Assessment of Weed Classification Using Hyperspectral Reflectance and Optimal Multispectral UAV Imagery
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Data
2.2. Spectral Separability Procedures
Classification Procedures
2.3. Band Identification
2.4. Multispectral Imagery Data Collection
2.5. Object Based Image Analysis (OBIA) Procedures
3. Results and Discussion
3.1. Classification of Weeds Species
3.2. Classification and Validation
3.3. Band Identification
3.4. Results of Image Processing
3.5. Weed Map Analysis
3.6. Accuracy Assessment
Confusion Matrix Accuracy
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mennan, H.; Jabran, K.; Zandstra, B.H.; Pala, F. Non-chemical weed management in vegetables by using cover crops: A review. Agronomy 2020, 10, 257. [Google Scholar] [CrossRef]
- Hutto, K.C.; Shaw, D.R.; Byrd, J.D.; King, R.L. Differentiation of turfgrass and common weed species using hyperspectral radiometry. Weed Sci. 2006, 54, 335–339. [Google Scholar] [CrossRef]
- Izquierdo, J.; Milne, A.E.; Recasens, J.; Royo-Esnal, A.; Torra, J.; Webster, R.; Baraibar, B. Spatial and Temporal Stability of Weed Patches in Cereal Fields under Direct Drilling and Harrow tillage. Agronomy 2020, 10, 452. [Google Scholar] [CrossRef]
- De Baerdemaeker, J. Future adoption of automation in weed control. In Automation: The Future of Weed Control in Cropping Systems; Springer: Berlin/Heidelberg, Germany, 2014; pp. 221–234. [Google Scholar]
- FAO and UNEP. Global Assessment of Soil Pollution: Report; FAO and UNEP: Rome, Italy, 2021. [Google Scholar] [CrossRef]
- De Castro, A.I.; Torres-Sánchez, J.; Peña, J.M.; Jiménez-Brenes, F.M.; Csillik, O.; López-Granados, F. An automatic random forest-OBIA algorithm for early weed mapping between and within crop rows using UAV imagery. Remote Sens. 2018, 10, 285. [Google Scholar] [CrossRef]
- Okamoto, H.; Suzuki, Y.; Noguchi, N. Field applications of automated weed control: Asia. In Automation: The Future of Weed Control in Cropping Systems; Springer: Berlin/Heidelberg, Germany, 2014; pp. 189–200. [Google Scholar]
- Kiani, S.; Jafari, A. Crop Detection and Positioning in the Field Using Discriminant Analysis and Neural Networks Based on Shape Features. J. Agric. Sci. Technol. (JAST) 2012, 14, 755–765. [Google Scholar]
- Gutjahr, C.; Gerhards, R. Decision rules for site-specific weed management. In Precision Crop Protection-The Challenge and Use of Heterogeneity; Springer: Berlin/Heidelberg, Germany, 2010; pp. 223–239. [Google Scholar]
- Torres-Sánchez, J.; López-Granados, F.; De Castro, A.I.; Peña-Barragán, J.M. Configuration and specifications of an unmanned aerial vehicle (UAV) for early site specific weed management. PLoS ONE 2013, 8, e58210. [Google Scholar] [CrossRef]
- Arafat, S.M.; Aboelghar, M.A.; Ahmed, E.F. Crop discrimination using field hyper spectral remotely sensed data. Adv. Remote Sens. 2013, 2, 63–70. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, J.; Cen, H.; Lu, Y.; Yu, X.; He, Y.; Pieters, J.G. Automated spectral feature extraction from hyperspectral images to differentiate weedy rice and barnyard grass from a rice crop. Comput. Electron. Agric. 2019, 159, 42–49. [Google Scholar] [CrossRef]
- Scherrer, B.; Sheppard, J.; Jha, P.; Shaw, J. Hyperspectral imaging and neural networks to classify herbicide-resistant weeds. J. Appl. Remote Sens. 2019, 13, 044516. [Google Scholar] [CrossRef]
- Che’Ya, N.N. Site-Specific Weed Management Using Remote Sensing. Ph.D. Thesis, The University of Queensland, Gatton, QLD, Australia, 7 October 2016. [Google Scholar] [CrossRef]
- Furlanetto, R.H.; Moriwaki, T.; Falcioni, R.; Pattaro, M.; Vollmann, A.; Junior, A.C.S.; Antunes, W.C.; Nanni, M.R. Hyperspectral reflectance imaging to classify lettuce varieties by optimum selected wavelengths and linear discriminant analysis. Remote Sens. Appl. Soc. Environ. 2020, 20, 100400. [Google Scholar] [CrossRef]
- Huang, H.; Lan, Y.; Yang, A.; Zhang, Y.; Wen, S.; Deng, J. Deep learning versus Object-based Image Analysis (OBIA) in weed mapping of UAV imagery. Int. J. Remote Sens. 2020, 41, 3446–3479. [Google Scholar] [CrossRef]
- ASD. ASD HandHeld 2: Hand-Held VNIR Spectroradiometer. 2021. Available online: https://www.malvernpanalytical.com/en/support/product-support/asd-range/fieldspec-range/handheld-2-hand-held-vnir-spectroradiometer#manuals (accessed on 15 June 2021).
- ASD. FieldSpec® HandHeld 2™ Spectroradiometer User Manual; ASD Inc.: Boulder, CO, USA, 2010; Volume 1, pp. 1–93. [Google Scholar]
- Congalton, R.G. A review of assessing the accuracy of classifications of remotely sensed data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Kamal, M.; Phinn, S.; Johansen, K. Object-based approach for multi-scale mangrove composition mapping using multi-resolution image datasets. Remote Sens. 2015, 7, 4753–4783. [Google Scholar] [CrossRef]
- Aziz, A.A. Integrating a REDD+ Project into the Management of a Production Mangrove Forest in Matang Forest Reserve, Malaysia. Ph.D Thesis, The University of Queensland, Brisbane, QLD, Australia, 2014. [Google Scholar] [CrossRef]
- Phinn, S.R.; Roelfsema, C.M.; Mumby, P.J. Multi-scale, object-based image analysis for mapping geomorphic and ecological zones on coral reefs. Int. J. Remote Sens. 2012, 33, 3768–3797. [Google Scholar] [CrossRef]
- Thenkabail, P.S. Remotely Sensed Data Characterization, Classification, and Accuracies; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Liu, B.; Li, R.; Li, H.; You, G.; Yan, S.; Tong, Q. Crop/Weed discrimination using a field Imaging spectrometer system. Sensors 2019, 19, 5154. [Google Scholar] [CrossRef] [PubMed]
- Pott, L.P.; Amado, T.J.; Schwalbert, R.A.; Sebem, E.; Jugulam, M.; Ciampitti, I.A. Pre-planting weed detection based on ground field spectral data. Pest Manag. Sci. 2020, 76, 1173–1182. [Google Scholar] [CrossRef]
- Li, Y.; Al-Sarayreh, M.; Irie, K.; Hackell, D.; Bourdot, G.; Reis, M.M.; Ghamkhar, K. Identification of weeds based on hyperspectral imaging and machine learning. Front. Plant Sci. 2021, 11, 2324. [Google Scholar] [CrossRef]
- Carvalho, S.; Schlerf, M.; van Der Putten, W.H.; Skidmore, A.K. Hyperspectral reflectance of leaves and flowers of an outbreak species discriminates season and successional stage of vegetation. Int. J. Appl. Earth Obs. Geoinf. 2013, 24, 32–41. [Google Scholar] [CrossRef]
- Kodagoda, S.; Zhang, Z. Multiple sensor-based weed segmentation. Proc. Inst. Mech. Eng. Part I J. Syst. Control Eng. 2010, 224, 799–810. [Google Scholar] [CrossRef]
- Liu, B.; Bruch, R. Weed Detection for Selective Spraying: A Review. Curr. Robot. Rep. 2020, 1, 19–26. [Google Scholar] [CrossRef]
- Buitrago, M.F.; Skidmore, A.K.; Groen, T.A.; Hecker, C.A. Connecting infrared spectra with plant traits to identify species. ISPRS J. Photogramm. Remote Sens. 2018, 139, 183–200. [Google Scholar] [CrossRef]
- Ahmad, S.; Pandey, A.C.; Kumar, A.; Lele, N.V. Potential of hyperspectral AVIRIS-NG data for vegetation characterization, species spectral separability, and mapping. Appl. Geomat. 2021, 1–12. [Google Scholar] [CrossRef]
- De Souza, M.F.; do Amaral, L.R.; de Medeiros Oliveira, S.R.; Coutinho, M.A.N.; Netto, C.F. Spectral differentiation of sugarcane from weeds. Biosyst. Eng. 2020, 190, 41–46. [Google Scholar] [CrossRef]
- Gray, C.J.; Shaw, D.R.; Bruce, L.M. Utility of hyperspectral reflectance for differentiating soybean (Glycine max) and six weed species. Weed Technol. 2009, 23, 108–119. [Google Scholar] [CrossRef]
- Zhang, N.; Ning, W.; John, K.; Floyd, D. Potential use of plant spectral characteristics in weed detection. In Proceedings of the American Society of Association Executives (ASAE) Annual International Meeting; Disney’s Coronado Springs Resorts, Orlando, FL, USA, 12–16 July 1998. Available online: https://www.ars.usda.gov/ARSUserFiles/30200525/264%20Potential_UsePlantFD.pdf (accessed on 13 March 2021).
