Evaluation of Resistance Sources of Tomato (Solanum lycopersicum L.) to Phylotype I Strains of Ralstonia solanacearum Species Complex in Benin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Development of F5 Lines Derived from Servo
2.2. Evaluation of the F5 Lines and Checks for BW Resistance
2.3. Trial Sites
2.4. Experimental Design and Agronomic Practices
2.5. Data Collection
2.6. Characterization/Identification of R. solanacearum Complex Species and Phylotypes
2.6.1. Collection of Samples
2.6.2. Species and Phylotype Identification
2.7. Data Analysis
3. Results
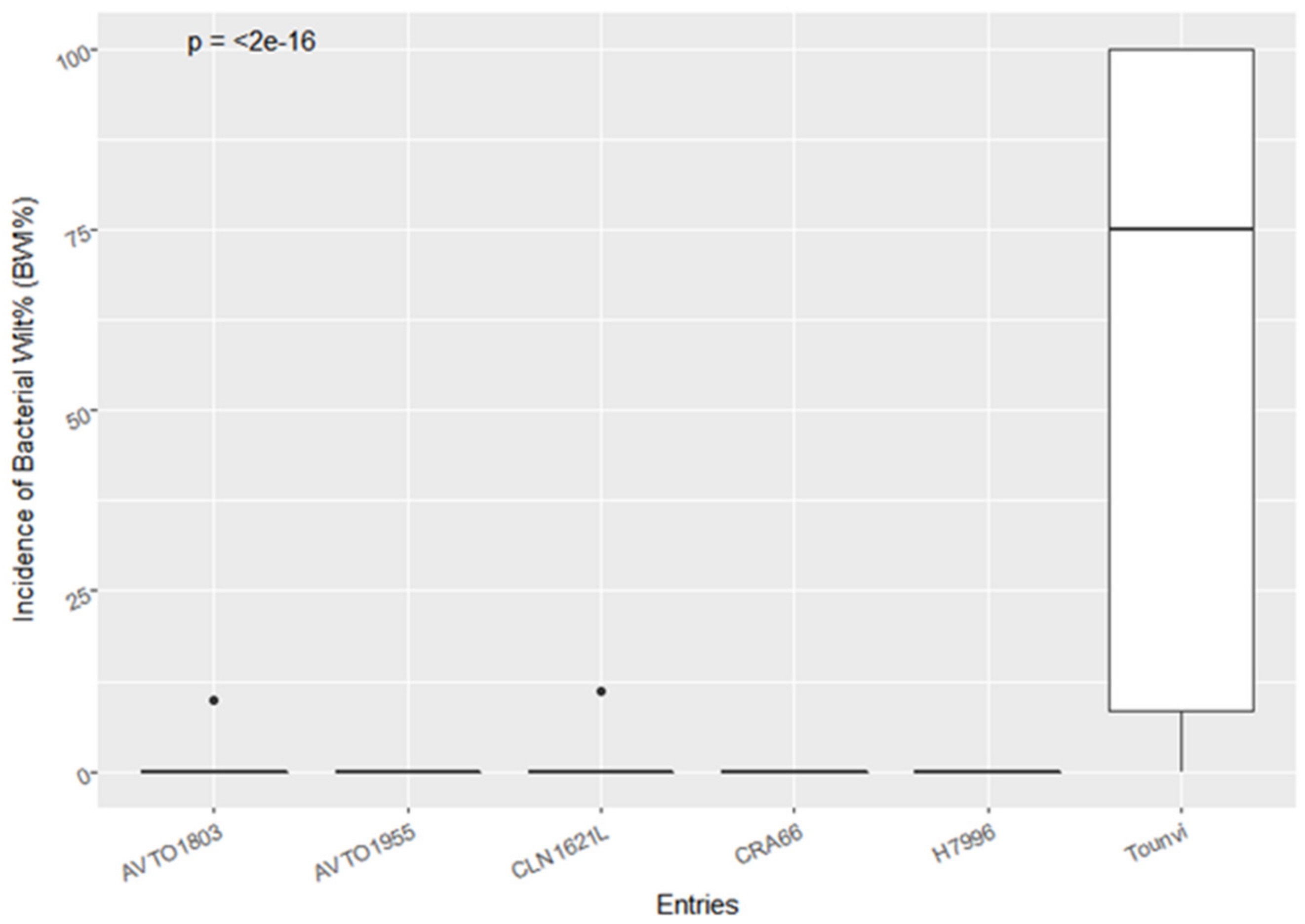
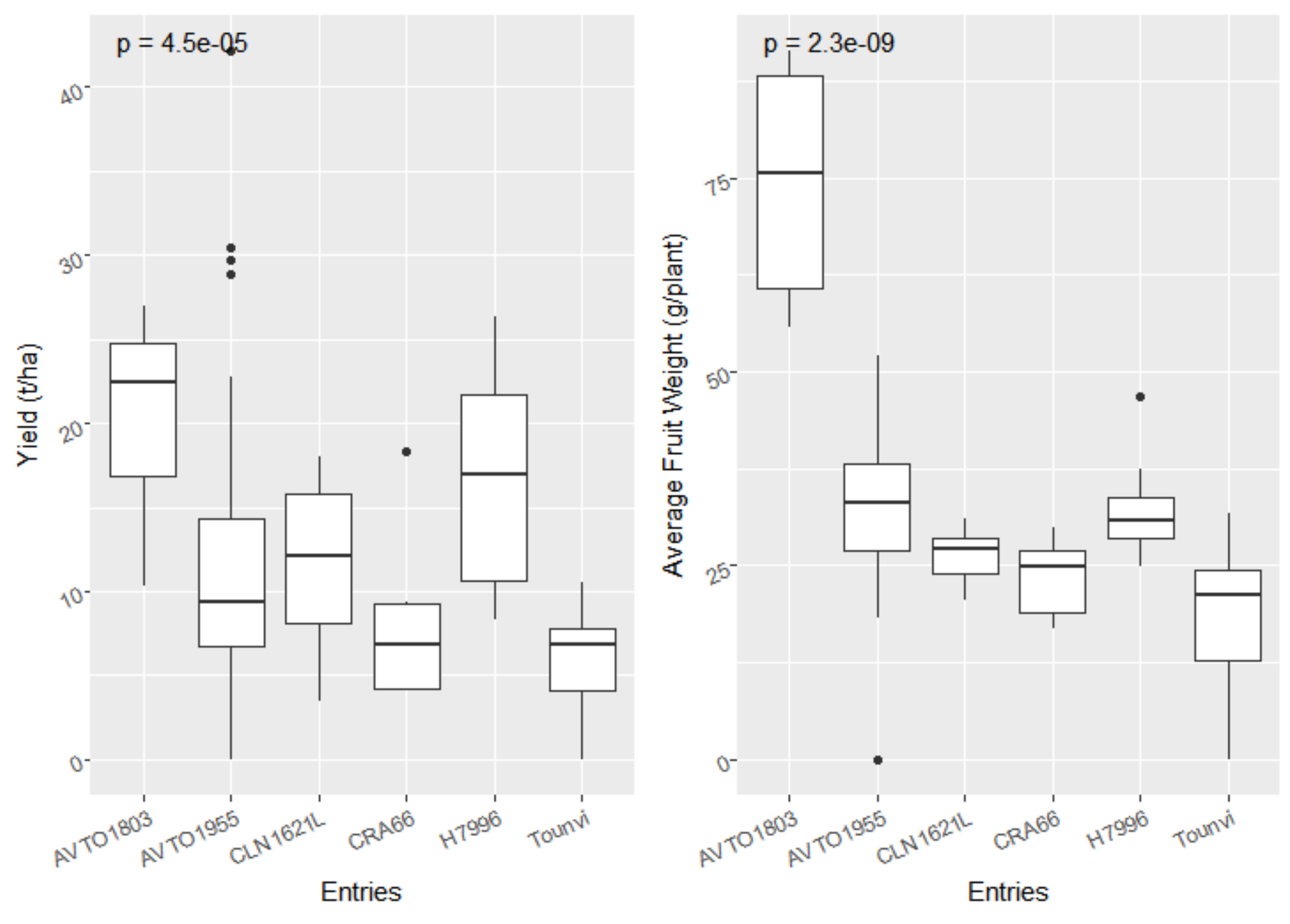
3.1. Bacterial Wilt Incidence and Agronomic Performance of the F5 Lines and Checks
3.2. Characterization/Identification of R. solanacearum Complex Species and Phylotypes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Database, F. Crops. Food and Agriculture Organization of the United Nation. 2020. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 6 April 2021).
