. Food Chem. 2001, 49, 2327–2332. [Google Scholar] [CrossRef]
- Ruiz, K.B.; Khakimov, B.; Engelsen, S.B.; Bak, S.; Biondi, S.; Jacobsen, S.E. Quinoa seed coats as an expanding and sustainable source of bioactive compounds: An investigation of genotypic diversity in saponin profiles. Ind. Crop. Prod. 2017, 104, 156–163. [Google Scholar] [CrossRef]
- Jacobsen, S.E. Quinoa Quality. Available online: https://www.quinoaquality.com/contact-quinoa-quality (accessed on 5 May 2021).
- de Mendiburu, F. Agricolae: Statistical Procedures for Agricultural Research. Available online: https://CRAN.R-project.org/package=agricolae (accessed on 7 April 2021).
- Bates, D.; Maechler, M.; Bolker, B.; Walker, S.; Christensen, R.H.B.; Singmann, H.; Dai, B.; Grothendieck, G.; Green, P. Package ‘lme4’: Linear Mixed-Effects Models Using ‘Eigen’ and S4. Available online: https://github.com/lme4/lme4/ (accessed on 16 April 2021).
- Verza, S.G.; Kaiser, S.; de Resende, P.E.; Ortega, G.G. Determination and Characterization of Chenopodium quinoa Willd. Saponins. Lat. Am. J. Pharm. 2017, 36, 326–331. [Google Scholar]
- Miranda, M.; Vega-Galvez, A.; Quispe-Fuentes, I.; Rodriguez, M.J.; Maureira, H.; Martinez, E.A. Nutritional aspects of six quinoa (Chenopodium quinoa Willd.) ecotypes from three geographical areas of Chile. Chil. J. Agric. Res. 2012, 72, 175–181. [Google Scholar] [CrossRef] [Green Version]
- Ng, K.G.; Price, K.R.; Fenwick, G.R. A TLC method for the analysis of quinoa (Chenopodium quinoa) saponins. Food Chem. 1994, 49, 311–315. [Google Scholar] [CrossRef]
- Ando, H.; Chen, Y.C.; Tang, H.J.; Shimizu, M.; Watanabe, K.; Mitsunaga, T. Food components in fractions of quinoa seed. Food Sci. Technol. Res. 2002, 8, 80–84. [Google Scholar] [CrossRef] [Green Version]
- Mroczek, A. Phytochemistry and bioactivity of triterpene saponins from Amaranthaceae family. Phytochem. Rev. 2015, 14, 577–605. [Google Scholar] [CrossRef]
- Christensen, S.A.; Pratt, D.B.; Pratt, C.; Nelson, P.T.; Stevens, M.R.; Jellen, E.N.; Coleman, C.E.; Fairbanks, D.J.; Bonifacio, A.; Maughan, P.J. Assessment of genetic diversity in the USDA and CIP-FAO international nursery collections of quinoa (Chenopodium quinoa Willd.) using microsatellite markers. Plant Genet. Resour. 2007, 5, 82–95. [Google Scholar] [CrossRef] [Green Version]
- Fuentes, F.F.; Martinez, E.A.; Hinrichsen, P.V.; Jellen, E.N.; Maughan, P.J. Assessment of genetic diversity patterns in Chilean quinoa (Chenopodium quinoa Willd.) germplasm using multiplex fluorescent microsatellite markers. Conserv. Genet. 2009, 10, 369–377. [Google Scholar] [CrossRef]