- Wilson, J.H.; Zhang, C.; Kovacs, J.M. Separating crop species in northeastern Ontario using hyperspectral data. Remote Sens. 2014, 6, 925–945. [Google Scholar] [CrossRef]
- Vrindts, E.; De Baerdemaeker, J.; Ramon, H. Weed detection using canopy reflection. Precis. Agric. 2002, 3, 63–80. [Google Scholar] [CrossRef]
- Borregaard, T.; Nielsen, H.; Nørgaard, L.; Have, H. Crop–weed discrimination by line imaging spectroscopy. J. Agric. Eng. Res. 2000, 75, 389–400. [Google Scholar] [CrossRef]
- Li, X.; Lee, W.S.; Li, M.; Ehsani, R.; Mishra, A.R.; Yang, C.; Mangan, R.L. Spectral difference analysis and airborne imaging classification for citrus greening infected trees. Comput. Electron. Agric. 2012, 83, 32–46. [Google Scholar] [CrossRef]
- Manevski, K.; Manakos, I.; Petropoulos, G.P.; Kalaitzidis, C. Discrimination of common Mediterranean plant species using field spectroradiometry. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 922–933. [Google Scholar] [CrossRef]
- Thorp, K.; Tian, L. A review on remote sensing of weeds in agriculture. Precis. Agric. 2004, 5, 477–508. [Google Scholar] [CrossRef]
- Lopez-Granados, F. Weed detection for site-specific weed management: Mapping and real-time approaches. Weed Res. 2011, 51, 1–11. [Google Scholar] [CrossRef]