- Agossou, G.; Ahouansou, T.; Aly, D.; Assogba-Komlan, F. Etude sur la Promotion de la Filière des Cultures Maraîchères au Bénin. Rapport Principal; Ministry of Agriculture, Livestocks and Fisheries of Republic of Benin (MAEP): Cotonou, Benin, 2001; p. 102.
- James, B.; Atcha-Ahowé, C.; Godonou, I.; Baimey, H.; Goergen, H.; Sikirou, R.; Toko, M. Integrated Pest. Management in Vegetable Production: A Guide for Extension Workers in West. Africa; International Institue of Tropical Agriculture (IITA): Ibadan, Oyo, Nigeria, 2010; p. 120. [Google Scholar]
- Sikirou, R.; Beed, F.; Ezin, V.; Hoteigni, J.; Miller, S.A. Distribution, pathological and biochemical characterization of Ralstonia solanacearum in Benin. Ann. Agric. Sci. 2017, 62, 83–88. [Google Scholar] [CrossRef]
- Kunwar, S.; Hsu, Y.C.; Lu, S.F.; Wang, J.F.; Jones, J.B.; Hutton, S.; Paret, M.; Hanson, P. Characterization 3333 of tomato (Solanum lycopersicum) accessions for resistance to phylotype I and phylotype II strains of the Ralstonia solanacearum species complex under high temperatures. Plant Breed. 2020, 139, 389–401. [Google Scholar] [CrossRef] [Green Version]
- Lebeau, A.; Daunay, M.-C.; Frary, A.; Palloix, A.; Wang, J.-F.; Dintinger, J.; Chiroleu, F.; Wicker, E.; Prior, P. Bacterial wilt resistance in tomato, pepper, and eggplant: Genetic resources respond to diverse strains in the Ralstonia solanacearum species complex. Phytopathology 2011, 101, 154–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prior, P.; Fegan, M. Diversity and molecular detection of Ralstonia solanacearum race 2 strains by multiplex PCR. In Bacterial Wilt Disease and the Ralstonia Solanacearum Species Complex; American Phytopathological Society (APS Press): St. Paul, MN, USA, 2005; pp. 405–414. [Google Scholar]
- Safni, I.; Cleenwerck, I.; De Vos, P.; Fegan, M.; Sly, L.; Kappler, U. Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: Proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstoniapseudosolanacearum sp. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 3087–3103. [Google Scholar]
- Prior, P.; Ailloud, F.; Dalsing, B.L.; Remenant, B.; Sanchez, B.; Allen, C. Genomic and proteomic evidence supporting the division of the plant pathogen Ralstonia solanacearum into three species. BMC Genom. 2016, 17, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Chesneau, T.; Maignien, G.; Boyer, C.; Cheron, J.-J.; Roux-Cuvelier, M.; Vanhuffel, L.; Poussier, S.; Prior, P. Sequevar diversity and virulence of Ralstonia solanacearum phylotype I on Mayotte Island (Indian Ocean). Front. Plant Sci. 2018, 8, 2209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanju., K.; Bamazi, B.; Banito, A.; Carter, M.; Weinstein, S.; Steidl, O.; Hayes, M.M.; Allen, C.; Paret, M. First Report of Bacterial Wilt Disease of Tomato, Pepper, and Gboma Caused by Ralstonia solanacearum Species Complex in Togo. Plant Dis. 2020. [Google Scholar] [CrossRef]
- Bihon, W.; Chen, J.-R.; Kenyon, L. Identification and characterization of Ralstonia spp. causing bacterial wilt disease of vegetables in Mali. J. Plant Pathol. 2020, 102, 1029–1039. [Google Scholar] [CrossRef]