| Quinoa Line | Accession Name | Type | Seed Source | Collection Region | Quinoa Line | Accession Name | Type | Seed Source | Collection Region | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Latitude | Longitude | Latitude | Longitude | ||||||||
| AZ-1 | Javi | Selfed line | INIA | −34.49778 | −72.00444 | AZ-27 | CHENO 047 | Selfed line | INIA | −34.70000 | −72.01667 |
| AZ-2 | Javi | Selfed line | INIA | −34.49778 | −72.00444 | AZ-29 | CHENO 207 | Selfed line | INIA | −34.49750 | −72.02111 |
| AZ-3 b | Cancosa | Selfed line | INIA | −20.49083 | −69.32917 | AZ-30 | CHENO 207 | Selfed line | INIA | −34.49750 | −72.02111 |
| AZ-4 | Cáhuil | Selfed line | INIA | −34.28111 | −71.85722 | AZ-31 | FARO | Selfed line | INIA | −34.46778 | −71.82583 |
| AZ-5 b | Cancosa | Selfed line | INIA | −20.49083 | −69.32917 | AZ-32 | FARO | Selfed line | INIA | −34.46778 | −71.82583 |
| AZ-6 | U de C9 | Selfed line | INIA | −35.73306 | −72.53306 | AZ-33 | EM10-1 | Selfed line | INIA | −34.53167 | −71.98722 |
| AZ-7 | Palmilla | Selfed line | INIA | −34.57722 | −71.38000 | AZ-34 | EM10-1 | Selfed line | INIA | −34.53167 | −71.98722 |
| AZ-8 | Palmilla | Selfed line | INIA | −34.57722 | −71.38000 | AZ-35 | EMPO 10-17 | Selfed line | INIA | −34.65083 | −71.89500 |
| AZ-9 a | R49 | Selfed line | INIA | −19.27639 | −68.64000 | AZ-36 | EMPO 10-15 | Selfed line | INIA | −34.64667 | −71.90806 |
| AZ-10 | U de C9 | Selfed line | INIA | −35.73306 | −72.53306 | AZ-37 | EMPO 10-14 | Selfed line | INIA | −34.52833 | −71.99278 |
| AZ-11 a | R49 | Drought tolerant line | INIA | −19.27639 | −68.64000 | AZ-38 | EMPO 10-14 | Selfed line | INIA | −34.52833 | −71.99278 |
| AZ-12 | Peñablanca-VI | Selfed line | INIA | −34.61139 | −71.64083 | AZ-39 | EMPO 10-10 | Selfed line | INIA | −34.51861 | −71.98028 |
| AZ-13 | -- | Selfed line | INIA | - | - | AZ-40 | EMPO 10-9 | Selfed line | INIA | −39.48194 | −72.13416 |
| AZ-14 a | Mix | Selfed line | INIA | −19.27639 | −68.64000 | AZ-41 | EMPO 10-8 | Selfed line | INIA | −34.51444 | −71.69722 |
| AZ-15 | Peñablanca-VI | Selfed line | INIA | −34.61139 | −71.64083 | AZ-42 | EMPO 10-7 | Selfed line | INIA | −34.64139 | −71.91194 |
| AZ-16 | Dorada P Paredones | Selfed line | INIA | −34.65750 | −71.97889 | AZ-43 | EMPO 10-6 | Selfed line | INIA | −34.65806 | −71.92889 |
| AZ-17 a | Mix | Selfed line | INIA | −19.27639 | −68.64000 | AZ-44 | EMPO 10-5 | Selfed line | INIA | −34.63667 | −71.95944 |
| AZ-18 | Cáhuil | Breeding line | INIA | −34.28111 | −71.85722 | AZ-45 | EMPO 10-4 | Selfed line | INIA | −34.62361 | −71.68778 |
| AZ-19 | Villarrica | Selfed line | INIA | −39.81944 | −73.24528 | AZ-46 | EMPO 10-2 | Selfed line | INIA | −34.69389 | −71.91917 |