- Steward, B.; Tian, L. Machine-vision weed density estimation for real-time, outdoor lighting conditions. Trans. ASAE 1999, 42, 1897. [Google Scholar] [CrossRef][Green Version]
- Gillespie, A.R.; Kahle, A.B.; Walker, R.E. Color enhancement of highly correlated images. I. Decorrelation and HSI contrast stretches. Remote Sens. Environ. 1986, 20, 209–235. [Google Scholar] [CrossRef]
- Lehmann, J.R.K.; Nieberding, F.; Prinz, T.; Knoth, C. Analysis of unmanned aerial system-based CIR images in forestry—A new perspective to monitor pest infestation levels. Forests 2015, 6, 594–612. [Google Scholar] [CrossRef]
- Slaughter, A.L. The Utility of Multispectral Imagery from an Unmanned Aircraft System for Determining the Spatial Distribution of Eragrostis Lehmanniana (Lehmann Lovegrass) in Rangelands. Ph.D. Thesis, New Mexico State University, Las Cruces, NM, USA, December 2014. Available online: http://www.worldcat.org/oclc/908844828 (accessed on 7 April 2021).
- Arroyo, L.A.; Johansen, K.; Phinn, S. Mapping land cover types from very high spatial resolution imagery: Automatic application of an object based classification scheme. In Proceedings of the GEOBIA 2010: Geographic Object-Based Image Analysis, Ghent, Belgium, 29 June–2 July 2010; Available online: https://www.isprs.org/proceedings/xxxviii/4-C7/pdf/arroyo_abstract.pdf (accessed on 10 April 2021).
- Peña, J.M.; Torres-Sánchez, J.; de Castro, A.I.; Kelly, M.; López-Granados, F. Weed mapping in early-season maize fields using object-based analysis of unmanned aerial vehicle (UAV) images. PLoS ONE 2013, 8, e77151. [Google Scholar] [CrossRef] [PubMed]
- Johansen, K.; Tiede, D.; Blaschke, T.; Arroyo, L.A.; Phinn, S. Automatic geographic object based mapping of streambed and riparian zone extent from LiDAR data in a temperate rural urban environment, Australia. Remote Sens. 2011, 3, 1139–1156. [Google Scholar] [CrossRef]
- Foody, G.M. Status of land cover classification accuracy assessment. Remote Sens. Environ. 2002, 80, 185–201. [Google Scholar] [CrossRef]
- Peña Barragán, J.M.; Kelly, M.; Castro, A.I.d.; López Granados, F. Object-Based Approach for Crop Row Characterization in UAV images for Site-Specific Weed Management. In Proceedings of the 4th GEOBIA, Rio de Janeiro, Brazil, 7–9 May 2012; pp. 426–430. [Google Scholar]