- N’Guessan, C.A.; Abo, K.; Fondio, L.; Chiroleu, F.; Lebeau, A.; Poussier, S.; Wicker, E.; Koné, D. So near and yet so far: The specific case of Ralstonia solanacearum populations from Cote d’Ivoire in Africa. Phytopathology 2012, 102, 733–740. [Google Scholar] [CrossRef] [Green Version]
- Adebayo, O.; Ekpo, E. Biovar of Ralstonia solanacearum causing bacterial wilt of tomato in Nigeria. Plant Dis. 2005, 89, 1129. [Google Scholar] [CrossRef]
- Scott, J.; Wang, J.; Hanson, P. Breeding tomatoes for resistance to bacterial wilt, a global view. In Proceedings of the I International Symposium on Tomato Diseases 695, Orlando, FL, USA, 4 June 2005. [Google Scholar]
- Hanson, P.M.; Wang, J.-F.; Licardo, O.; Mah, S.Y.; Hartman, G.L.; Lin, Y.-C.; Chen, J.-T. Variable reaction of tomato lines to bacterial wilt evaluated at several locations in Southeast Asia. HortScience 1996, 31, 143–146. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.-F.; Hanson, P.; Barnes, J. Worldwide evaluation of an international set of resistance sources to bacterial wilt in tomato. In Bacterial Wilt Disease; Prior, P., Allen, C., Elphinstone, J., Eds.; Springer: Heidelberg/Berlin, Germany, 1998; pp. 269–275. [Google Scholar]
- Scott, J. Tomato improvement for bacterial disease resistance for the tropics: A contemporary basis and future prospects. In Proceedings the First International Conference on the Processing Tomato and First International Symposium on Tropical Tomato Diseases; Pernambuco, Brazil, 18–21 November 1996, ASHS Press: Alexandria, VA, USA, 1997. [Google Scholar]
- Opena, R.; Hartman, G.; Chen, J.; Yang, C. Breeding for Bacterial wilt Resistance in Tropical Tomato; Asian Vegetable Reseacrh and Development Center: Tainan, Taiwan, 1990. [Google Scholar]
- Walter, J. Hereditary resistance to disease in tomato. Annu. Rev. Phytopathol. 1967, 5, 131–160. [Google Scholar] [CrossRef]
- Wang, J.-F.; Ho, F.-I.; Truong, H.T.H.; Huang, S.-M.; Balatero, C.H.; Dittapongpitch, V.; Hidayati, N. Identification of major QTLs associated with stable resistance of tomato cultivar ‘Hawaii 7996’ to Ralstonia solanacearum. Euphytica 2013, 190, 241–252. [Google Scholar] [CrossRef]
- Ho, F.; Chung, C.; Wang, J. Distribution of Major QTLs Associated with Resistance to Ralstonia Solanacearum Phylotype I Strain in a Global set of Resistant Tomato Accessions. In Report of the Tomato Genetics Cooperative; Hal Inrae: Ithaca, NY, USA, 2013; pp. 22–30. [Google Scholar]
- Hanson, P.; Lu, S.-F.; Wang, J.-F.; Chen, W.; Kenyon, L.; Tan, C.-W.; Tee, K.L.; Wang, Y.-Y.; Hsu, Y.-C.; Schafleitner, R. Conventional and molecular marker-assisted selection and pyramiding of genes for multiple disease resistance in tomato. Sci. Hortic. 2016, 201, 346–354. [Google Scholar] [CrossRef] [Green Version]