| AZ-20 | Villarrica | Selfed line | INIA | −39.81944 | −73.24528 | AZ-47 | EMPO 10-1 | Breeding line | INIA | −34.53639 | −71.96917 |
| AZ-21 | KM21 | Selfed line | INIA | −39.48194 | −72.13417 | AZ-48 | PJEV 029 | Selfed line | INIA | −36.05806 | −72.47306 |
| AZ-22 | KM23 | Selfed line | INIA | −39.48194 | −72.13417 | AZ-49 | PJEV 028 | Selfed line | INIA | −35.95444 | −72.42139 |
| AZ-23 | KM23 | Selfed line | INIA | −39.48194 | −72.13417 | AZ-50 | PJEV 027 | Selfed line | INIA | −35.93667 | −72.70639 |
| AZ-24 | KM30 | Selfed line | INIA | −39.48194 | −72.13417 | AZ-51 | PJEV 026 | Breeding line | INIA | −35.93528 | −72.70694 |
| AZ-25 | CHENO 042 | Selfed line | INIA | −34.78667 | −72.04917 | AZ-52 | PJEV 025 | Selfed line | INIA | −35.91444 | −72.68972 |
| AZ-26 | CHENO 046 | Breeding line | INIA | −34.70000 | −72.01667 | AZ-53 | PJEV 006 | Selfed line | INIA | −34.52778 | −71.94611 |
| AZ-54 | PJ001 | Selfed line | INIA | −34.70000 | −72.01667 | AZ-76 | PRJ3 | Selfed line | INIA | −34.49778 | −72.00444 |
| AZ-55 | PJ002 | Selfed line | INIA | −34.53917 | −71.92833 | AZ-77 | PRJ3 | Selfed line | INIA | −34.49778 | −72.00444 |
| AZ-56 | PJEV 007 | Selfed line | INIA | −34.53222 | −71.98556 | AZ-78 | PJEV 016 | Breeding line | INIA | −35.12694 | −71.91722 |
| AZ-57 | PJEV 008 | Selfed line | INIA | −34.49833 | −72.02278 | AZ-79 | PJEV 017 | Selfed line | INIA | −35.01667 | −71.91833 |
| AZ-58 | PJ003 | Selfed line | INIA | −34.52028 | −71.97722 | AZ-80 | Palmilla | Selfed line | INIA | −34.57722 | −71.38000 |
| AZ-59 | PJ005 | Selfed line | INIA | −34.53556 | −71.58583 | AZ-81 | PJEV 018 | Selfed line | INIA | −35.00889 | −71.92611 |
| AZ-60 | PJEV 009 | Selfed line | INIA | −34.68417 | −71.99667 | AZ-82 | PJEV 019 | Selfed line | INIA | −35.04583 | −71.91194 |
| AZ-61 | PJEV010 | Selfed line | INIA | −34.68583 | −72.00139 | AZ-83 | PJEV 020 | Selfed line | INIA | −35.50111 | −72.08278 |
| AZ-62 | EAM 1 | Breeding line | INIA | −34.76833 | −72.07556 | AZ-84 | Dorada y Paredones | Selfed line | INIA | −34.65750 | −71.97889 |
| AZ-54 | PJ001 | Selfed line | INIA | −34.70000 | −72.01667 | AZ-85 | PJEV 021 | Selfed line | INIA | −35.50361 | −72.08306 |
| AZ-55 | PJ002 | Selfed line | INIA | −34.53917 | −71.92833 | AZ-86 | PJEV 022 | Selfed line | INIA | −35.59028 | −72.60917 |
| AZ-56 | PJEV 007 | Selfed line | INIA | −34.53222 | −71.98556 | AZ-87 | Roja Paredones | Selfed line | INIA | −34.65750 | −71.97889 |
| AZ-57 | PJEV 008 | Selfed line | INIA | −34.49833 | −72.02278 | AZ-88 | PJEV 003 | Selfed line | INIA | −34.61278 | −71.65139 |