- Mesas-Carrascosa, F.-J.; Torres-Sánchez, J.; Clavero-Rumbao, I.; García-Ferrer, A.; Peña, J.-M.; Borra-Serrano, I.; López-Granados, F. Assessing optimal flight parameters for generating accurate multispectral orthomosaicks by UAV to support site-specific crop management. Remote Sens. 2015, 7, 12793–12814. [Google Scholar] [CrossRef]
- Torres-Sánchez, J.; Pena, J.M.; de Castro, A.I.; López-Granados, F. Multi-temporal mapping of the vegetation fraction in early-season wheat fields using images from UAV. Comput. Electron. Agric. 2014, 103, 104–113. [Google Scholar] [CrossRef]
- Hengl, T. Finding the right pixel size. Comput. Geosci. 2006, 32, 1283–1298. [Google Scholar]
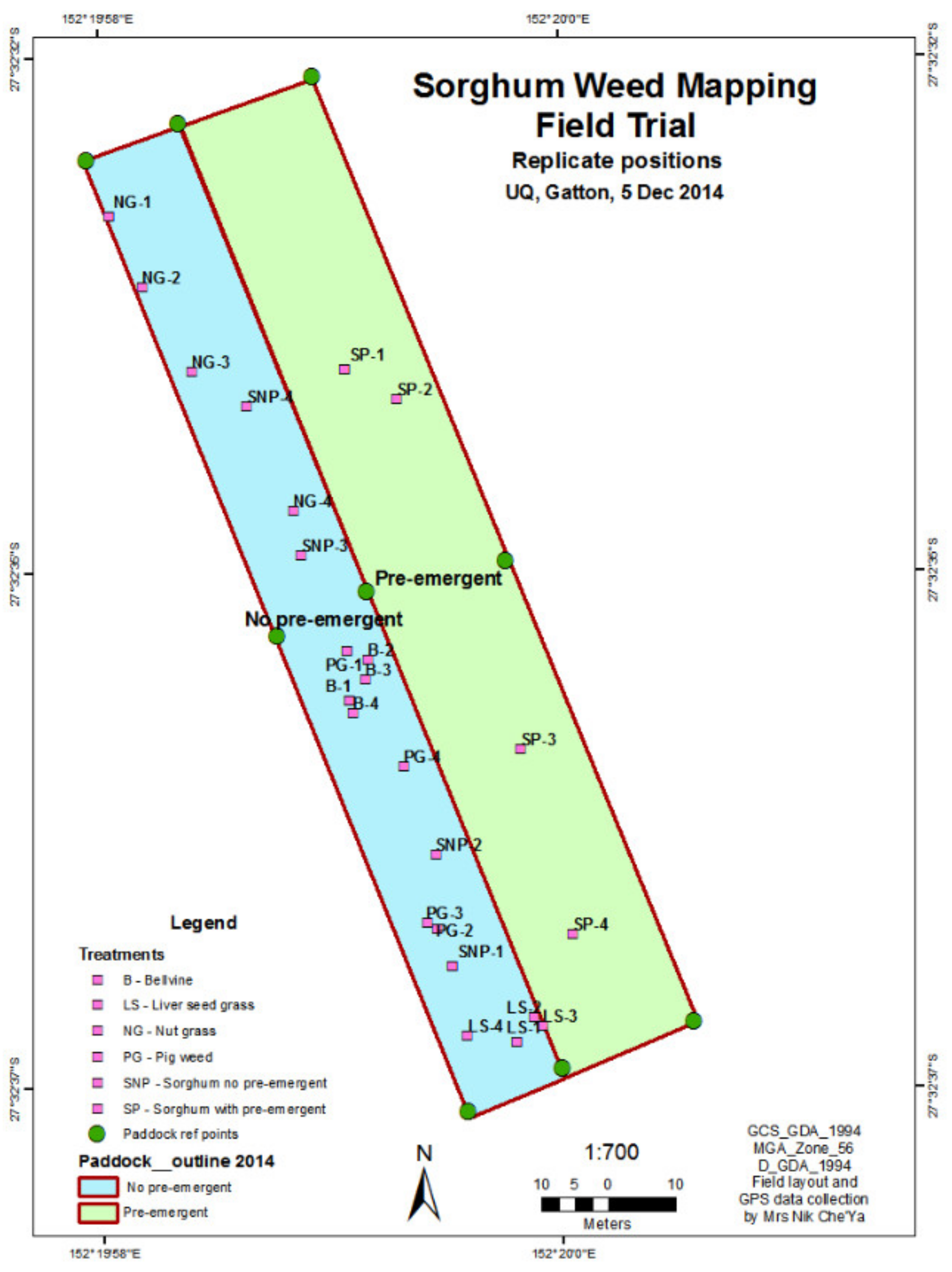
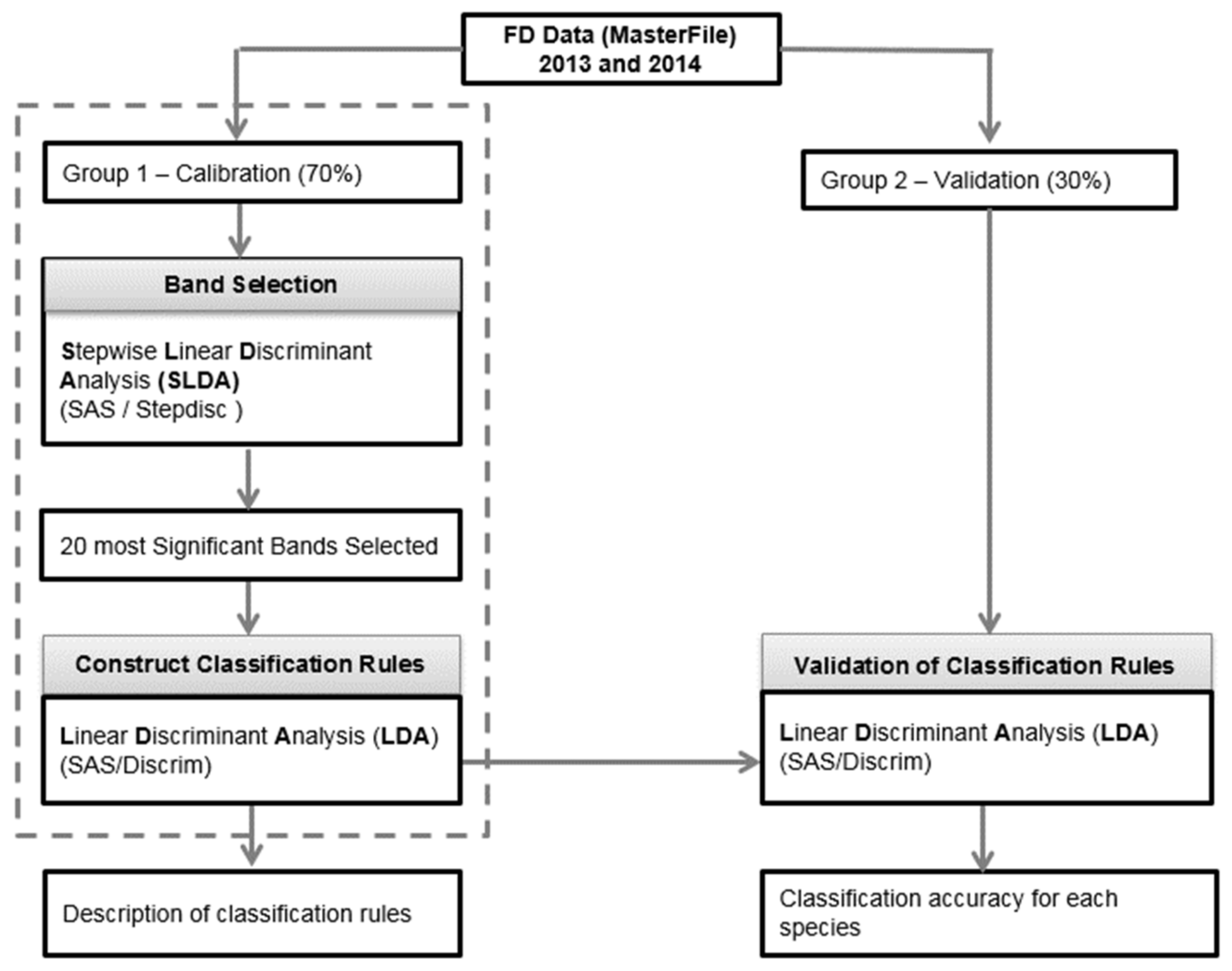
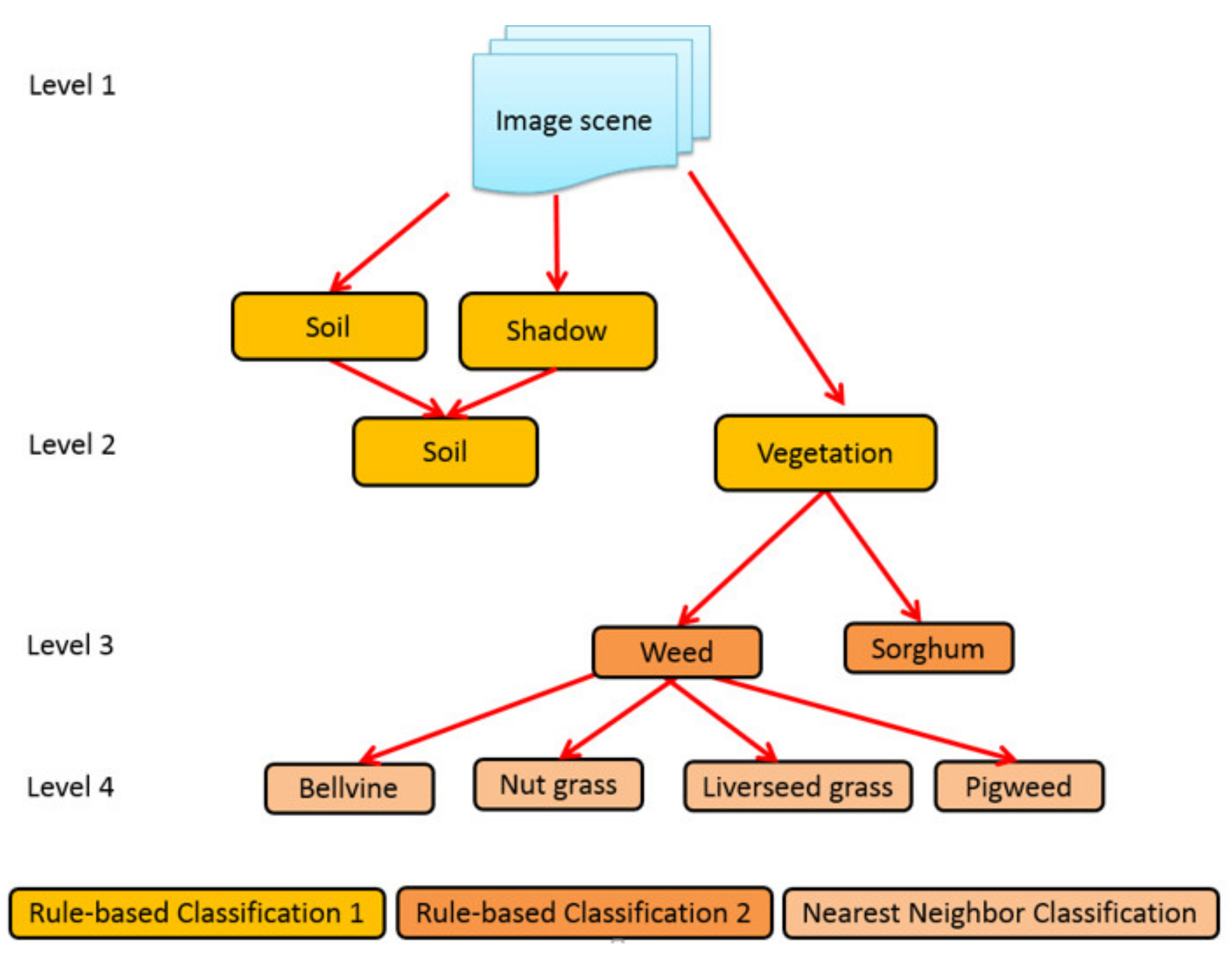
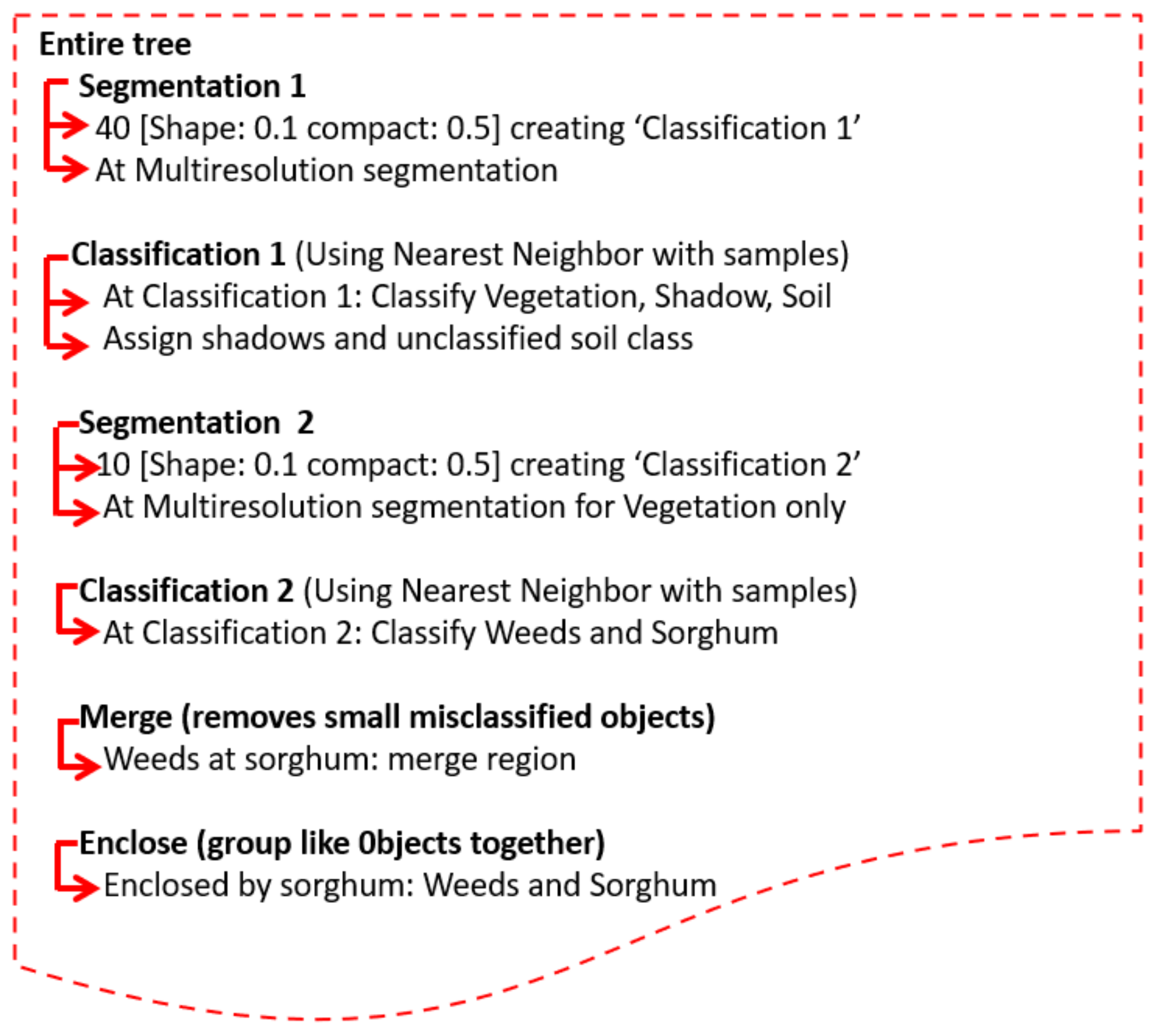
| No | Weeds Species | 2013 | 2014 |
|---|---|---|---|
| 1 | Amaranthus macrocarpus (AM) | ✓ | ✗ |
| 2 | Ipomea plebeia (B) | ✗ | ✓ |
| 3 | Malva sp. (MW) | ✓ | ✗ |
| 4 | Cyperus rotundus (NG) | ✓ | ✓ |
| 5 | Urochoa panicoides (LG) | ✓ | ✓ |
| 6 | Portulaca oleracea (PG) | ✗ | ✓ |
| Spectral Region | Bands | Sources |
|---|---|---|
| NIR | 850 nm | MCA 6, LDA |
| Red-edge | 750, 730, * 720 and * 710 nm | MCA 6, * LDA |
| Red | 680 nm | MCA 6, LDA |