- Safni, I.; Subandiyah, S.; Fegan, M. Ecology, epidemiology and disease management of Ralstonia syzygii in Indonesia. Front. Microbiol. 2018, 9, 419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burlakoti, R.R.; Hsu, C.-f.; Chen, J.-r.; Sheu, Z.-M.; Bihon, W.; Kenyon, L. Capture of Ralstonia solanacearum species complex strains directly from plant tissue sampled on FTA cards for molecular characterization. J. Plant Pathol. 2020, 102, 11–17. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Hothorn, T.; Bretz, F.; Westfall, P.; Heiberger, R.M.; Schuetzenmeister, A.; Scheibe, S.; Hothorn, M.T. Package ‘multcomp’. In Simultaneous Inference in General Parametric Models; Project for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Adegbola, Y.; Ahoyo Adjovi, N.; Adekambi, S.; Zossou, R.; Sonehekpon, E.; Assogba Komlan, F.; Djossa, E. Consumer preferences for fresh tomatoes in Benin using a conjoint analysis. J. Int. Food Agribus. Mark. 2019, 31, 1–21. [Google Scholar] [CrossRef]
- Traore, O.; Wonni, I.; Boro, F.; Somtore, E.; Zombré, C.T.; Dianda, O.Z.; Wicker, E.; Ilboudo, P.; Ouedraogo, L.S.; Somda, I. Evaluation of the 19 varieties and accessions of tomato against bacterial wilt in Bobo-Dioulasso, Burkina Faso. Int. J. Biol. Chem. Sci. 2020, 14, 2870–2879. [Google Scholar] [CrossRef]
- Baichoo, Z.; Jaufeerally-Fakim, Y. Ralstonia solanacearum upregulates marker genes of the salicylic acid and ethylene signaling pathways but not those of the jasmonic acid pathway in leaflets of Solanum lines during early stage of infection. Eur. J. Plant Pathol. 2017, 147, 615–625. [Google Scholar] [CrossRef]
- Shin, I.S.; Hsu, J.-C.; Huang, S.-M.; Chen, J.-R.; Wang, J.-F.; Hanson, P.; Schafleitner, R. Construction of a single nucleotide polymorphism marker based QTL map and validation of resistance loci to bacterial wilt caused by Ralstonia solanacearum species complex in tomato. Euphytica 2020, 216, 54. [Google Scholar] [CrossRef] [Green Version]
- Daunay, M.-C.; Laterrot, H.; Scott, J.; Hanson, P.; Wang, J. Tomato resistance to bacterial wilt caused by Ralstonia solanaearum EF Smith: Ancestry and peculiarities. In Report of the Tomato Genetics Cooperative; Hal Inrae: La Colle sur Loup, France, 2010. [Google Scholar]
- Ailloud, F.; Lowe, T.; Cellier, G.; Roche, D.; Allen, C.; Prior, P. Comparative genomic analysis of Ralstonia solanacearum reveals candidate genes for host specificity. BMC Genom. 2015, 16, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.H.; Tsai, K.C.; Prior, P.; Wang, J.F. Phylogenetic relationships and population structure of R alstonia solanacearum isolated from diverse origins in T aiwan. Plant Pathol. 2014, 63, 1395–1403. [Google Scholar] [CrossRef] [Green Version]
| Code | Entries | Bwr-12 | Bwr-6 | ||
|---|---|---|---|---|---|
| SLM6-17 | SLM6-94 | SLM6-110 | |||
| T1 | AVTO1955-1 | + | + | + | + |
| T2 | AVTO1955-2 | + | + | + | + |
| T3 | AVTO1955-3 | + | + | + | + |
| T4 | AVTO1955-5 | + | + | + | + |
| T5 | AVTO1955-6 | + | + | + | + |
| T6 | AVTO1955-9 | + | + | + | + |
| T7 | AVTO1955-10 | + | + | + | + |
| T8 | AVTO1955-11 | + | + | + | + |
| T9 | AVTO1955-12 | + | + | + | + |
| T10 | AVTO1955-14 | + | + | + | + |
| T11 | AVTO1955-15 | + | + | + | + |
| T12 | AVTO1955-16 | + | + | + | + |
| T13 | AVTO1955-17 | + | + | + | + |
| T14 | AVTO1955-18 | + | + | + | + |
| T15 | AVTO1955-19 | + | + | + | + |
| T16 | AVTO1955-20 | + | + | + | + |
| T17 | AVTO1955-21 | + | + | + | + |
| T18 | AVTO1955-22 | + | + | + | + |