| AZ-59 | PJ005 | Selfed line | INIA | −34.53556 | −71.58583 | AZ-86 | PJEV 022 | Selfed line | INIA | −35.59028 | −72.60917 |
| AZ-60 | PJEV 009 | Selfed line | INIA | −34.68417 | −71.99667 | AZ-87 | Roja Paredones | Selfed line | INIA | −34.65750 | −71.97889 |
| AZ-61 | PJEV010 | Selfed line | INIA | −34.68583 | −72.00139 | AZ-88 | PJEV 003 | Selfed line | INIA | −34.61278 | −71.65139 |
| AZ-62 | EAM 1 | Breeding line | INIA | −34.76833 | −72.07556 | AZ-89 | PJEV 003 | Selfed line | INIA | −34.61278 | −71.65139 |
| AZ-63 | EAM 1 | Selfed line | INIA | −34.76833 | −72.07556 | AZ-91 | PJEV 023 | Selfed line | INIA | −35.91083 | −72.68556 |
| AZ-64 | EAM 1 | Selfed line | INIA | −34.76833 | −72.07556 | AZ-92 | PJEV 23 | Selfed line | INIA | −35.91083 | −72.68556 |
| AZ-65 | EAM 2 | Selfed line | INIA | −34.98639 | −71.42750 | AZ-93 | PJEV 024 | Selfed line | INIA | −35.91028 | −72.68667 |
| AZ-66 | PJEV 011 | Selfed line | INIA | −34.76472 | −72.07806 | AZ-94 | PJEV 024 | Selfed line | INIA | −35.91028 | −72.68667 |
| AZ-67 | PJEV012 | Selfed line | INIA | −34.83667 | −72.05944 | AZ-95 | PJEV 05 | Selfed line | INIA | −34.53583 | −71.95694 |
| AZ-68 | EAM 3 | Selfed line | INIA | −34.49778 | −72.00444 | AZ-96 | PJEV 05 | Selfed line | INIA | −34.53583 | −71.95694 |
| AZ-69 | EAM 4 | Selfed line | INIA | -- | -- | AZ-97 | Plantas Verdes | Breeding line | INIA | -- | -- |
| AZ-70 | PJEV 013 | Selfed line | INIA | −34.84194 | −72.14194 | AZ-98 | Plantas Moradas | Breeding line | INIA | −39.81944 | −73.24528 |
| AZ-71 | EAM 5 | Selfed line | INIA | −34.53556 | −71.58583 | AZ-99 | Kinia | Breeding line | INIA | −39.48194 | −72.13417 |
| AZ-72 | PJEV 014 | Selfed line | INIA | −34.92806 | −72.17944 | AZ-100 | Javi | Selfed line | INIA | −39.48194 | −72.13417 |
| AZ-73 | PJEV 015 | Selfed line | INIA | −35.00944 | −71.91833 | AZ-101 | Javi | Selfed line | INIA | −39.48194 | −72.13417 |
| AZ-74 | JML01 | Selfed line | INIA | −35.86611 | −71.59694 | AZ-102 | Cancosa | Selfed line | INIA | −34.57722 | −71.38000 |
| AZ-103 | Cáhuil | Selfed line | INIA | −34.65750 | −71.978888 | AZ-113 | Peñablanca-VI | Selfed line | INIA | −34.57722 | −71.38000 |
| AZ-104 | Cáhuil | Selfed line | INIA | −35.91083 | −72.68555 | AZ-114 a | MIX | Selfed line | INIA | −34.57722 | −71.38000 |
| AZ-105 | Javi | Selfed line | INIA | −34.49777 | −72.00444 | AZ-115 | Dorada P Paredones | Selfed line | INIA | −34.65750 | −71.97888 |
| AZ-107 | Palmilla | Selfed line | INIA | −34.57722 | −71.38000 | AZ-129 | CHEN0207 | Selfed line | INIA | −34.49750 | −72.02111 |
| AZ-108 a | R49 | Selfed line | INIA | −34.57722 | −71.38000 | Cq-1 | Vikinga | Variety | Uni. of Copenhagen | -- | -- |