| Green | 560 nm | MCA 6, LDA |
| Blue | 440 nm | MCA 6, LDA |
| Species | Calibration Data (%) | Validation Data (%) | ||||
|---|---|---|---|---|---|---|
| Wk 2 | Wk 3 | Wk 4 | Wk 2 | Wk 3 | Wk 4 | |
| Amaranthus macrocarpus (AM) | 100 | 100 | 100 | 100 | 83 | 80 |
| Urochoa panicoides (LS) | 100 | 100 | 100 | 100 | 100 | 100 |
| Malva sp. (MW) | 88 | 100 | 100 | 75 | 100 | 100 |
| Cyperus rotundus (NG) | 100 | 100 | 100 | 100 | 67 | 100 |
| Sorghum bicolur (L.) Moench (SG) | 92 | 92 | 100 | 67 | 75 | 71 |
| Mean | 96 | 98 | 100 | 88 | 85 | 90 |
| High accuracy (>80); Wk = Week | ||||||
| Species | Calibration Data (%) | Validation Data (%) | ||||
|---|---|---|---|---|---|---|
| Wk 2 | Wk 3 | Wk 4 | Wk 2 | Wk 3 | Wk 4 | |
| Ipomea plebeia (B) | 100 | 100 | 100 | 100 | 100 | 100 |
| Urochoa panicoides (LS) | 100 | 100 | 100 | 89 | 100 | 100 |
| Cyperus rotundus (NG) | 100 | 100 | 100 | 100 | 100 | 100 |
| Portulaca oleracea (PG) | 100 | 100 | 100 | 100 | 100 | 100 |
| Mean | 100 | 100 | 100 | 97 | 100 | 100 |
| High accuracy (>80), Wk = Week | ||||||
| Combination Number | The Band Combinations (nm) | AM (%) | LS (%) | MW (%) | NG (%) | Mean (%) |
|---|---|---|---|---|---|---|
| 1 | 440, 560, 680, 710, 720, 730 | 43 | 100 | 100 | 100 | 86 |