| T19 | CLN1621L | + | – | – | – |
| T20 | Tounvi | – | – | – | – |
| T21 | Hawaii7996 | + | + | + | + |
| T22 | CRA66 | + | + | + | + |
| T23 | AVTO1803 | + | + | + | + |
| Weather Parameters | April | May | June | July | August | September |
|---|---|---|---|---|---|---|
| Total Rainfall (mm) | 119 | 123.8 | 561.9 | 44.9 | 0 | 203.6 |
| Total Evaporation (mm) | 145.34 | 136.03 | 91.86 | 98.24 | 129.65 | 93.2 |
| Average Rainfall (mm/day) | 3.97 | 3.99 | 18.73 | 1.44 | 0 | 6.78 |
| Average Min. Temp (°C) | 25.47 | 24.91 | 23.75 | 24.05 | 23.05 | 23.65 |
| Average Max. Temp (°C) | 32.10 | 31.44 | 29.50 | 28.05 | 28.06 | 28.51 |
| Average Min. Rel./Hum (%) | 65.84 | 70.33 | 77.37 | 76.82 | 70.36 | 73.31 |
| Average Max. Rel./Hum | 92.58 | 94.12 | 96.22 | 93.5 | 89.79 | 90.75 |
| Entry | Days to 50% Flowering | Fruit Set % | Total Yield (t/ha) | Marketable (t/ha) | Fruit Length (mm) | Fruit Width (mm) | Ave Fruit Weight (g) | °Brix | Bacterial Wilt Incidence (BWI%) | Growth Habit | Fruit Shape |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AVTO1955-1 | 56 bc | 43 a | 8.15 ac | 8.00 ac | 40 bfg | 33 ab | 26 ab | 5.25 | 0.00 | SD | Oblong |
| AVTO1955-10 | 57 bc | 63 ab | 11.00 acd | 10.87 ace | 51 d | 42 bc | 46 e | 5.01 | 0.00 | SD | Oblong |
| AVTO1955-11 | 56 bc | 53 ab | 10.28 acd | 10.10 ace | 49 df | 40 ab | 40 ce | 5.43 | 0.00 | I | Oblong |
| AVTO1955-12 | 54 bc | 45 a | 13.15 ae | 13.08 acd | 41 bdf | 34 ab | 33 bc | 4.53 | 0.00 | I | Oblong |
| AVTO1955-14 | 62 b | 60 ab | 14.83 bce | 14.31 bcd | 44 cdef | 36 ab | 31 bc | 5.30 | 0.00 | I | Oblong |
| AVTO1955-15 | 56 bc | 75 b | 14.75 bce | 14.65 bcd | 48 cdef | 39 ab | 40 ce | 5.14 | 0.00 | SD | Oblong |
| AVTO1955-16 | 46 ab | 52 ab | 8.25 ac | 8.14 ac | 48 def | 36 ab | 32 bc | 5.33 | 0.00 | SD | Oblong |
| AVTO1955-17 | 56 bc | 63 ab | 7.11 ab | 6.97 ab | 46 cdef | 36 ab | 32 bc | 5.28 | 0.00 | SD | Oblong |
| AVTO1955-18 | 49 ab | 64 ab | 10.25 acd | 10.14 ace | 49 def | 37 ab | 34 bc | 4.95 | 0.00 | SD | Oblong |
| AVTO1955-19 | 60 b | 51 ab | 10.29 acd | 10.08 ace | 46 cdef | 34 ab | 31 bc | 5.30 | 0.00 | SD | Oblong |
| AVTO1955-2 | 49 ab | 39 a | 5.31 a | 5.22 a | 37 bc | 32 ab | 23 ab | 5.24 | 0.00 | SD | Oblong |
| AVTO1955-20 | 43 ab | 45 a | 9.77 acd | 9.41 ace | 49 def | 36 ab | 32 bc | 5.08 | 0.00 | SD | Oblong |
| AVTO1955-21 | 60 b | 55 ab | 10.24 acd | 10.03 ace | 46 cdef | 36 ab | 33 bc | 4.73 | 0.00 | SD | Oblong |
| AVTO1955-22 | 54 bc | 62 ab | 9.08 acd | 8.89 ac | 44 bdf | 35 ab | 31 bc | 5.16 | 0.00 | SD | Oblong |
| AVTO1955-3 | 45 ab | 65 ab | 16.44 ce | 16.32 cd | 43 bdf | 36 ab | 29 ac | 5.57 | 0.00 | SD | Oblong |
| AVTO1955-5 | 57 bc | 59 ab | 12.05 ae | 11.90 acd | 38 be | 34 ab | 25 ab | 5.21 | 0.00 | SD | Oblong |
| AVTO1955-6 | 44 ab | 59 ab | 17.66 de | 17.51 de | 46 cdef | 36 ab | 31 bc | 5.17 | 0.00 | SD | Oblong |
| AVTO1955-9 | 58 bc | 60 ab | 10.22 acd | 10.16 ace | 48 cdef | 39 ab | 39 cde | 4.97 | 0.00 | SD | Oblong |
| CLN1621L | 37 ac | 70 ab | 12.42 ae | 12.29 acd | 40 bdf | 37 ab | 27 abd | 4.34 | 0.00 | D | Round |
| Tounvi | 29 a | 49 ab | 5.71 a | 5.62 a | 23 a | 29 a | 18 a | 4.31 | 57.64 | I | Round |