| AZ-110 | Palmilla | Selfed line | INIA | −34.65750 | −71.97888 | Cq-2 | Titicaca | Variety | Uni. of Copenhagen | -- | -- |
| AZ-111 a | R49 | Selfed line | INIA | −35.91083 | −72.68555 | Cq-3 | ATLAS | Variety | INIA | -- | -- |
| AZ-112 | Dorada P Paredones | Selfed line | INIA | −34.49777 | −72.00444 | ||||||
| LC-FTICR-MS | LC-MS/MS | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | Saponin a | tR b (min) | m/z Experimental | Rel. Int. (%) | Composition (M-H)− | Δ c [ppm] | Molecular Formula | m/z | Lit. |
| 1 | 3-O-Hex-Hex-PA 28-O-Hex | 5.36 | 1001.49539 | 100 | C49 H77 O21 | −0.89 | C49H78O21 | 1001 [M-H]−, 839 [M-H-Hex]−, 677 [M-H-2 Hex]−, 515 [M-H-3 Hex]− or [PA-H]− | [16,21,22] |
| 1002.49931 | 59.07 | C48 [13]C H77 O21 | −0.33 | ||||||
| 1003.50333 | 14.02 | C47 [13]C2 H77 O21 | 0.34 | ||||||
| 2 | 3-O-Hex-Pent-AG485 28-O-Hex | 5.53 | 941.43785 | 100 | C46 H69 O20 | −0.98 | C46H70O20 | 941 [M-H]−, 779 [M-H-Hex]−, 617 [M-H-2 Hex]−, 485 [M-H-2 Hex-Pent]− or [AG485 -H]− | [16,24] |
| 942.44187 | 52.49 | C45 [13]C H69 O20 | −0.27 | ||||||
| 943.44482 | 15.17 | C44 [13]C2 H69 O20 | −0.7 | ||||||
| 3 | 3-O-HexA-Pent-PA 28-O-Hex | 5.75 | 985.46400 | 100 | C48 H73 O21 | −1 | C48H74O21 | 985 [M-H]−, 823 [M-H-Hex]−, 647 [M-H-Hex- HexA]−, 515 [M-H-Hex-HexA-Pent]− or [PA-H]− | [24] |
| 986.46742 | 60.18 | C47 [13]C H73 O21 | −0.93 | ||||||
| 987.47110 | 8.02 | C46 [13]C2 H73 O21 | −0.6 | ||||||
| 4 | 3-O-Hex-Pent-PA 28-O-Hex | 6.03 | 971.48585 | 100 | C48 H75 O20 | 0.14 | C48H76O20 | (1017 [M + FA-H]−) d, 971 [M-H]−,809 [M-H-Hex]−, 647 [M-H-2 Hex]−, 515 [M-H-2 Hex-Pent]− or [PA-H]− | [16,17,18,19,21,22] |
| 972.48963 | 52.7 | C47 [13]C H75 O20 | 0.57 | ||||||
| 973.49181 | 17.3 | C46 [13]C2 H75 O20 | −0.63 | ||||||
| 5 | 3-O-Hex-Hex-SA 28-O-Hex | 6.63 | 985.46519 | 100 | C48 H73 O21 | 0.2 | C48H74O21 | (1031 [M + FA-H]−) d, 985 [M-H]−, 823 [M-H-Hex]−, 661 [M-H-2 Hex]−, 499 [M-H-3 Hex]− or [SA-H]− | [24] |
| 986.46874 | 54.6 | C47 [13]C H73 O21 | 0.41 | ||||||
| 987.47249 | 11.4 | C46 [13]C2 H73 O21 | 0.79 | ||||||
| 6 | 3-O-Hex-Pent-SA 28-O-Hex | 7.03 | 955.49035 | 100 | C48 H75 O19 | −0.47 | C48H76O19 | (1001 [M + FA-H]−) d, 955 [M-H]−, 793 [M-H-Hex]−, 631 [M-H-2 Hex]−, 499 [M-H-2 Hex-Pent]− or [SA-H]− | [16,18] |
| 956.49438 | 56.18 | C47 [13]C H75 O19 | 0.23 | ||||||
| 957.49710 | 16.08 | C46 [13]C2 H75 O19 | −0.43 | ||||||
| 7 | 3-O-HexA-Hed 28-O-Hex | 7.14 | 809.43306 | 100 | C42 H65 O15 | 0.2 | C42H66O15 | 809 [M-H]−, 647 [M-H-Hex]−, 471 [M-H–Hex-HexA]− or [Hed-H]− | [16] |
| 810.43673 | 43.43 | C41 [13]C H65 O15 | 0.59 | ||||||
| 811.43956 | 10.93 | C40 [13]C2 H65 O15 | −0.05 | ||||||