| 2 | 440, 560, 680, 710, 720, 750 | 43 | 100 | 100 | 100 | 86 |
| 3 | 440, 560, 680, 710, 720, 850 | 71 | 100 | 100 | 100 | 93 |
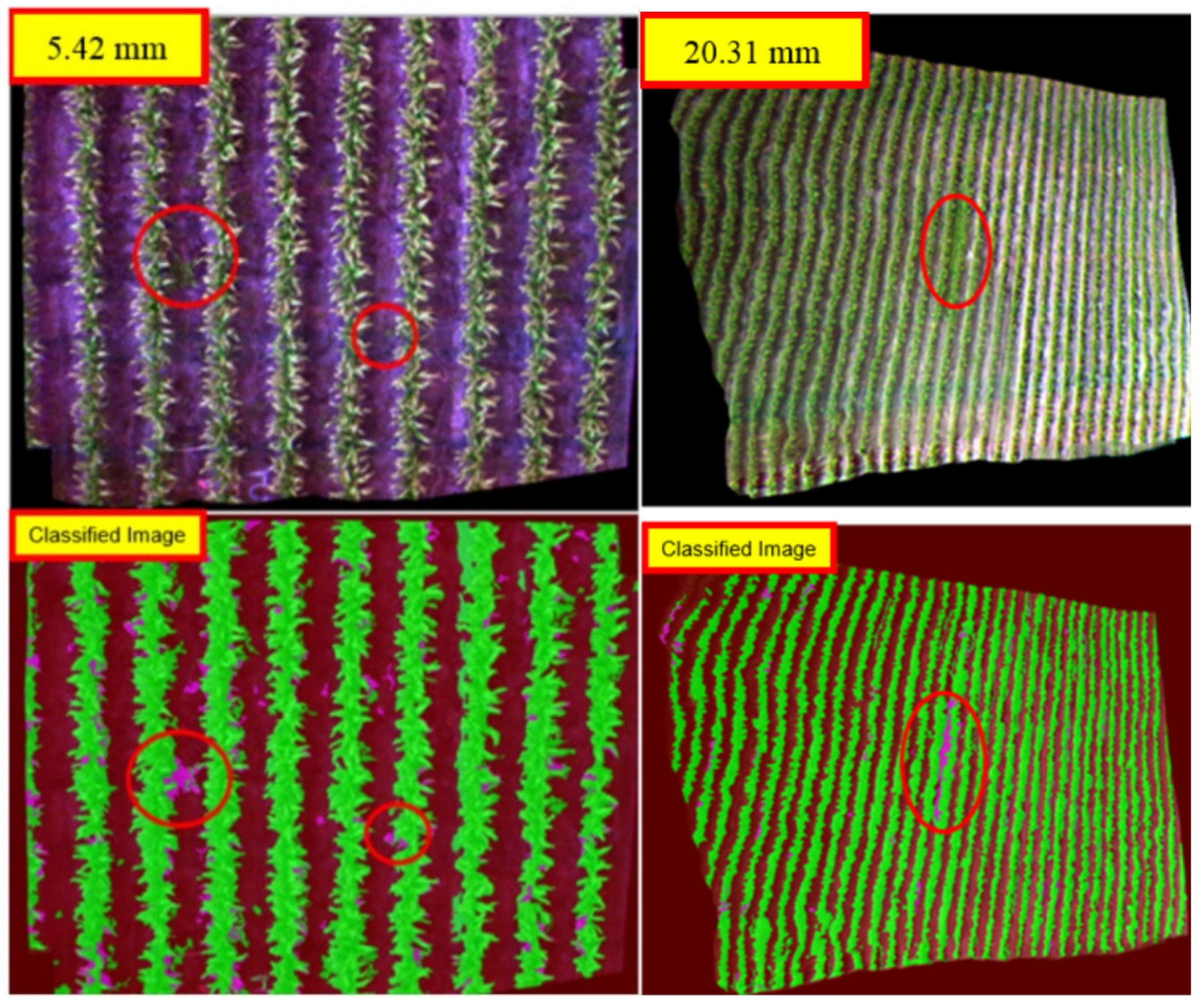
| Image Height: 1.6 m (RGB) | Week 3 (GSD: 10.83 mm) | Week 3 (GSD: 20.31 mm) |
|---|---|---|
| 17th December 2014 | 17th December 2014 | |
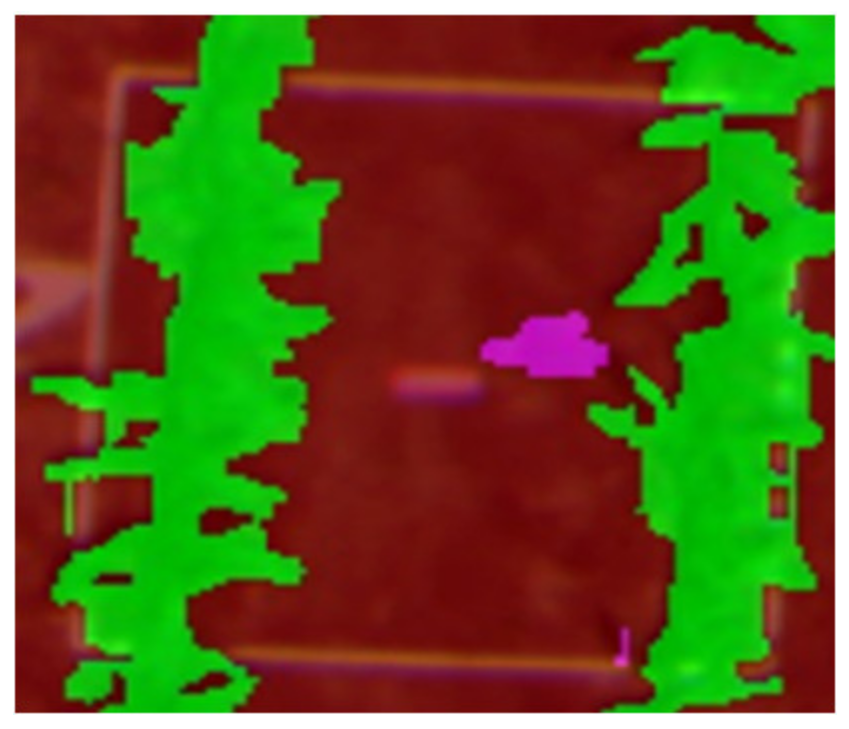
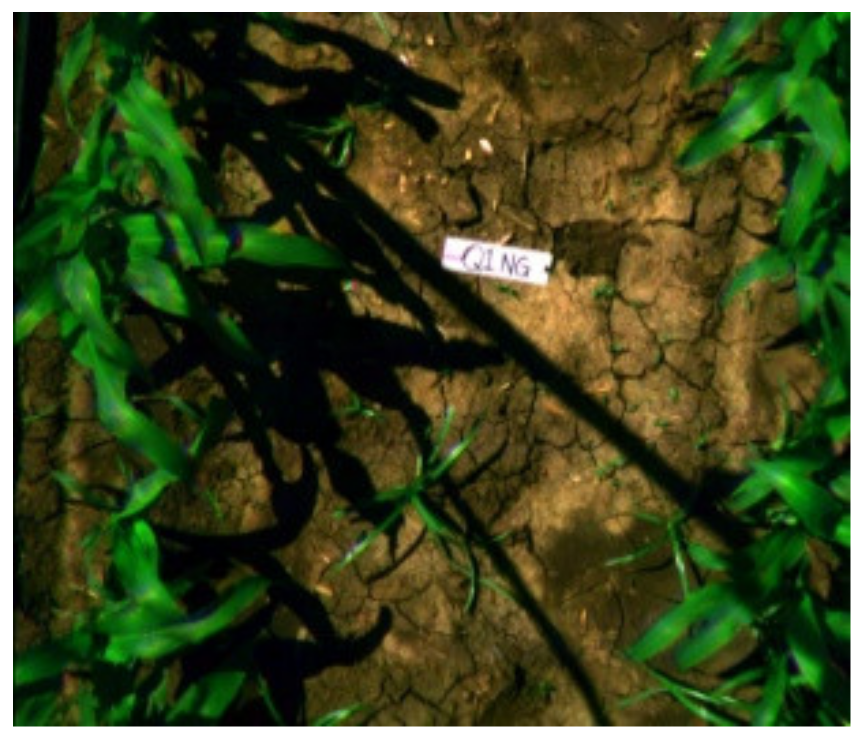 | 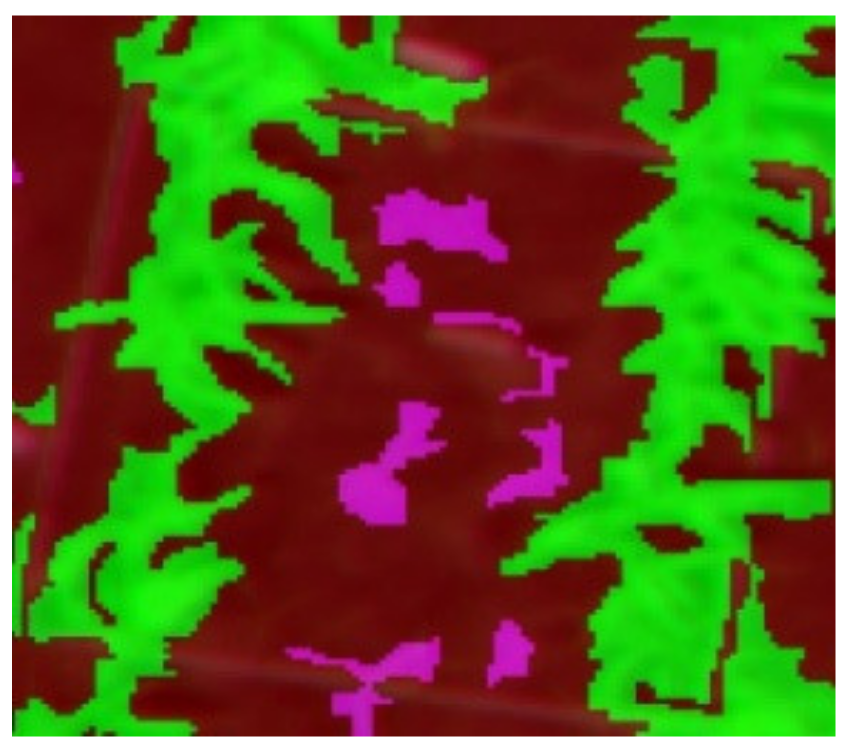 |  |
| Q1NG (Cyperus rotundus) | Correctly classified | Misclassified as Sorghum (in the circle) |
 |  | 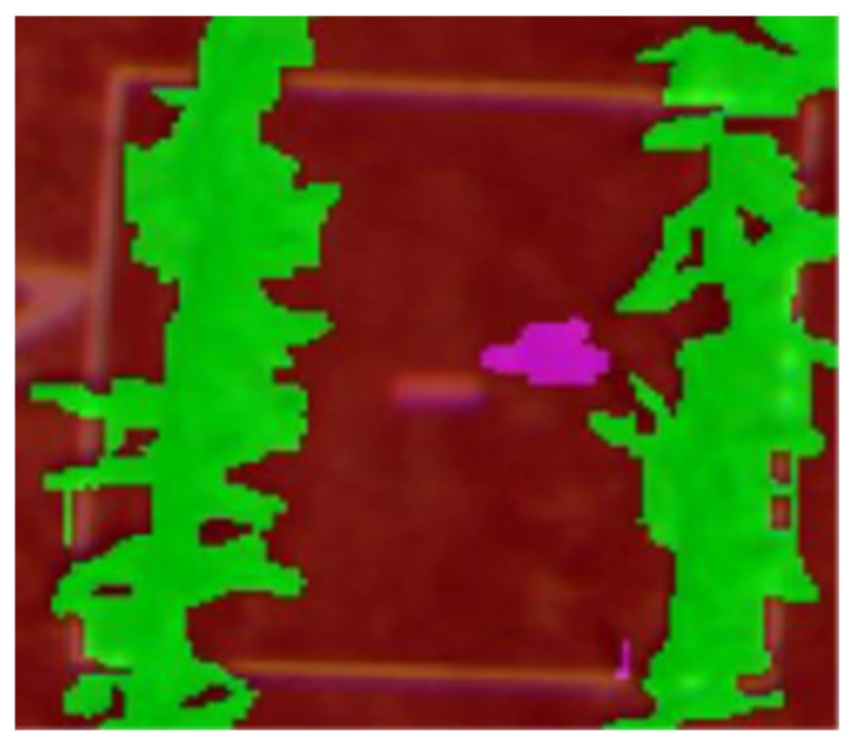 |
| Q1PG (Portulaca oleracea) | Correctly classified | Correctly classified |
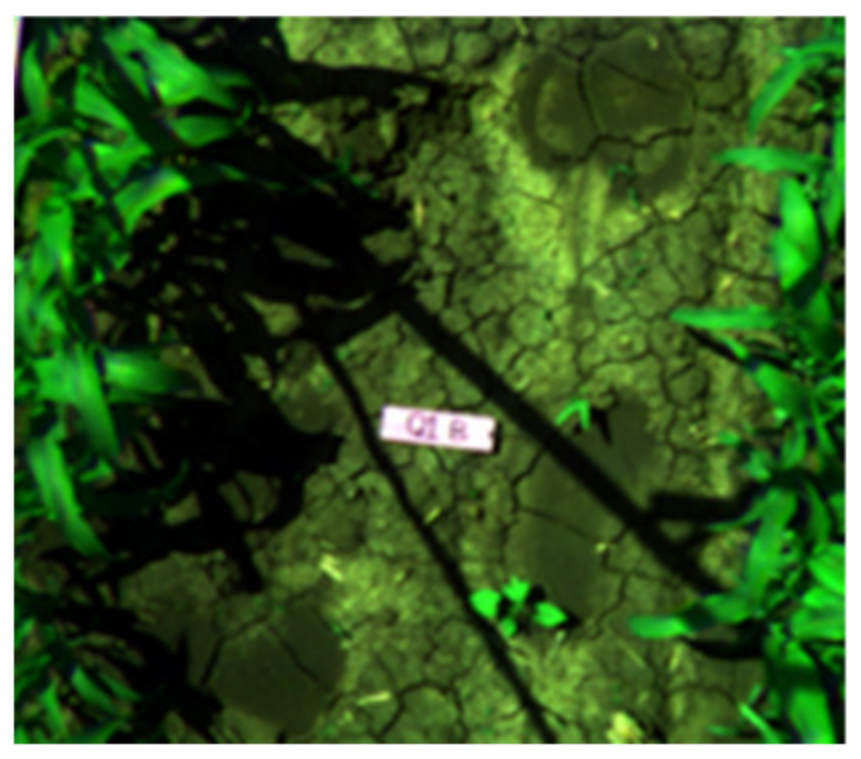 | 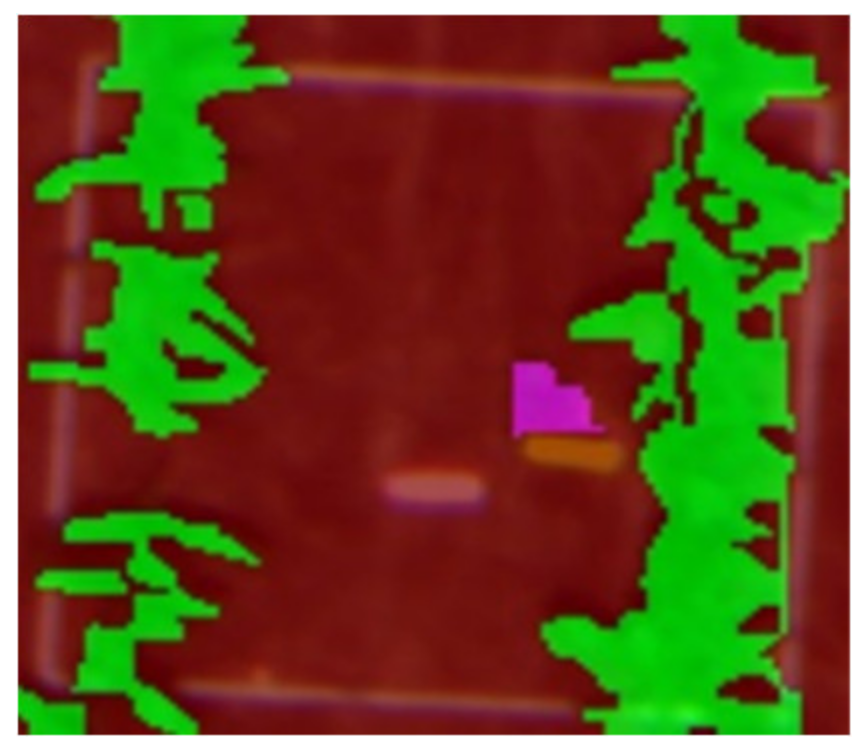 | 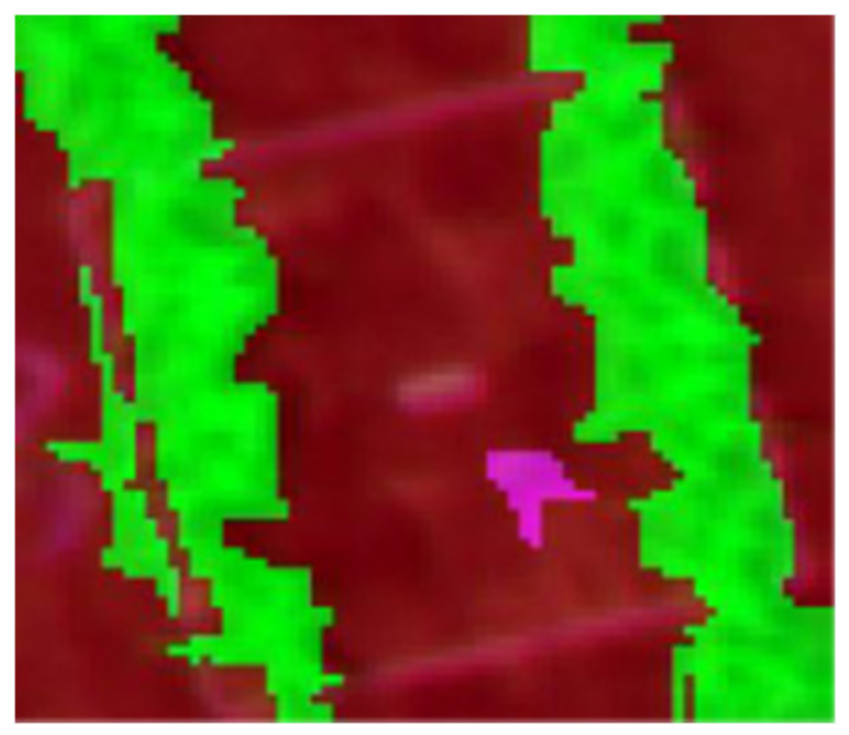 |
| Q1B (Ipomea plebeian) | Correctly classified | Correctly classified |
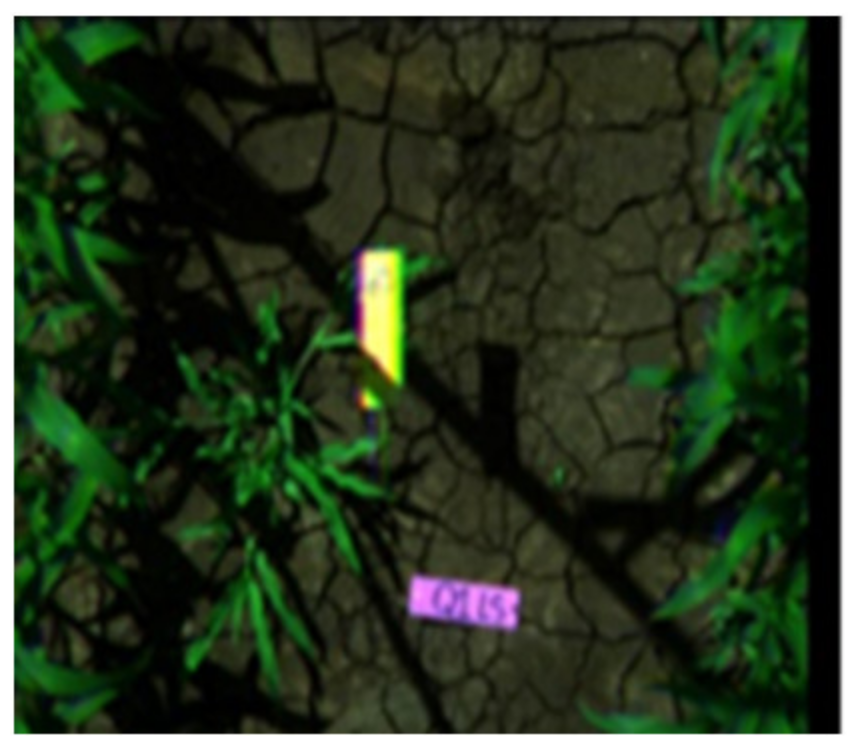 | 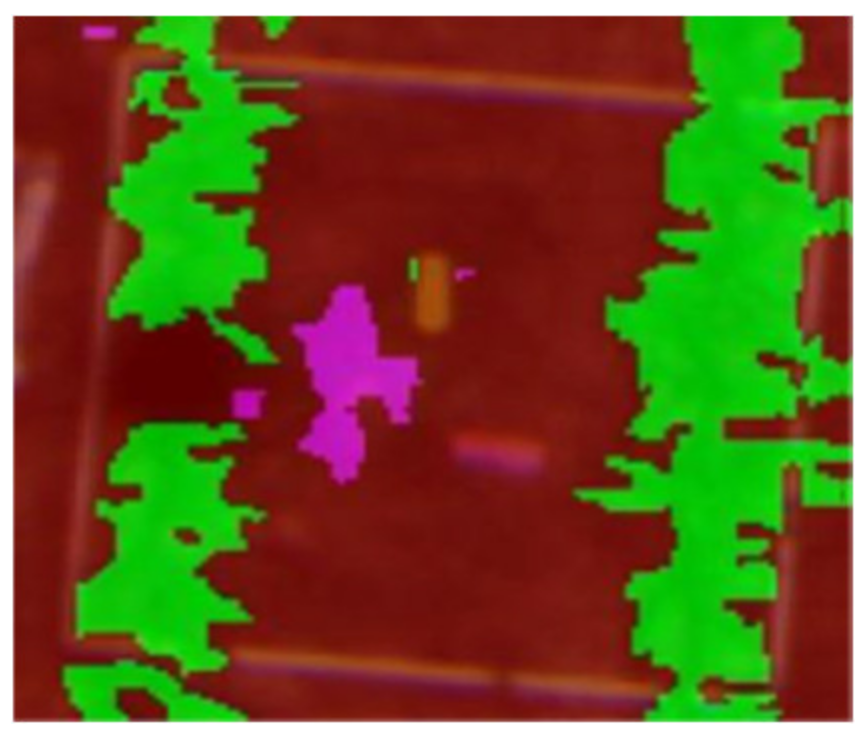 | 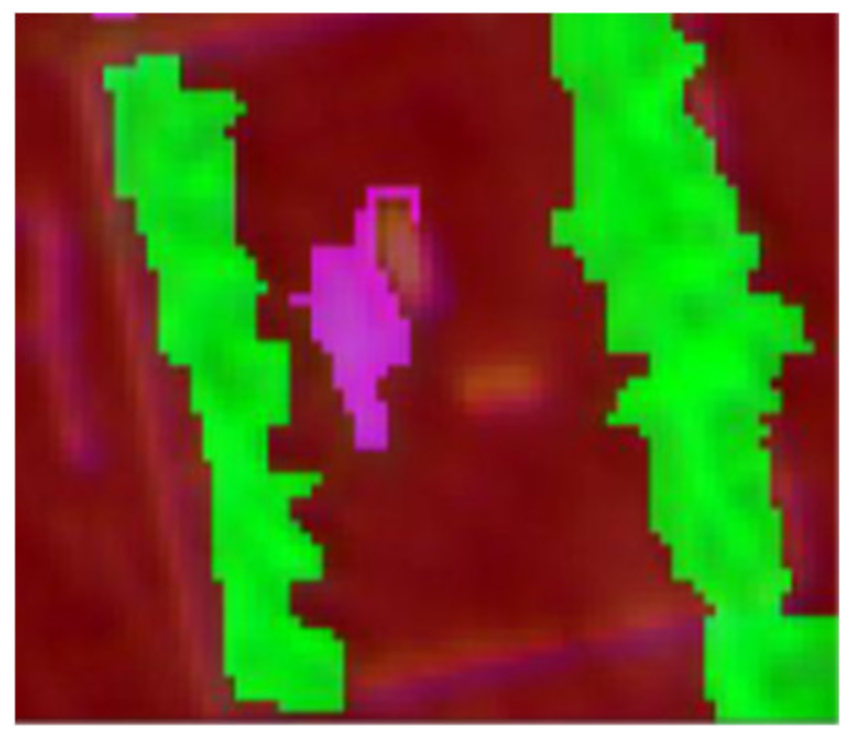 |
| Q1LS (Urochoa panicoides) | Correctly classified | Correctly classified |
| Spatial Resolution | Confusion Matrix | Weed (%) | Soil (%) | Sorghum (%) |
|---|---|---|---|---|
| 5.42 mm | Producer Accuracy (PA) | 81 | 100 | 90 |