| H7996 | 42 ab | 42 a | 17.07 de | 15.04 bce | 39 bf | 38 ab | 32 bc | 5.26 | 0.00 | SD | Round |
| CRA66 | 59 b | 43 a | 7.88 ac | 7.76 ac | 33 ab | 36 ab | 24 ab | 5.40 | 0.00 | I | Round |
| CLN4018I | 52 bc | 66 ab | 20.29 e | 19.55 d | 50 dg | 53 c | 75 f | 4.64 | 1.25 | SD | Round |
| Mean | 51 | 55 | 11.72 | 11.01 | 43 | 37 | 33 | 5.08 | 2.62 | - | - |
| Entry mean square | *** | *** | *** | *** | *** | ** | *** | ns | ns | - | - |
| CV% | 21.64 | 27.98 | 42.31 | 42.16 | 12.81 | 14.01 | 20.52 | 16.24 | 24.05 | - | - |
| LSD (p = 0.05) | 10.95 | 15.30 | 4.75 | 4.62 | 5.73 | 5.29 | 6.72 | 0.85 | 0.03 |
| Location | Crop | Entry Code | Plant Status at Collection | Organ | Tobacco Test | BW Positive | Phylotype PCR | Sequevar | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | WorldVeg-Benin | Tomato | CRA66 | Not Wilted | collar | + | + | I | 31 |
| 2 | WorldVeg-Benin | Tomato | CLN1621L | Wilted | Collar/Diseased plant | + | + | I | 14 * |
| 3 | WorldVeg-Benin | Tomato | CLN1621L | Wilted | Collar | + | + | I | 14 * |
| 4 | WorldVeg-Benin | Tomato | AVTO1955-5 | Not Wilted | Collar/health plant | + | + | I | 14 * |
| 5 | WorldVeg-Benin | Tomato | AVTO1955-5 | Not Wilted | Collar/health plant | + | + | I | 14 * |
| 6 | University of Abomey-Calavi | Tomato | Tounvi 1 | Wilted | Collar/Diseased plant | + | + | I | 18 * |
| 7 | University of Abomey-Calavi | Tomato | Tounvi 1 | Wilted | Pure culture | + | + | I | 18 * |
| 8 | University of Abomey-Calavi | Tomato | Tounvi 2 | Wilted | Collar/Diseased plant | + | + | I | 14 * |
| 9 | University of Abomey-Calavi | Tomato | Tounvi 2 | Wilted | Pure culture | + | + | I | 14 * |
| 10 | WorldVeg-Benin (off trial site)(Check) | Tomato | ENZA 2 | Wilted | collar | + | + | I | 14 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zohoungbogbo, H.; Quenum, A.; Honfoga, J.; Chen, J.-R.; Achigan-Dako, E.; Kenyon, L.; Hanson, P. Evaluation of Resistance Sources of Tomato (Solanum lycopersicum L.) to Phylotype I Strains of Ralstonia solanacearum Species Complex in Benin. Agronomy 2021, 11, 1513. https://doi.org/10.3390/agronomy11081513
Zohoungbogbo H, Quenum A, Honfoga J, Chen J-R, Achigan-Dako E, Kenyon L, Hanson P. Evaluation of Resistance Sources of Tomato (Solanum lycopersicum L.) to Phylotype I Strains of Ralstonia solanacearum Species Complex in Benin. Agronomy. 2021; 11(8):1513. https://doi.org/10.3390/agronomy11081513
Chicago/Turabian StyleZohoungbogbo, Herbaud, Adonis Quenum, Judith Honfoga, Jaw-Rong Chen, Enoch Achigan-Dako, Lawrence Kenyon, and Peter Hanson. 2021. "Evaluation of Resistance Sources of Tomato (Solanum lycopersicum L.) to Phylotype I Strains of Ralstonia solanacearum Species Complex in Benin" Agronomy 11, no. 8: 1513. https://doi.org/10.3390/agronomy11081513
APA StyleZohoungbogbo, H., Quenum, A., Honfoga, J., Chen, J.-R., Achigan-Dako, E., Kenyon, L., & Hanson, P. (2021). Evaluation of Resistance Sources of Tomato (Solanum lycopersicum L.) to Phylotype I Strains of Ralstonia solanacearum Species Complex in Benin. Agronomy, 11(8), 1513. https://doi.org/10.3390/agronomy11081513