| 8 | 3-O-HexA-SA 28-O-Hex | 7.27 | 837.42806 | 100 | C43 H65 O16 | 0.3 | C43H66O16 | 837 [M-H]−, 675 [M-H-Hex]−, 499 [M-H-Hex-HexA]− or [SA-H]− | |
| 838.43165 | 57.59 | C42 [13]C H65 O16 | 0.58 | ||||||
| 839.43421 | 11.22 | C41 [13]C2 H65 O16 | −0.37 | ||||||
| 9 | 3-O-Hex-Pent-Hed 28-O-Hex | 7.30 | 927.43863 | 100 | C47 H75 O18 | 2.96 | C47H76O18 | (973 [M + FA-H]−) d 927 [M-H]−, 765 [M-H-Hex]−, 603 [M-H-2 Hex]−, 471 [M-H-2 Hex-Pent]− or [Hed-H]− | [18,22,24] |
| 928.50152 | 44.27 | C46 [13]C H75 O18 | 2.45 | ||||||
| 10 | 3-O-HexA-OA 28-O-Hex | 8.30 | 793.43723 | 100 | C42 H65 O14 | −0.94 | C42H66O14 | 793 [M-H]−, 631 [M-H-Hex]−, 455 [M-H-Hex-HexA]− or [OA-H]− | [16,18,20,21,22] |
| 794.44077 | 48.31 | C41 [13]C H65 O14 | −0.71 | ||||||
| 795.44454 | 9.08 | C40 [13]C2 H65 O14 | −0.19 | ||||||
| 11 | 3-O-Pent-HexA-Hed 28-O-Hex | 8.38 | 941.44005 | 100 | C46 H69 O20 | 1.28 | C46H70O20 | 941 [M-H]−, 809 [M-H-Pent]−, 647 [M-H-Pent-Hex]−, 471 [M-H-Pent-Hex-HexA]− or [Hed-H]− | [16,17,18,19,21,22] |
| 942.44388 | 50.0 | C45 [13]C H69 O20 | 1.75 | ||||||
| 943.44724 | 16.2 | C44 [13]C2 H69 O20 | 1.77 | ||||||
| 12 | 3-O-Pent-HexA-OA 28-O-Hex | 9.60 | 925.47997 | 100 | C47 H73 O18 | −0.29 | C47H74O18 | 925 [M-H]−, 793 [M-H-Pent]−, 631 [M-H-Pent-Hex]−, 455 [M-H-Pent-Hex-HexA]− or [OA-H]− | [16,17,18,19,21,22] |
| 926.48382 | 53.69 | C46 [13]C H73 O18 | 0.24 | ||||||
| 927.48743 | 15.5 | C45 [13]C2 H73 O18 | 0.52 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandya, A.; Thiele, B.; Zurita-Silva, A.; Usadel, B.; Fiorani, F. Determination and Metabolite Profiling of Mixtures of Triterpenoid Saponins from Seeds of Chilean Quinoa (Chenopodium quinoa) Germplasm. Agronomy 2021, 11, 1867. https://doi.org/10.3390/agronomy11091867
Pandya A, Thiele B, Zurita-Silva A, Usadel B, Fiorani F. Determination and Metabolite Profiling of Mixtures of Triterpenoid Saponins from Seeds of Chilean Quinoa (Chenopodium quinoa) Germplasm. Agronomy. 2021; 11(9):1867. https://doi.org/10.3390/agronomy11091867
Chicago/Turabian StylePandya, Archis, Björn Thiele, Andres Zurita-Silva, Björn Usadel, and Fabio Fiorani. 2021. "Determination and Metabolite Profiling of Mixtures of Triterpenoid Saponins from Seeds of Chilean Quinoa (Chenopodium quinoa) Germplasm" Agronomy 11, no. 9: 1867. https://doi.org/10.3390/agronomy11091867
APA StylePandya, A., Thiele, B., Zurita-Silva, A., Usadel, B., & Fiorani, F. (2021). Determination and Metabolite Profiling of Mixtures of Triterpenoid Saponins from Seeds of Chilean Quinoa (Chenopodium quinoa) Germplasm. Agronomy, 11(9), 1867. https://doi.org/10.3390/agronomy11091867