| User Accuracy (UA) | 81 | 100 | 90 | |
| Overall Accuracy (OA) | 92 | |||
| KHAT (K) | 97 | |||
| 20.31 mm | Producer Accuracy (PA) | 56 | 98 | 77 |
| User Accuracy (UA) | 61 | 95 | 77 | |
| Overall Accuracy (OA) | 84 | |||
| KHAT (K) | 74 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Che’Ya, N.N.; Dunwoody, E.; Gupta, M. Assessment of Weed Classification Using Hyperspectral Reflectance and Optimal Multispectral UAV Imagery. Agronomy 2021, 11, 1435. https://doi.org/10.3390/agronomy11071435
Che’Ya NN, Dunwoody E, Gupta M. Assessment of Weed Classification Using Hyperspectral Reflectance and Optimal Multispectral UAV Imagery. Agronomy. 2021; 11(7):1435. https://doi.org/10.3390/agronomy11071435
Chicago/Turabian StyleChe’Ya, Nik Norasma, Ernest Dunwoody, and Madan Gupta. 2021. "Assessment of Weed Classification Using Hyperspectral Reflectance and Optimal Multispectral UAV Imagery" Agronomy 11, no. 7: 1435. https://doi.org/10.3390/agronomy11071435
APA StyleChe’Ya, N. N., Dunwoody, E., & Gupta, M. (2021). Assessment of Weed Classification Using Hyperspectral Reflectance and Optimal Multispectral UAV Imagery. Agronomy, 11(7), 1435. https://doi.org/10.3390/agronomy11071435






